Abstract
Objective
Given that the molecular diagnosis of autosomal dominant polycystic kidney disease (ADPKD) is complicated, we aim to apply blocker displacement amplification (BDA) on the mutational screening of PKD1 and PKD2.
Methods
A total of 35 unrelated families with ADPKD were recruited from the Center for Reproductive Medicine, Women and Children’s Hospital of Chongqing Medical University (Chongqing, China), from October 2018 to October 2021. Long-range PCR followed by next-generation sequencing were applied for resequencing of PKD1 and PKD2, and the putatively disease-causative variants were verified with BDA. The effects of ADPKD on male and female infertility and the factors influencing the clinical outcomes of preimplantation genetic testing (PGT) for ADPKD were investigated.
Results
A total of 26 PKD1 variants and 5 PKD2 variants were identified, of which 13 were newly discovered. The BDA system worked effectively for eliminating the interference of pseudogenes in genetic testing of PKD1 (1–33 exons) with different concentrations of genome DNA. The females with ADPKD have no specific infertility factors, while 68.2% of the affected men were with abnormal sperm concentration and/or motility with an indefinite genotype–phenotype relationship. As for PGT, the fertilization rate of couples with the male partner having ADPKD was relatively lower compared to those with the female partner being affected. The ADPKD patients receiving PGT usually achieved high rates of live births.
Conclusion
These findings expanded the variant spectrum of PKD genes and emphasized the application prospect of blocker displacement amplification on PKD1-related genetic diagnosis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Autosomal dominant polycystic kidney disease (ADPKD) is a late-onset systemic disorder manifested by progressive development and enlargement of kidney cysts, hypertension, and renal insufficiency [1, 2]. In the meantime, ADPKD is often accompanied with multiple extrarenal complications, including liver cysts, intracranial aneurysms, cardiac valvular disease, pancreatic cysts, and seminal vesicle cysts. The prevalence of ADPKD was estimated to be 1/400 to 1/1000 live births [3, 4]. It is one of the most frequent genetic causes of end-stage kidney disease (ESKD) , with up to 50% of cases progressing to ESKD by the age of 60 [5].
Systematic genetic studies have identified a general genotype–phenotype correlation between the PKD gene variants and the clinical phenotypes of ADPKD patients. The major causative genes of ADPKD included PKD1 (MIM 601313) on chromosome 16p13 [6], PKD2 (MIM 173910) on chromosome 4q22 [7], GANAB (MIM 104160) on chromosome 11q13 [8], and DNAJB11 (MIM 611341) on chromosome 3q27 [9], associating with 78%, 15%, 0.3%, and 0.1% of all reported ADPKD cases, respectively. The pathogenicity of the rest 7% ADPKD was attributed to unknown genetic factors [10, 11]. The pathogenic variants in PKD1 could induce a more severe disease compared with PKD2 variants, such as the onset of ESRD at an earlier age (58.1 years versus 79.7 years) [12,13,14]. The non-truncating variants (missense and in-frame insertion/deletion) of PKD1 induced a lower risk of ESKD compared with the truncating variants (nonsense and frameshift) (67.9 years versus 55.6 years) [1, 14]. About 90% of individuals with ADPKD have a positive family history, and at least 10% of patients can be traced to a de novo pathogenic variant [15, 16]. Each child of an affected individual has a 50% chance of inheriting the pathogenic variant.
The complexity of the genomic structure that the 5′ region of the PKD1 gene (exons 1-33) is duplicated in six pseudogenes on chromosome 16 which share 97.7% sequence identity with the GC-rich PKD1 gene and the genetic heterogeneity of ADPKD have long been obstacles to routine genetic testing [10, 17]. Nowadays, long-range PCR (LR-PCR) followed by next-generation sequencing (NGS) has offered a cost- and time-effective method for the molecular diagnosis of ADPKD [18, 19]. Once the disease-causative variant in a ADPKD family has been identified, preimplantation genetic testing for monogenic disorder (PGT-M) provides an invasive prenatal diagnosis that involves the biopsy of a single or few cells from in vitro fertilized embryos and testing of the biopsied samples for genetic aberrations, followed by the selective transfer of unaffected embryos under specific conditions [20, 21].
However, the direct amplification of PKD1 gene in genome DNA remained laborious and the factors influencing the clinical outcome of PGT for ADPKD still need to be explored. Previous studies have shown that the blocker displacement amplification (BDA) method can achieve distinctly differential hybridization yield based on differences in the hybridization affinity of probes to variant and wild-type templates [22], thereby providing good sequence selectivity for PKD1 in genetic diagnosis. In this study, we performed mutational screening of PKD1 and PKD2 using LR-PCR combined with next-generation sequencing in 35 unrelated Chinese families with ADPKD, as well as describing the application of BDA on PKD1 genetic testing. Herein, a total of 31 likely pathogenic or pathogenic variants were identified including 13 novel variants in PKD genes. We also investigated the effects of ADPKD on male infertility, especially the genotype–phenotype correlation between variants of PKD genes and male semen quality, and the factors influencing the clinical outcome of PGT cycles for ADPKD. These findings expanded the variant spectrum of PKD genes and highlighted the application prospect of BDA on PKD1-related genetic diagnosis and PGT.
Methods and materials
Subjects and ethical approval
A total of 35 unrelated families were recruited from the Center for Reproductive Medicine, Women and Children’s Hospital of Chongqing Medical University (Chongqing, China), from October 2018 to October 2021, including 27 patients with a positive family history of ADPKD. These patients came to the clinic either for genetic counselling to avoid delivering a baby with ADPKD or treatment for infertility which then were diagnosed with ADPKD based on ultrasonic examination before undergoing assisted reproductive technology (ART) treatment. All diagnoses were confirmed by ultrasonic examination according to previously described criteria: (1) the presence of three or more (unilateral or bilateral) renal cysts in an individual aged 15–39 years, or (2) the presence of two or more cysts in each kidney in an individual aged 40–59 years, or (3) large echogenic kidneys without distinct macroscopic cysts in an infant/child at 50% risk of ADPKD [23, 24].
All clinical assessments, including ultrasonography and ART, were performed in the related clinical departments. All individuals signed a written informed consent form and blood samples were collected from all probands and their family members whenever possible. This study was approved by the Clinical Application and Ethics Committee of Human Assisted Reproductive Technology of Chongqing Health Center for Women and Children.
Resequencing and variant analysis of PKD1 and PKD2
Genomic DNA (gDNA) was extracted from peripheral blood samples using a QIAamp® DNA blood midi kit (QIAGEN, Germany) according to the manufacturer’s protocol. The entire DNA sequences of PKD1 and PKD2 were amplified in a total of ten distinct long-range PCR (LR-PCR) reactions. The primers combined with rarely differential bases from the pseudogenes are listed in Table S1. LR-PCR amplification reaction system and amplification conditions for the various LR-PCR fragments are described in Table S2 and Table S3, respectively. The LR-PCR products were purified with Agencourt AMPure XP Beads (Beckman, USA), quantified, and fragmented by Q800R ultrasonic crusher (Qsonica, USA). A sequencing library was constructed by KAPA HTP Library Preparation Kit (KAPA Biosystems, USA), and then, LR-PCR products were sequenced with Illumina Hiseq X10 (Illumina, Inc., USA). All sequencing reads were mapped to the PKD1/2 reference genome with the NextGENe software (SoftGenetics, USA), with the reference sequences being NM_001009944.2 for PKD1 and NM_000297.2 for PKD2. Then, the data were exported as fastq files.
Variants identified in PKD1 gene (exons 1-33) as putatively disease-causative were verified with blocker displacement amplification (BDA), preventing the false positivity due to the interference of pseudogenes or mismatch in long-segment amplification. The primers and blockers used in BDA-based PCR are listed in Table S4, and the amplification reaction system and amplification conditions are described in Table S5 and Table S6, respectively. Variants in PKD1 gene (exons 34-46) or PKD2 gene were verified with conventional PCR. And the family members were subjected to the inheritance history analysis, providing clues for the preimplantation genetic testing (PGT). The list of probands and their family members is attached as Table S7.
Evaluation of the pathogenicity of variants
The PKDB (http://pkdb.mayo.edu), VarSome (https://varsome.com/) [25], Human Gene Mutation Database (HGMD, http://www.hgmd.cf.ac.uk/ac/index.php), and gnomAD database (http://gnomad-sg.org/) were searched for previously reported ADPKD-associated genetic variants. Frameshift variants, typical splicing, nonsense, and in-frame changes of two or more amino acids were defined as pathogenic factors for ADPKD [26]. A novel mutation was defined as one that had not been described in PKDB or HGMD or reported in ADPKD patients. The potential pathogenicity of all identified missense variants, indicated by a frequency below 1% in the Asian population of the gnomAD database, was evaluated by pedigree analysis and in silico analysis using VarCards (http://varcards.biols.ac.cn/) [27]. All variants were classified into five categories: ‘pathogenic,’ ‘likely pathogenic,’ ‘uncertain significance,’ ‘likely benign,’ and ‘benign,’ according to the standards and guidelines of the American College of Medical Genetics and Genomics (ACMG) for the interpretation of variants [28].
Semen analysis
Semen was collected by masturbation into a sterile container after 2–7 days of abstinence and subjected to routine biological examination in the source laboratories according to the World Health Organization (WHO, 2021) guidelines [29, 30]. Briefly, within 1 h after ejaculation, the samples were liquefied and analyzed for semen volume, sperm concentration, round cells, normal morphology, and sperm motility. Sperm count was determined via a hemocytometer under a light microscope, and sperm mobility was assessed via a computer-assisted sperm analysis (CASA) system. Asthenozoospermia was defined by reduced or absent sperm motility in the fresh ejaculate (progressive motility < 32%) [31]. Oligozoospermia specifically referred to the condition in which sperm concentration was below the lower reference limit of 15 million sperm/mL of ejaculate. Azoospermia referred to the condition in which no sperm could be detected in the ejaculate during semen analysis. Asthenozoospermia and oligozoospermia might exist at the same time, which was collectively called oligoasthenozoospermia.
Assisted reproductive therapies and PGT-M procedure
Most individuals with a molecular diagnosis of PKD variants chose differential approaches to conceive offspring, including natural pregnancy, in vitro fertilization (IVF), intracytoplasmic sperm injection (ICSI), and ICSI combined with preimplantation genetic testing (PGT). For the PGT-M procedure, whole-genome amplification of each embryo biopsy sample was performed using MALBAC WGA kit (Yikon Genomics, China), following the manufacturer’s instructions. A total of 60 single-nucleotide polymorphism (SNP) markers linked to the variant alleles were selected for linkage analysis. The variant site and SNPs were amplified with specific primer pairs, and the MALBAC WGA amplification products were sequenced. The chromosomal copy number, variant site, and SNPs were analyzed, as published previously [32].
Statistical data analysis
Continuous data were presented as mean ± standard deviation (SD). Mann–Whitney U test was used to compare the differences in female age, male age, sperm concentration, and sperm motility. Pearson chi-square test was used to compare categorical data such as the number of fertilized oocytes.
Results
Blocker displacement amplification of PKD1 gene
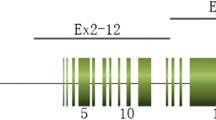
As shown by Fig. 1a, the entire DNA sequences of PKD1 and PKD2, including most 5′ and 3′ untranslated regions and the exon–intron regions, were amplified in a total of ten distinct long-range PCR (LR-PCR) reactions (five reactions for PKD1 gene and five reactions for PKD2 gene). LR-PCR products were analyzed with agarose gel electrophoresis, and the results suggested that the primers combined with the rarely differential bases from the pseudogenes were highly effective in capturing the targeted sequences (Fig. 1b). After high-throughput screening with next-generation sequencing, a pair of primers and a blocker containing several bases overlapping with the forward primer and covering at least one differential nucleotide base between PKD1 and the pseudogenes were designed. Blocker displacement amplification (BDA) of the putatively disease-causative variants in PKD1 1-33 exons was performed for variant verification and pedigree analysis (Fig. 1c). Variants in PKD1 gene (exons 34-46) or PKD2 gene were verified with conventional PCR. The BDA system worked effectively for different concentrations of genome DNA as low as 1 ng (Fig. 1d). Furthermore, Sanger sequencing demonstrated that the pseudogenes concealed the PKD1 gene without using BDA, while the sequence of PKD1 gene was identified with specificity and its variants were determined after applying BDA, implying the efficacy of BDA in eliminating the interference of pseudogenes for the genetic testing of PKD1 (1-33 exons) (Fig. 1e).
Mutation analysis of PKD1 and PKD2. (a) The diagrammatic images of PKD1 and PKD2, and the location of primers for long-range PCR. (b) The result of agarose gel electrophoresis of LR-PCR products. (c) Primer and blocker sequences designed for BDA to detect the sequence of exon 15 of PKD1 gene. The location of reverse primer is highlighted in yellow. The PKD1 gene bears a cytosine nucleotide, and the six pseudogenes bear a thymine nucleotide. The 4 A sequence at the 3′ end of the blocker serves as a termination sequence to prevent the blocker from being elongated by the Taq polymerase during PCR. (d) The ΔRn of BDA-based PCR for different doses of genomic DNA. (e) The results of Sanger sequence for BDA-PCR products. The red nucleotide bases and arrowheads showed the variants of PKD1 gene in ADPKD patients, while the blue nucleotide bases and arrowheads showed the differential bases between PKD1 gene and the pseudogenes
The variant spectrum of PKD1 and PKD2 identified in ADPKD patients
Finally, a total of 31 genetic variants were identified in this study, comprising 26 and 5 variants in PKD1 and PKD2, respectively (Table 1). They all have extremely rare frequencies (< 10−4) or are absent in the gnomAD database (Table S8). We evaluated the potential pathogenicity of all identified variants according to the ACMG standards and guidelines for the interpretation of variants (Table S8). Among the PKD1 variants, there were 15 pre-truncated variants (9 nonsense and 6 frameshift), resulting in the truncation of PKD1 protein and associated with severe clinical phenotypes. Additionally, 2 in-frame variants, 7 missense variants, and 2 splice variants affecting the PKD1 mRNA splicing during transcription were also identified. The 5 PKD2 gene variants consisted of 3 pre-truncated variants (2 nonsense and 1 frameshift), 1 missense variant, and 1 in-frame variant. Among these variants, 13 are novel and have not been described in the ADPKD variant database or HGMD or been reported in ADPKD patients. Among the 31 probands, there are 23 male patients and 8 female patients, which is deviated from the theoretical 1 : 1 ratio. Based on the genetic hereditary analysis, 51.6% (16/31) of the proband were inherited from the mother, 35.5% (11/31) of the proband were inherited from the father, and 12.9% (4/31) were de novo, which was consistent with the previously reported ratio [15, 16].
Infertility factor and semen quality of the ADPKD patients
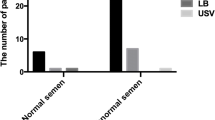
Previous studies showed that male ADPKD patients have an increased risk for infertility due to cyst formation in the reproductive tract and/or structural abnormalities of the spermatozoa [33, 34]. To analyze the effect of ADPKD on male and female fertility, we analyzed the variant type, infertility factor, and the semen quality of the ADPKD-affected couples. Among the 31 unrelated ADPKD probands, 23 were male and 22 provided semen specimens for analysis. The results showed that semen from 15 (15/22, 68.2%) male individuals were abnormal with azoospermia (4), asthenozoospermia (4), and oligoasthenozoospermia (7) (Table 1). These males carried 14 PKD1 variants and 1 PKD2 variant, including 8 pre-truncated variants, 2 splice variants, 2 in-frame indel variants, and 3 missense variants, respectively. The ADPKD males with normal semen quality carried 5 PKD1 variants (2 pre-truncated, 2 missense and 1 in-frame deletion) and 2 PKD2 pre-truncated variants. Testicular aspiration for the 4 azoospermia individuals (3 missense and one frameshift) successfully obtained sperm for intracytoplasmic sperm injection (ICSI) in the assisted reproductive technology (ART) process, implying obstruction of the vas deferens. The females with ADPKD have no specific infertility factors as only one patient (family 16) was affected with the obstruction of fallopian tube. Thus, the PKD variants affected the sperm concentration and motility with an indefinite genotype–phenotype relationship, as the variant type was not different between the ADPKD male individuals with normal or abnormal semen parameters.
Reproductive and PGT outcomes in females and males with ADPKD
Currently, a total of 23 couples has chosen different approaches to conceive (Table 1). The flowchart (Fig. 2) presents the number of couples at pre-clinical and clinical stages and the reasons for dropout from the study. Three couples (including two male probands with normal semen and one female proband) conceived naturally, and three couples (one with oligoasthenozoospermia and two with asthenozoospermia) conceived through ICSI without PGT. The other 17 couples chose ICSI combined with PGT to avoid the delivery of PKD variants.
The 17 couples undergoing assisted reproductive technology (ART) and PGT for reproduction were classified into two groups: Group A, the male partner was affected (n = 11) and Group B, the female partner was affected (n = 6). The details of the PGT cycles are shown in Table 2. For Group A, 81.8% (9/11) of the couples underwent one PGT cycle, one couple underwent two PGT cycles, and the other couple underwent six oocyte retrieval cycles with three cycles without blastocyst formation. For Group B, 83.3% (5/6) of the couples underwent one PGT cycle, while the other couple underwent two PGT cycles. Overall, a diagnosis was obtained in 97.8% (90/92) of the blastocysts. 53.3% (48/90) of the blastocysts were unsuitable for implantation due to affected with PKD variants (42/48) or chromosome aneuploidy (6/48). Additionally, 46.7% (42/90) of the blastocysts were normal and 17 cycles of frozen embryo transfer (FET) were performed for 14 couples. A clinical pregnancy was obtained in 13 cycles (76.5%), which led to a live birth in 9 cycles. There were 9 singleton deliveries for 9 couples, 3 continuing pregnancies for 3 couples and one premature rupture of membranes was also reported.
There were not significant differences in female age, male age, number of cumulus-oocyte complexes and mature oocytes submitted to ICSI between Group A and Group B (all P > 0.05). The sperm concentration and sperm motility in Group A were significantly lower than those in Group B (both P < 0.05), which had a significant effect on the fertilization rate (65.4% in Group A versus 82.1% in Group B, P < 0.001). However, the clinical pregnancy rate and live birth rate did not differ between the two groups.
Discussion
In this study, we described a new strategy for PKD1 genetic testing based on blocker displacement amplification. And we analyzed the genetic variants of 31 unrelated ADPKD families, the semen quality of 22 male patients, and the clinical outcomes of PGT cycles for 17 couples from October 2018 to October 2021. A total of 26 PKD1 variants and 5 PKD2 variants were identified, of which 13 were newly discovered. The analysis of male infertility showed that ADPKD affected the sperm concentration and motility, but the genotype–phenotype relationship could not be defined. The ADPKD patients receiving PGT usually achieved high rates of live births.
PKD1 and PKD2 located on chromosome 16p13.3 and 4q21.1, encode polycystin-1 (PC-1) and PC-2, respectively [35]. So far, a total of 2790 clinical known PKD1 variants were observed, of which 1226 were pathogenic ((including 894 of pathogenic and 332 of likely pathogenic) (Varsome, version: 27-Jun-2022). Most of these variants were frameshift (412) and nonsense (367), and the others were missense (288), splice junction loss (97), in-frame indel (44) , and synonymous or non-coding (18). In addition, 602 clinical known PKD2 variants were detected, of which 273 were pathogenic (including 213 of pathogenic and 60 of likely pathogenic). These PKD2 variants were missense (49), nonsense (90), frameshift (92), splice junction loss (35), in-frame indel (3), and non-coding (4). The PKD1 gene is located within an CpG-rich 750 kb segment of chromosome 16p13.3, and the design of primers for PKD1 amplification and the co-amplification of multiple amplicons remain tricky [36]. To the best of our knowledge, this is the first report of blocker displacement amplification applied in ADPKD genetic diagnosis. In our study, a total of 31 PKD1 and PKD2 variants were identified, comprising 18 protein-truncating, 8 missenses, 2 splice junction loss, and 3 in-frame indels. The positive detection rate was 88.6% (31/35), and 41.9% (13/31) were novel variants. The proportion of ADPKD patients with a family history of the disease accounted for 87.1% (27/31) of all probands, which was comparable with the previously published data [12].
PKD1 and PKD2 have been reported to play a pivotal role in the development and maintenance of male reproductive tract [33, 34]. Male patients with ADPKD usually suffer from infertility, resulting from abnormal semen induced by ejaculatory duct cysts, seminal vesicle cysts, or ultrastructural flagellar defects [37, 38]. However, the correlation between the variant-type of PKD genes and semen quality remains unclear. In the present study, 15 male patients were found to have abnormal semen (68.2%, 15/22) with the decreased sperm concentration and/or motility, while the variant type was not different between the ADPKD male individuals with normal or abnormal semen parameters. Furthermore, after the treatment of ICSI, 11 male individuals with semen abnormalities all achieved paternity or at least obtained embryos. Therefore, we suggested that male patients with ADPKD should achieve paternity as young as possible, and ICSI combined with PGT should be considered for the treatment of patients suffering from low semen quality [30, 31].
In this study, among 17 couples who started a PGT treatment, 14 couples achieved one live birth or obtained transferable embryos. Although the small sample size of this data set limited the interpretation of the results, this study showed that PGT application for ADPKD achieved good reproductive outcomes. The fertilization rate of couples with the male partner having ADPKD was relatively lower compared to those with the female partner being affected. However, the female age is the only well-established determinant factor for treatment outcome, while male infertility induced by ADPKD does not substantially affect the live birth delivery rate [21].
Since our clinic specializes in reproductive and genetic disorders, most of the subjects included in the study came to seek treatment for infertility. The number of male patients exceeded that of female patients and the proportion of ADPKD males with abnormal sperm quality is not a true reflection of the ADPKD population.
Data availability
All data generated or analyzed during this study are included in the article and the supplementary information files.
References
Cornec-Le Gall E, Alam A, Perrone RD. Autosomal dominant polycystic kidney disease. Lancet. 2019;393(10174):919–35.
Saini AK, Saini R, Singh S. Autosomal dominant polycystic kidney disease and pioglitazone for its therapy: a comprehensive review with an emphasis on the molecular pathogenesis and pharmacological aspects. Mol Med. 2020;26(1):128.
Lanktree MB, et al. Prevalence estimates of polycystic kidney and liver disease by population sequencing. J Am Soc Nephrol. 2018;29(10):2593–600.
Willey CJ, et al. Prevalence of autosomal dominant polycystic kidney disease in the European Union. Nephrol Dial Transplant. 2017;32(8):1356–63.
Nowak KL, Hopp K. Metabolic reprogramming in autosomal dominant polycystic kidney disease: evidence and therapeutic potential. Clin J Am Soc Nephrol. 2020;15(4):577–84.
Raj S, Singh RG, Das P. Mutational screening of PKD1 and PKD2 in Indian ADPKD patients identified 95 genetic variants. Mutat Res. 2020;821:111718.
He WB, et al. Novel mutations of PKD genes in Chinese patients suffering from autosomal dominant polycystic kidney disease and seeking assisted reproduction. BMC Med Genet. 2018;19(1):186.
Porath B, et al. Mutations in GANAB, encoding the glucosidase iialpha subunit, cause autosomal-dominant polycystic kidney and liver disease. Am J Hum Genet. 2016;98(6):1193–207.
Huynh VT, et al. Clinical spectrum, prognosis and estimated prevalence of DNAJB11-kidney disease. Kidney Int. 2020;98(2):476–87.
Cornec-Le Gall E, Torres VE, Harris PC. Genetic complexity of autosomal dominant polycystic kidney and liver diseases. J Am Soc Nephrol. 2018;29(1):13–23.
Lanktree MB, et al. Insights into autosomal dominant polycystic kidney disease from genetic studies. Clin J Am Soc Nephrol. 2020;16:790.
Hwang YH, et al. Refining genotype-phenotype correlation in autosomal dominant polycystic kidney disease. J Am Soc Nephrol. 2016;27(6):1861–8.
Cornec-Le Gall E, et al. The PROPKD score: a new algorithm to predict renal survival in autosomal dominant polycystic kidney disease. J Am Soc Nephrol. 2016;27(3):942–51.
Cornec-Le Gall E, et al. Type of PKD1 mutation influences renal outcome in ADPKD. J Am Soc Nephrol. 2013;24(6):1006–13.
Chebib FT, Torres VE. Autosomal dominant polycystic kidney disease: core curriculum 2016. Am J Kidney Dis. 2016;67(5):792–810.
Shi H, et al. A novel monogenic preimplantation genetic testing strategy for sporadic polycystic kidney caused by de novo PKD1 mutation. Clin Genet. 2021;99(2):250–8.
Cornec-Le Gall E, et al. Genetics and pathogenesis of autosomal dominant polycystic kidney disease: 20 years on. Hum Mutat. 2014;35(12):1393–406.
Li W, et al. The mutation-free embryo for in vitro fertilization selected by MALBAC-PGD resulted in a healthy live birth from a family carrying PKD 1 mutation. J Assist Reprod Genet. 2017;34(12):1653–8.
Bullich G, et al. A kidney-disease gene panel allows a comprehensive genetic diagnosis of cystic and glomerular inherited kidney diseases. Kidney Int. 2018;94(2):363–71.
De Rycke M, Berckmoes V. Preimplantation genetic testing for monogenic disorders. Genes (Basel). 2020;11(8).
Berckmoes V, et al. Factors influencing the clinical outcome of preimplantation genetic testing for polycystic kidney disease. Hum Reprod. 2019;34(5):949–58.
Wu LR, et al. Multiplexed enrichment of rare DNA variants via sequence-selective and temperature-robust amplification. Nat Biomed Eng. 2017;1:714–23.
Pei Y, et al. Imaging-based diagnosis of autosomal dominant polycystic kidney disease. J Am Soc Nephrol. 2015;26(3):746–53.
Pei Y, et al. Unified criteria for ultrasonographic diagnosis of ADPKD. J Am Soc Nephrol. 2009;20(1):205–12.
Kopanos C, et al. VarSome: the human genomic variant search engine. Bioinformatics. 2019;35(11):1978–80.
Kataoka H, et al. Prediction of renal prognosis in patients with autosomal dominant polycystic kidney disease using PKD1/PKD2 mutations. J Clin Med. 2020;9(1).
Li J, et al. VarCards: an integrated genetic and clinical database for coding variants in the human genome. Nucleic Acids Res. 2018;46(D1):D1039–48.
Richards S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405–24.
Baldi E, et al. Extended semen examinations in the sixth edition of the WHO Laboratory Manual for the Examination and Processing of Human Semen: contributing to the understanding of the function of the male reproductive system. Fertil Steril. 2022;117(2):252–7.
Organization, GWH, WHO laboratory manual for the examination and processing of human semen, sixth edition. License: CC BY-NC-SA 3.0 IGO., 2021.
Tu C, et al. Genetic underpinnings of asthenozoospermia. Best Pract Res Clin Endocrinol Metab. 2020;34(6):101472.
Huang L, et al. Single-cell whole-genome amplification and sequencing: methodology and applications. Annu Rev Genomics Hum Genet. 2015;16:79–102.
Nie X, Arend LJ. Pkd1 is required for male reproductive tract development. Mech Dev. 2013;130(11-12):567–76.
Nie X, Arend LJ. Novel roles of Pkd2 in male reproductive system development. Differentiation. 2014;87(3-4):161–71.
Su Q, et al. Structure of the human PKD1-PKD2 complex. Science. 2018;361(6406).
The European Polycystic Kidney Disease Consortium. The polycystic kidney disease 1 gene encodes a 14 kb transcript and lies within a duplicated region on chromosome 16. Cell. 1994;77(6):881–94.
Kim JA, Blumenfeld JD, Prince MR. Seminal vesicles in autosomal dominant polycystic kidney disease. In: Li X, editor. Polycystic kidney disease. Brisbane (AU); 2015.
Houston BJ, et al. A systematic review of the validated monogenic causes of human male infertility: 2020 update and a discussion of emerging gene-disease relationships. Hum Reprod Update. 2021;28(1):15–29.
Funding
This study was supported by grants from the Natural Science Foundation of Chongqing (CSTC2021JCYJ-MSXMX0722) and the Chongqing Health Center for Women and Children (2020YJMS01).
Author information
Authors and Affiliations
Contributions
T.T Lin, J.F Luo, D.Y Liu, and G.N Huang conceived and designed the study. B.H Dong, W Zhang, and K Chen carried out the experiments. H.B Yu and Y.Z Xiang provided the clinical samples. D.Y Liu conducted the PGT cycle. T.T Lin wrote the manuscript. D.Y Liu and G.N Huang critically commented on and edited the manuscript. All authors read and approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Ethics statement
The studies involving human participants were reviewed and approved by the Clinical Application and Ethics Committee of Human Assisted Reproductive Technology of Chongqing Health Center for Women and Children.
Consent
Informed consent was obtained from the proband/participants.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Lin, T., Luo, J., Yu, H. et al. Blocker displacement amplification-based genetic diagnosis for autosomal dominant polycystic kidney disease and the clinical outcomes of preimplantation genetic testing. J Assist Reprod Genet 40, 783–792 (2023). https://doi.org/10.1007/s10815-023-02722-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10815-023-02722-1