Abstract
In this study, the correlation between the specific growth rate of Phaeodactylum tricornutum and fucoxanthin (fucoxanthin) biosynthesis was investigated both in batch and in semi-continuous cultures. Fucoxanthin content from P. tricornutum biomass showed a positive correlation with specific growth rate during the normal culture period. The maximum yield of fucoxanthin (2.42 mg g−1 dry biomass) was observed at day 5, and the highest specific growth rate (1.91) was observed at day 4, both of which were during the exponential phase. Upon treatment with growth inhibitors (AZD-8055 or norflurazon) during the exponential phase, fucoxanthin content decreased with growth rate in a concentration-dependent manner. In a semi-continuous culture of P. tricornutum with dilution rates from 0.1 to 0.5 day−1, we found a positive correlation between specific growth rate and fucoxanthin biosynthesis in a steady-state condition. Based on the transcriptome analysis results, different metabolic and key regulatory genes were active at different growth phases. Particularly, during the exponential growth phase, various genes related to regulatory mechanisms, such as cell growth and replication, were expressed, whereas in the stationary phase, their expression was reduced. In the fucoxanthin biosynthesis pathway, 1-deoxy-d-erythritol 2,4-cyclodiphosphate synthase (ISPD) and violaxanthin deepoxidase (VDE) genes showed higher expression levels in the exponential phase, thus indicating that they are critical genes for the regulation of fucoxanthin biosynthesis in P. tricornutum. Our results are valuable to our understanding of the basic mechanism of fucoxanthin biosynthesis, thus providing theoretical guidance for the commercial production of fucoxanthin derived from P. tricornutum.
Graphical abstract

Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Phaeodactylum tricornutum has been used as a model in diatom research. The sequencing of its genome was completed in 2008, and many biotechnological methods have been developed using this microalga, including genetic transformation (Niu et al. 2012), gene knockout tools (Madhuri et al. 2019), and CRISPR/Cas9 for gene-targeted mutation (Serif et al. 2018). Phaeodactylum tricornutum is considered an important microalgal cell factory for the production of useful foreign materials, such as antibody proteins, antibiotics, and medicinal proteins (Desbois et al. 2008; Hempel et al. 2011; Butler et al. 2020). In the industry, this microalga has been used as feedstock for marine organisms in the early life stage (Sørensen et al. 2016). Owing to its high lipid content (20–30% of dry cell biomass) and fast growth, its potential for biofuel production has been extensively investigated (Branco-Vieira et al. 2018). In the food industry, P. tricornutum has been used as a source of healthy nutritional substances, such as omega-3 fatty acids, including docosahexaenoic acid and eicosapentaenoic acid (Xu et al. 2017; Cui et al. 2019). Recently, it has been suggested as a new source for the commercial production of the marine carotenoid fucoxanthin (Conceição et al. 2020).
Fucoxanthin is the most abundant carotenoid in nature, accounting for more than 10% of the total carotenoid production in the ecosystem (Peng et al. 2011). Fucoxanthin has also been reported to possess numerous biological activities, including anticancer, anti-obesity, antidiabetic, antiangiogenic, antioxidant, and antimalarial (Zarekarizi et al. 2019). Among these, its anti-obesity effect is well studied. Fucoxanthin products are now commercially available, most of which are derived from Laminaria japonica and Undaria pinnatifida, in the form of a simple extract. However, it is economically difficult to isolate high-purity fucoxanthin from this species, thus limiting its application in the food and cosmetic industries (Dai and Kim 2016). Microalgae, including Isochrysis aff. galbana, Nitzschia laevis, Odontella aurita, Chaetoceros gracilis, and P. tricornutum, are emerging as new sources for fucoxanthin production as their fucoxanthin concentrations are at least 10 times more than those of macroalgae (w/w). In addition, several industrial processes to isolate high-purity fucoxanthin in these species have been developed (Kim et al. 2012; Zarekarizi et al. 2019). Among these species, P. tricornutum has been extensively investigated as fucoxanthin-producing biomass, and its standardized extract containing fucoxanthin, has been developed for functional food (Kim et al. 2012; Koo et al. 2019).
To produce P. tricornutum biomass, various studies have been conducted using different cultivation systems, such as open (raceway) ponds and photobioreactors (tubular, column, and flat-panel types). In addition to the cultivation systems, cultivation conditions (photoautotrophic, heterotrophic, mixotrophic, and photoheterotrophic cultivation), environmental factors (light, CO2 concentration, and nutrients), and the culture method (batch, semi-continuous, and continuous culture) have been explored to understand the physiology of microalgae growth and determine the best condition for its production (Vuppaladadiyam et al. 2018; Guler et al. 2019). In the culture of microalgae, growth rate is one of the most important parameters affecting biomass production. Particularly in microorganisms, the specific growth rate (SGR; the number of divisions per cell per unit time) is a key parameter to evaluate culture growth (Pérez et al. 2008). Generally, the growth of bacteria or other microorganisms, such as protozoa, yeasts, and microalgae, follows a sigmoidal growth curve with four phases (i.e., lag, exponential, stationary, and death phase), and the maximum SGR occurs in the exponential phase (Rodríguez-León et al. 2018). In several studies of microalgae culture, the results of the growth kinetic model are expressed in SGR and this is used to explain the effect of various environmental or nutritional factors, or the correlation between the growth rate and various parameters (Li et al. 2014; Li et al. 2017). In particular, many studies have shown the negative correlation between lipid metabolism and microalgal growth. In fucoxanthin-producing microalgae such as I. galbana and P. tricornutum, the carotenoid or fucoxanthin contents were analyzed according to the growth phases, which have different SGRs, and different levels of these compounds were detected (Gómez-Loredo et al. 2016). Thus, fucoxanthin biosynthesis might be correlated with SGR in the culture period, but there have been no studies focusing on the correlation between fucoxanthin biosynthesis and the SGR in the fucoxanthin-producing microalgae.
In this study, the correlation between the SGR and fucoxanthin biosynthesis in P. tricornutum was investigated to understand fucoxanthin biosynthesis during culture and determine the optimal culture conditions based on SGR regulation for biomass production. We found a positive relationship between the SGR of P. tricornutum and the fucoxanthin content of the biomass in batch culture. This relationship was further confirmed after treatment of the culture with a growth inhibitor and through semi-continuous culture at different dilution rates. In addition, transcriptome sequencing of P. tricornutum was performed to identify the key genes involved in the regulation of fucoxanthin biosynthesis.
Materials and methods
Materials and reagents
ATP competitive mTOR kinase inhibitor (AZD-8055), norflurazon, and fucoxanthin analytical standards were from Sigma-Aldrich (USA). Methanol and water used for high-performance liquid chromatography (HPLC) analysis and ethanol for extraction were purchased from Daejung Chemicals & Metals (Korea). The F/2 medium was prepared as described previously (Guillard 1975).
P. tricornutum strain and culture conditions
Phaeodactylum tricornutum UTEX 646 was obtained from the University of Texas collection (https://utex.org/, USA). In the normal batch culture, P. tricornutum was grown in 2 L of sterile, natural seawater enriched with the F/2 medium, incubated at 20 ± 1 °C under a continuous 40 W cool fluorescent lamp (150 μmol photons m−2 s−1), and aerated with 0.03% (v/v air) CO2. A semi-continuous subculture in a 2-L photobioreactor was started on day 9, and new medium was added to the culture daily after harvesting the same volume of the “old” culture solution according to the dilution rate. Dilution rate was defined as the ratio of the volume of the new medium added to the whole culture volume and ranged from 0.1 to 0.5. The semi-continuous culture was maintained for 11 days after the start of the experiment.
Treatment by growth inhibitors
AZD-8055 and norflurazon were added to the culture solution under normal conditions on day 3. The stock solution was prepared by dissolving the inhibitor in ethanol and passing it through a 0.4-μm Millipore filter. The AZD-8055 solution was added to the culture at final concentrations of 10, 20, and 50 μM. Norflurazon was administered at final concentrations of 0.1, 0.5, and 2.0 μM. Cell concentration and SGR were measured daily, and fucoxanthin content was analyzed during the exponential (day 4) and stationary (day 10) growth phases.
Measurement of cell concentration and specific growth rate
The number of P. tricornutum cells was counted daily using a light microscope and a hemocytometer. The daily SGR (μ) of P. tricornutum during the culture period was calculated as follows:
where N1 and N2 are the cell concentrations at the beginning and end of the experiment, respectively, with 1-day intervals (t2 − t1).
Fucoxanthin analysis
Culture solution (10 mL) was collected and centrifuged for 10 min at 3950×g. The supernatant was discarded, and the microalgal biomass was freeze-dried for 2 days. Then, 10 mL ethanol was added to the dried biomass and fucoxanthin was extracted by sonication (model UCP-10, 300 W, 40 kHz, JEIO Technology, Korea) for 1 h at 24–30 °C. The supernatant (1 mL) was filtered through a 0.22-μm syringe filter for use in HPLC analysis. An Agilent 1200 HPLC system (Agilent Technologies, USA) consisting of a G1312A binary pump, a G1367B auto sampler, a G1315D PDA detector, and a G1316A column oven was used for fucoxanthin quantification. The mobile phase, i.e., methanol and water, was eluted at a flow rate of 0.7 mL min−1 at 35 °C. The YMC carotenoid column (250 mm length × 4.6 mm i.d.; 5-μm particle size; Waters, USA) was used for separation under the following gradient program: methanol was increased from 90 to 100% for 20 min, held maintained at 100% for the next 5 min, decreased to 90% for 5 min, and then held at 90% for 5 min. The sample solution (10 μL) was injected, and the chromatogram was recorded at 445 nm. Fucoxanthin was quantified based on a calibration curve in the concentration range 0.5–50 μg mL−1.
Transcriptome sequencing
Phaeodactylum tricornutum cells were harvested during the exponential (day 4; D4) and stationary (day 14; D14) growth phases under normal culture conditions. They were washed with the distilled water and centrifuged. The cell pellets were frozen at − 80 °C for 24 h. Then, the frozen cell pellets were ground using liquid nitrogen and total RNA was extracted using a Qiagen Plant RNeasy Mini kit (Qiagen, Germany) according to the manufacturer’s instructions. RNA concentration and purity were measured using a NanoDrop ND-1000 UV/Vis spectrophotometer (Thermo Fisher Scientific, USA). The cDNAs were synthesized and amplified from the purified RNA to generate the libraries according to the Illumina RNA-Seq protocol. The libraries were sequenced using Illumina HiSeq 2000 according to the manufacturer’s instructions. The sequencing results were pre-processed using the DynamicTrim and LengthSort programs from the SolexaQA package. Gene expression was given as the total number of sequencing reads analyzed by the HTseq software after mapping using HISAT2 and normalized using the DESeq library. Raw and processed data in the form of sequence reads and assembled transcriptomes were deposited in GenBank under the accession numbers BioProject PRJNA625589 (transcriptome assembly) and SAMN14641819~24 (sequence read archive).
Transcriptome data analysis
The sequence information of P. tricornutum (GCF_000150955.2_ASM15095v2_genomic.fan) obtained from the NCBI database was used as the reference sequence for gene annotation. Gene annotation was based on the reference sequence, the amino acid sequence from the NR (Stramenopiles) database of the NCBI, and the BLASTX result with e-value ≤ 1e-10. Differentially expressed genes (DEGs) were selected using the 2-fold change and binomial test methods with p value ≤ 0.1. Gene ontology of DEGs was performed by alignment with a p value ≤ 0.05 and classified into BP (biological process), CC (cellular component), and MF (molecular function) according to the functional category. DEG levels related to terpenoid and carotenoid biosynthetic pathways were selected using the TBLASTN tool with an e-value ≤ 1e-10. The metabolic pathway analysis of the DEGs was performed using the KEGG (http://www.genome.jp/kegg) database (Kanehisa and Goto 2000).
Statistical analysis
Data are expressed as the means ± standard deviation based on triplicate experiments. Statistical analysis was performed using IBM SPSS Software Statistics v20 (IBM, USA).
Data were analyzed using one-sided t test or two-way ANOVA, followed by Tukey’s multiple comparisons test. P values ≤ 0.05 or ≤ 0.01 were considered significant.
Results
Specific growth rate and fucoxanthin biosynthesis under normal culture conditions
Figure 1 a shows the growth curve of P. tricornutum and its daily SGR under normal culture conditions. Its growth follows a sigmoidal curve, reaching the stationary phase at day 7. The daily SGR increased steeply until the exponential phase (day 4) and gradually decreased, achieving a maximum SGR of 1.91 day−1. The fucoxanthin content ranged from 1.13 to 2.42 mg g−1 dry biomass (Fig. 1b) and had a quite similar trend as SGR. Fucoxanthin content increased until day 5 and gradually decreased thereafter. Thus, these findings indicate that the SGR of P. tricornutum is positively correlated with fucoxanthin biosynthesis.
Effect of growth inhibitors on fucoxanthin biosynthesis
To further investigate the correlation between SGR and fucoxanthin biosynthesis in P. tricornutum, growth inhibitors, either AZD-8055 or norflurazon, were added to cultures on day 3 at a range of concentrations (AZD-8055, 10–50 μM; norflurazon, 0.1–2.0 μM). Subsequently, the cell count and SGR were examined daily, and fucoxanthin content was determined on days 4 and 10.
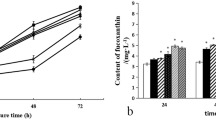
As shown in Fig. 2a, the growth of P. tricornutum decreased when the concentration of the growth inhibitor was increased. Growth inhibition was seen just after inhibitor treatment, and different SGRs could be determined from day 4. From day 4, the SGR showed a decreasing trend as the concentration of inhibitors was increased, and by day 10, SGR was at the minimum (Fig. 2b). This result indicates that the SGR during the exponential phase could be changed by growth inhibitors. At these points, fucoxanthin content was analyzed, and Fig. 2c shows that it decreased as SGR decreased at day 4, and at day 10, fucoxanthin content was also at the minimum. From these results, we can confirm the positive correlation between the SGR and fucoxanthin biosynthesis in P. tricornutum.
Effect of AZD-8055 and norflurazon on a cell concentration, b specific growth rate, and c fucoxanthin content of Phaeodactylum tricornutum. The specific growth rate and fucoxanthin content were analyzed at days 4 and 10 from P. tricornutum cultures treated with growth inhibitors under normal culture conditions. Error bars represent standard deviation from three independent replicates. ** indicates a significant difference with control at p ≤ 0.01. ND means not detected
Specific growth rate and fucoxanthin biosynthesis under semi-continuous culture conditions
As a production system for microalgae, the semi-continuous culture has several benefits over batch culture, such as higher automation, easier to scale up, high volumetric biomass productivity, and no time lost to the preparation of fresh culture systems (Zhu 2015). In this study, semi-continuous culture was applied to P. tricornutum biomass production. The dilution rate, which is the proportion of new medium used to intermittently replenish the culture solution, was varied from 0.1 to 0.5, with a 1-day interval. The semi-continuous culture of P. tricornutum was started from day 9 and was maintained for 11 days thereafter. SGR and fucoxanthin were measured daily during steady-state conditions (days 10–20).
As shown in Fig. 3a, a steady state was established in the culture after day 10 and the average cell concentration was reduced as dilution rate was increased. The average daily SGR ranged from 0.1 to 0.8 and showed a positive correlation with the dilution rate (Fig. 3b), indicating that the growth rate of P. tricornutum is proportional to the dilution rate in a semi-continuous culture. Daily biomass productivity, based on the daily harvest volume and cell concentration, increased at higher dilution rates (Fig. 3c). Interestingly, fucoxanthin biosynthesis was decreased when dilution rate was decreased from 0.5 to 0.1 (Fig. 3d). When the dilution rate was 0.5, the fucoxanthin amount remained almost the same as that during the start of continuous culture. However, when the dilution rate was 0.1, fucoxanthin serially decreased to the basal levels at the end of the continuous culture period. From these results, we confirmed that fucoxanthin biosynthesis is highly correlated with the SGR of P. tricornutum (Fig. 3d).
Semi-continuous cultures of P. tricornutum. A semi-continuous culture was started from day 9 with a cell counts having different dilution rates. b The specific growth rate, c daily cell productivity, and d fucoxanthin content were analyzed based on the dilution rate. Error bars represent standard deviation from three independent replicates. Different letters indicate significant differences at p ≤ 0.05
Transcriptome analysis of P. tricornutum
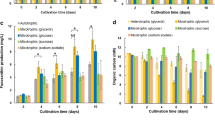
Illumina HiSeqX RNA sequencing was carried out in order to examine the gene expression profile and to identify the regulation mechanisms of fucoxanthin biosynthesis during the exponential (day 4, D4) and stationary phase (day 14, D14) of P. tricornutum under normal culture conditions. Fucoxanthin biosynthesis might be differently regulated by the large difference in SGR in these two growth phases. Table S1 summarizes the results of transcriptome sequencing. The number and the volcano plots of the up- and downregulated genes in the exponential phase in comparison with those in the stationary phase are shown in Fig. S1. Among the 10,145 unigenes annotated with functional description, 1689 were twofold upregulated and 1993 were twofold downregulated; these data are shown as the differences in the average log2 expression (Fig. S1a). These up- or downregulated genes are displayed in a volcano plot, with a statistical value of p < 0.05 (Fig. S1b). Based on sequence homology, these unigenes were classified into three Gene Ontology (GO) categories, namely BP, MF, and CC (Fig. 4). In the exponential growth phase, 1627 genes were assigned to BP (72.25 %), 611 to MF (27.13 %), and only 14 to CC (0.13 %). Among the unigenes in the BP group, biological regulation proteins were the most abundantly encoded (293 genes). However, in the stationary phase, only 179 genes related to BP were upregulated (6.12 %), and the other two groups were upregulated similarly (1431 genes for CC, 49.92 %; 1315 for MF, 44.96 %). Thus, this indicates that many genes related to regulatory mechanisms were expressed during the exponential growth phase, and their expression was reduced during the stationary phase. This result was confirmed by the KEGG classification of DEGs (Fig. 5). In the exponential growth phase, the replication and repair groups, which are highly related to cell growth, included more DEGs than those in the stationary growth phase. However, DEGs related to lipid, carbohydrate, and energy metabolism were more abundant in the stationary phase (Fig. 5a). In particular, DEGs related to various lipid metabolisms were more abundant in the stationary phase (Fig. 5b). There have been several reports of a negative correlation between lipid production and cell growth in P. tricornutum (Bauer et al. 2019). This was also seen in our study, by transcriptome sequencing in two different growth phases with different SGRs.
Gene Ontology (GO) annotation of the differentially expressed genes (DEGs) from P. tricornutum at day 4 (D4, exponential phase) and day 14 (D14, stationary phase), under normal culture conditions. a Upregulated genes and b downregulated genes at D4 were classified into three functional categories, namely molecular function (MP), cellular component (CP), and biological processes (BP)
KEGG pathway classifications of the differentially expressed genes (DEGs) from P. tricornutum at day 4 (D4, exponential phase) and day 14 (D14, stationary phase) under normal culture conditions. Among the DEGs related to lipid metabolism, more genes were downregulated than upregulated in a major classification of the KEGG pathway and b subclassification of lipid metabolism
The relative expression of genes involved in the fucoxanthin biosynthetic pathway is shown in Fig. 6 as analyzed using KEGG. Generally, most genes were highly expressed during the exponential growth phase compared with the stationary growth phase. In particular, in the upstream step of the MEP (2-C-methyl-d-erythritol 4-phosphate) pathway, which uses the substrates pyruvate and glyceraldehyde 3-phosphate to generate IPP (isopentenyl diphosphate) and DMAPP (dimethylallyl diphosphate) in 7 steps, the ISPD (1-deoxy-d-erythritol 2,4-cyclodiphosphate synthase) gene showed the highest difference (among 7 genes) in expression level between the two growth phases. In the downstream step, the VDE (violaxanthin deepoxidase) gene was the most highly expressed in the exponential phase.
Heatmap of genes related to the fucoxanthin biosynthesis pathway expressed between D4 and D14. Colors indicate the expression levels at D4 compared with D14 and expressed as log2 fold change. DXS, 1-deoxy-d-xylulose 5-phosphate synthase; DXR, 1-deoxy-d-erythritol 2,4-cyclodiphosphate synthase; ISPD, 2-C-methyl-d-erythritol 4-phosphate cytidylyltransferase; CMK, 4-diphosphocytidyl-2c-methyl-d-erythritol kinase; ISPF, 2-C-methyl-d-erythritol 2,4-cyclodiphosphate synthase; HDS, 1-hydroxy-2-methyl-2-(E)-butenyl-4-diphosphate synthase; HDR, hydroxymethylbutenyl diphosphate reductase; GGPS, farnesyltranstransferase; PSY, phytoene synthase; PDS, phytoene desaturase; Z-ISO, zeta-carotene isomerase; ZDS, ζ-carotene desaturase; CRTISO, carotenoid isomerase; carotene 7,8-desaturase; LCYE, lycopene ε-cyclase; LCYB, lycopene β-cyclase; BCH, β-carotene hydroxylase; LUT1, ε-ring hydroxylase; BCH, β-carotene hydroxylase; VDE, violaxanthin deepoxidase; ZEP, zeaxamthin eposidase; NXS, neoxanthin synthase; CYP707A, carotene ε-hydroxylase
Discussion
In this study, we demonstrated the correlation between fucoxanthin biosynthesis and the SGR of P. tricornutum using batch (Figs. 1 and 2) and semi-continuous culture production (Fig. 3). Fucoxanthin biosynthesis of P. tricornutum was increased along with SGR. Thus, the maximum level of fucoxanthin biosynthesis occurred in mid-exponential phase (Fig. 1b), which had the highest SGR in the batch culture. In semi-continuous culture, this was achieved at 0.5 dilution rate, which had the highest daily average SGR (Fig. 3c). Microalgae are important as lipid producers for bioethanol and omega-3 fatty acids. Thus, numerous studies have investigated the correlation between microalgal growth and lipid metabolism. In these reports, lipid metabolism was generally negatively correlated with microalgal growth (Xin et al. 2011; Conceição et al. 2020). However, there are only a few studies on the relationship between microalgal growth and pigment biosynthesis (Saoudi-Helis et al. 1994; Yi et al. 2019). In particular, in microalgae-producing fucoxanthin, only a few studies have been conducted to investigate the fucoxanthin content based on the growth phase. In I. galbana and P. tricornutum, their carotenoid or fucoxanthin contents were higher in the exponential phase than in the stationary phase (Gómez-Loredo et al. 2016). However, this study did not focus on the correlation between SGR and fucoxanthin biosynthesis. Thus, to the best of our knowledge, this is the first study to report on the correlation between fucoxanthin biosynthesis and SGR in P. tricornutum. Here, two growth inhibitors (AZD-8055 and norflurazon) were added to the P. tricornutum culture to change the SGR (Fig. 2). Fucoxanthin biosynthesis in the exponential and stationary phases was also changed based on the SGR decrease induced by the increasing concentration of the two inhibitors (Fig. 2b, c). These inhibitor experiments further supported the positive correlation between fucoxanthin biosynthesis and the SGR of P. tricornutum. Meanwhile, the decreasing trend in fucoxanthin contents during the stationary phase was slightly different between the two inhibitor treatments. In Fig. 2c, at day 10, fucoxanthin content was reduced to the minimum after treatment with either inhibitor. However, fucoxanthin content was serially decreased to almost zero by increased norflurazon concentration, whereas it was maintained at the minimum in the AZD-8055-treated groups. Although AZD-8055 and norflurazon are both growth inhibitors that act on plants and microalgae, their inhibition mechanisms are quite different. On the one hand, AZD-8055 is a specific inhibitor of the target of rapamycin (TOR) kinase, which is important for cell growth, proliferation, metabolism, and angiogenesis (Imamura et al. 2016). Thus, this inhibitor is known to inhibit cell proliferation in P. tricornutum in a dose-dependent manner (Prioretti et al. 2017). On the other hand, norflurazon is a herbicide that blocks chloroplast biogenesis by inhibiting carotenoid synthesis via the enzyme phytoene desaturase (PDS) (Undabeytia et al. 2000; Taparia et al. 2019). This inhibitor works on plants and various microalgae, including P. tricornutum, Haematococcus pluvialis, and Chlorella zofingiensis (Steinbrenner and Sandmann 2006; Liu et al. 2010; Prioretti et al. 2017). Thus, their varying results on inhibiting fucoxanthin biosynthesis during the stationary phase may be due to their different inhibition mechanisms.
There have been a few studies on the semi-continuous culture of P. tricornutum. Cerón-García et al. (2013) compared the semi-continuous culture and the fed-batch culture in mixotrophic conditions. The optimal dilution rate and nutrient concentration for the production of P. tricornutum have also been examined in a semi-continuous culture system (Fabregas et al. 1996) (Wang et al. 2018). However, the correlations among fucoxanthin biosynthesis, SGR, and dilution rate have not yet been investigated until now. In particular, the strategy for producing valuable components can be different from that for biomass production.
Based on our results a fast growth rate should be maintained to obtain the maximum fucoxanthin and biomass contents. However, sometimes, biomass production and component production are not proportional to the growth rate; for example, lipid production is inversely correlated with the growth rate of microalgae. Thus, understanding the relationship between fucoxanthin biosynthesis and the growth rate of P. tricornutum is important to maximize fucoxanthin production in various culture systems. In continuous culture, a 1-day interval was used to replenish the medium (Fig. 3a) and a steady state of culture was established with decreased cell concentration along with dilution rate. However, at a dilution rate of 0.5, when enough time (for example, 2–3 days) passes to reach the maximum cell concentration before the addition of the new medium, P. tricornutum biomass productivity might increase, thus maintaining high fucoxanthin levels during semi-continuous culture. The time to reach maximum cell concentration would be different with culture volume. Therefore, more optimization studies should be conducted to maximize the production of biomass and fucoxanthin from P. tricornutum.
In addition, the optimal harvest time of P. tricornutum biomass for fucoxanthin production would be different in fed-batch and semi-continuous cultures. In the fed-batch culture, the end of the exponential phase (days 5–6 in this study) is the optimal harvest time because the highest fucoxanthin content and cell concentration were achieved here. Meanwhile, in the semi-continuous culture, the dilution rate should be maintained at 0.5 because the highest cell productivity with higher fucoxanthin content (Fig. 3c, d). As mentioned above, the biomass of P. tricornutum can be harvested at the maximum cell concentration after the replenishment of the medium with 2–3-day interval.
From Illumina HiSeqX RNA sequencing of P. tricornutum in the exponential and stationary phase under normal culture condition, it was confirmed that many genes related to regulatory mechanism were expressed more in the exponential phase compared to the stationary phase (Fig. 4a). Especially, among the DEGs, the genes belonged to the replication and repair group, which is highly related to cell growth, were more abundant in the exponential phase (Fig. 5a). Thus, we speculate that the positive correlation between fucoxanthin biosynthesis and the SGR during culture was associated with regulatory mechanisms related to cell growth. In algae, fucoxanthin functions as an antenna pigment in the thylakoid membrane. Fucoxanthin and chlorophyll comprise light-harvesting complexes that transfer energy to the photosynthetic electron transport chain and offer protection against excess light (Petrushkina et al. 2017; Zarekarizi et al. 2019). In other words, fucoxanthin biosynthesis in microalgae is closer to primary metabolic process, unlike in higher organisms such as plants, in which carotenoid biosynthesis belongs to secondary metabolism. Thus, fucoxanthin biosynthesis might be regulated together with the cell growth in P. tricornutum, and that is why fucoxanthin biosynthesis is positively correlated with SGR in P. tricornutum.
Several key regulatory genes involved in fucoxanthin biosynthesis have been suggested in microalgae. In the MEP pathway, 1-deoxy-d-xylulose 5-phosphate synthase (DXS) is the first enzyme and considered a key enzyme for the rate-limiting step in the regulation of this pathway (Yang and Wei 2020). In addition, PSY (phytone synthase) found in all photosynthetic organisms is often considered a rate-limiting enzyme in the carotenoid biosynthetic pathway. Thus, these two enzymes are targets for metabolic engineering of the carotenoid biosynthetic pathway in many organisms, even in P. tricornutum. Eilers et al. (2016) have reported that the transformation of these two genes in the P. tricornutum genome increased fucoxanthin biosynthesis by 1.8- to 2.4-fold.
Here (Fig. 6), DXS was similarly expressed in both growth phases, but PSY was more significantly expressed in the exponential phase. However, ISPD and VDE showed greater increases in gene expression in the exponential phase than DXS and PSY, indicating that ISPD and VDE may be key genes in the regulation of fucoxanthin biosynthesis, according to the cell growth rate. Among the DEGs, 35 genes were assigned as transcription factors (Fig. S2). Among them, the heat shock protein (HSF) family was the largest group (18 genes), followed by the MYB (DNA-binding domain) family (8 genes). In a previous study, transcription factors were predicted from the genome sequence of 7 microalgae (Thiriet-Rupert et al. 2016). Unlike other microalgae, P. tricornutum possesses a higher number of transcription factors belonging to the HSF (67 genes, 34.18 %) and MYB (27 genes, 13.77 %) families. Our results were similar to those previously reported. In addition to these two families, we identified transcription factors of the bZIP (basic leucine zipper), C-Myb R2R3 (cellular MYB R2R3), C2H2-ZF (C2H2 Zinc finger protein), homeodomain, transcription initiation factor TFIID subunit), and WDR (WD40 repeat protein) groups as DEGs. Among them, the transcription factor (XM_002180741.1) belonging to C-Myb R2R3 showed the largest difference in gene expression in the exponential phase. Further studies are required to assess the relationship between this transcription factor and fucoxanthin biosynthesis.
Conclusion
In this study, a positive correlation between fucoxanthin biosynthesis and the SGR of P. tricornutum was demonstrated by three different culture conditions: normal, inhibited, and semi-continuous culture. The maximum fucoxanthin content in the normal culture was achieved during the exponential phase, in which the growth rate of cells increased gradually. In the semi-continuous culture, high level of fucoxanthin was maintained with high dilution rate, which promoted higher cell growth rate. Thus, we speculate that fucoxanthin biosynthesis is highly related with the growth rate of P. tricornutum. From the results of transcriptome analysis, in the exponential and stationary growth phases, fucoxanthin biosynthesis is closely related to cell growth metabolism, rather than lipid and carbohydrate metabolism, and ISPD and VDE are suggested as key regulatory genes for fucoxanthin biosynthesis during the exponential phase. These results provide valuable information for the increased understanding of fucoxanthin biosynthetic metabolism and for the commercial production of functional ingredients from P. tricornutum.
References
Bauer C, Schmitz C, Corrêa R, Herrera C, Ramlov F, Oliveira E, Pizzato A, Varela L, Cabral D, Yunes R (2019) In vitro fucoxanthin production by the Phaeodactylum tricornutum diatom. Stud Nat Prod Chem 63:211–242
Branco-Vieira M, San Martin S, Agurto C, Santos MA d, Freitas MA, Mata TM, Martins AA, Caetano NS (2018) Potential of Phaeodactylum tricornutum for biodiesel production under natural conditions in Chile. Energies 11:54
Butler T, Kapoore RV, Vaidyanathan S (2020) Phaeodactylum tricornutum: a diatom cell factory. Trends Biotechnol 38:606–622
Cerón-García M, Fernández-Sevilla J, Sánchez-Mirón A, García-Camacho F, Contreras-Gómez A, Molina-Grima E (2013) Mixotrophic growth of Phaeodactylum tricornutum on fructose and glycerol in fed-batch and semi-continuous modes. Bioresour Technol 147:569–576
Conceição D, Lopes R, Derner R, Cella H, Carmo A, D’Oca M, Petersen R, Passos M, Vargas J, Galli-Terasawa L, Kava V (2020) The effect of light intensity on the production and accumulation of pigments and fatty acids in Phaeodactylum tricornutum. J Appl Phycol 32:1017–1025
Cui Y, Thomas-Hall SR, Schenk PM (2019) Phaeodactylum tricornutum microalgae as a rich source of omega-3 oil: progress in lipid induction techniques towards industry adoption. Food Chem 297:124937–124946
Dai J, Kim JC (2016) In vivo anti-obesity efficacy of fucoxanthin-loaded emulsions stabilized with phospholipid. J Pharm Investig 46:669–675
Desbois AP, Lebl T, Yan L, Smith VJ (2008) Isolation and structural characterization of two antibacterial free fatty acids from the marine diatom, Phaeodactylum tricornutum. Appl Environ Microbiol 81:755–764
Eilers U, Bikoulis A, Breitenbach J, Büchel C, Sandmann G (2016) Limitations in the biosynthesis of fucoxanthin as targets for genetic engineering in Phaeodactylum tricornutum. J Appl Phycol 28:123–129
Fabregas J, Patino M, Morales ED, Cordero B, Otero A (1996) Optimal renewal rate and nutrient concentration for the production of the marine microalga Phaeodactylum tricornutum in semicontinuous cultures. Appl Environ Microbiol 62:266–268
Gómez-Loredo A, Benavides J, Rito-Palomares M (2016) Growth kinetics and fucoxanthin production of Phaeodactylum tricornutum and Isochrysis galbana cultures at different light and agitation conditions. J Appl Phycol 28:849–860
Guillard RR (1975) Culture of phytoplankton for feeding marine invertebrates. In: Smith WL, Chanley MH (eds) Culture of marine invertebrate animals. Springer, Boston, pp 29–60
Guler B, Deniz I, Demirel Z, Oncel S, Imamoglu E (2019) Transition from start-up to scale-up for fucoxanthin production in flat plate photobioreactor. J Appl Phycol 31:1525–1533
Hempel F, Lau J, Klingl A, Maier UG (2011) Algae as protein factories: expression of a human antibody and the respective antigen in the diatom Phaeodactylum tricornutum. PloS One 6:e28424. doi:
Imamura S, Kawase Y, Kobayashi I, Shimojima M, Ohta H, Tanaka K (2016) TOR (target of rapamycin) is a key regulator of triacylglycerol accumulation in microalgae. Plant Signal Behav 11:e1149285
Kanehisa M, Goto S (2000) KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res 28:27–30
Kim SM, Jung YJ, Kwon ON, Cha KH, Um BH, Chung D, Pan CH (2012) A potential commercial source of fucoxanthin extracted from the microalga Phaeodactylum tricornutum. Appl Environ Microbiol 166:1843–1855
Koo SY, Hwang JH, Yang SH, Um JI, Hong KW, Kang K, Pan CH, Hwang KT, Kim SM (2019) Anti-obesity effect of standardized extract of microalga Phaeodactylum tricornutum containing fucoxanthin. Mar Drugs 17:311–326
Li Y, Xu J, Gao K (2014) Light-modulated responses of growth and photosynthetic performance to ocean acidification in the model diatom Phaeodactylum tricornutum. PloS One 9:e96173
Li F, Beardall J, Collins S, Gao K (2017) Decreased photosynthesis and growth with reduced respiration in the model diatom Phaeodactylum tricornutum grown under elevated CO2 over 1800 generations. Glob Chang Biol 23:127–137
Liu J, Zhong Y, Sun Z, Huang J, Sandmann G, Chen F (2010) One amino acid substitution in phytoene desaturase makes Chlorella zofingiensis resistant to norflurazon and enhances the biosynthesis of astaxanthin. Planta 232:61–67
Madhuri S, Bártulos CR, Serif M, Lepetit B, Kroth PG (2019) A strategy to complement PtAUREO1a in TALEN knockout strains of Phaeodactylum tricornutum. Algal Res 39:101469–101478
Niu YF, Yang ZK, Zhang MH, Zhu CC, Yang WD, Liu JS, Li HY (2012) Transformation of diatom Phaeodactylum tricornutum by electroporation and establishment of inducible selection marker. Biotechniques 52:1–3
Peng J, Yuan JP, Wu CF, Wang JH (2011) Fucoxanthin, a marine carotenoid present in brown seaweeds and diatoms: metabolism and bioactivities relevant to human health. Mar Drugs 9:1806–1828
Pérez EB, Pina IC, Rodríguez LP (2008) Kinetic model for growth of Phaeodactylum tricornutum in intensive culture photobioreactor. Biochem Eng J 40:520–525
Petrushkina M, Gusev E, Sorokin B, Zotko N, Mamaeva A, Filimonova A, Kulikovskiy M, Maltsev Y, Yampolsky I, Guglya E (2017) Fucoxanthin production by heterokont microalgae. Algal Res 24:387–393
Prioretti L, Avilan L, Carrière F, Montane MH, Field B, Grégori G, Menand B, Gontero B (2017) The inhibition of TOR in the model diatom Phaeodactylum tricornutum promotes a get-fat growth regime. Algal Res 26:265–274
Rodríguez-León JA, de Carvalho JC, Pandey A, Soccol CR, Rodríguez-Fernández DE (2018) Kinetics of the solid-state fermentation process. In: Pandey A, Larroche C, Soccol C (eds) Current developments in biotechnology and bioengineering: current advances in solid-state fermentation. Elsevier, Amsterdam, pp 57–82
Saoudi-Helis L, Dubacq JP, Marty Y, Samain JF, Gudin C (1994) Influence of growth rate on pigment and lipid composition of the microalga Isochrysis aff. galbana clone T. iso. J Appl Phycol 6:315–322
Serif M, Dubois G, Finoux AL, Teste MA, Jallet D, Daboussi F (2018) One-step generation of multiple gene knock-outs in the diatom Phaeodactylum tricornutum by DNA-free genome editing. Nat Commun 9:1–10
Sørensen M, Berge GM, Reitan KI, Ruyter B (2016) Microalga Phaeodactylum tricornutum in feed for Atlantic salmon (Salmo salar)—effect on nutrient digestibility, growth and utilization of feed. Aquaculture 460:116–123
Steinbrenner J, Sandmann G (2006) Transformation of the green alga Haematococcus pluvialis with a phytoene desaturase for accelerated astaxanthin biosynthesis. Appl Environ Microbiol 72:7477–7484
Taparia Y, Zarka A, Leu S, Zarivach R, Boussiba S, Khozin-Goldgerg I (2019) A novel endogenous selection marker for the diatom Phaeodactylum tricornutum based on a unique mutation in phytoene desaturase 1. Sci Rep 9:8217–8229
Thiriet-Rupert S, Carrier G, Chénais B, Trottier C, Bougaran G, Cadoret JP, Schoefs B, Saint-Jean B (2016) Transcription factors in microalgae: genome-wide prediction and comparative analysis. BMC Genom 17:282–298
Undabeytia T, Nir S, Tel-Or E, Rubin B (2000) Photostabilization of the herbicide norflurazon by using organoclays. J Agric Food Chem 48:4774–4779
Vuppaladadiyam AK, Prinsen P, Raheem P, Lique R, Zhao M (2018) Microalgae cultivation and metabolites production: a comprehensive review. Biofuels Bioprod Bioref 12:304–324
Wang S, Verma SK, Said IH, Thomsen L, Ullrich MS, Kuhnert N (2018) Changes in the fucoxanthin production and protein profiles in Cylindrotheca closterium in response to blue light-emitting diode light. Microb Cell Factories 17:1–13
Xin L, Hong-Ying H, Yu-Ping Z (2011) Growth and lipid accumulation properties of a freshwater microalga Scenedesmus sp. under different cultivation temperature. Bioresour Technol 102:3098–3102
Xu J, Fan X, Li X, Fiu G, Zhang Z, Zhu Y, Fu Z, Qian H (2017) Effect of salicylic acid on fatty acid accumulation in Phaeodactylum tricornutum during stationary growth phase. J Appl Phycol 29:2801–2810
Yang R, Wei D (2020) Improving fucoxanthin production in mixotrophic culture of marine diatom Phaeodactylum tricornutum by LED light shift and nitrogen supplementation. Front Bioeng Biotechnol 8:820–834
Yi Z, Su Y, Cherek P, Nelson D, Lin J, Folfsson O, Wu H, Ashtiani K, Brynjolfsson S, Fu W (2019) Combined artificial high-silicate medium and LED illumination promote carotenoid accumulation in the marine diatome Phaeodactylum tricornutum. Microb Cell Fact 18:209–220
Zarekarizi A, Hoffmann L, Burritt D (2019) Approaches for the sustainable production of fucoxanthin, a xanthophyll with potential health benefits. J Appl Phycol 31:281–299
Zhu L (2015) Microalgal culture strategies for biofuel production: a review. Biofuel Bioprod Biorefin 9:801–814
Funding
This work was supported by the intramural grant (2Z06110) from the Korea Institute of Science and Technology (KIST), Republic of Korea.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
ESM 1
(DOCX 215 kb)
Rights and permissions
About this article
Cite this article
Kwon, D.Y., Vuong, T.T., Choi, J. et al. Fucoxanthin biosynthesis has a positive correlation with the specific growth rate in the culture of microalga Phaeodactylum tricornutum. J Appl Phycol 33, 1473–1485 (2021). https://doi.org/10.1007/s10811-021-02376-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10811-021-02376-5