Abstract
Microalgal lipid triacylglycerol (TAG) is a promising source for sustainable production of biofuels and edible oils. TAG biosynthesis in microalgae can be induced by nitrogen starvation (−N); however, regulation of the genes involved in this process is poorly known. To explore the regulation of gene encoding diacylglycerol acyltransferase 2 of oleaginous microalga Neochloris oleoabundans (NeoDGAT2) responsible for TAG biosynthesis, regulatory sequence of NeoDGAT2 gene (RDG) was cloned, and its functional regions were mapped by deletion analysis using the modified cyan fluorescent protein gene (CFP) as a reporter. The efficiency of CFP gene mTurquoise2 (Tq) without any intron, Tq1 and Tq2 with one and two copies of Chlamydomonas reinhardtii rbcS2 intron 1, respectively, was evaluated; Tq2 exhibited the highest CFP fluorescence activity in N. oleoabundans was therefore used as reporter for RDG deletion analysis. Deletion analysis of RDG revealed that the −N inducible region contained the predicted binding site of MYB transcription factor (MYB-bs). Specific binding between MYB-bs of RDG and the DNA-binding domain of MYB-related transcription factor ROC40 from C. reinhardtii was observed using electrophoretic mobility shift assay. Therefore, the corresponding MYB transcription factor in N. oleoabundans is probably the transcription factor regulating NeoDGAT2. The interaction between MYB transcription factor and the MYB-bs may play a role in regulating −N induced expression of NeoDGAT2, affecting TAG accumulation. MYB transcription factors can be the potential targets for engineering to increase TAG content. Increasing TAG content is essential for products derived from microalgal TAG to achieve economic viability.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Microalgal lipid triacylglycerol (TAG) is a promising source for sustainable production of biofuels (Chisti 2007; Mata et al. 2010) and edible oils (Draaisma et al. 2013; Klok et al. 2014). TAG accumulation in microalgae can be induced by abiotic stresses such as the deprivation of nutrients particularly nitrogen (Hu et al. 2008). However, nitrogen starvation (−N) eventually leads to inhibition of cell division and photosynthesis (Scragg et al. 2002; Song et al. 2013). Identifying the regulatory mechanisms that control the gene expression involved in TAG accumulation in microalgae under −N condition is one of the necessary steps prior to design novel solutions for development of products derived from microalgal TAG at a commercial scale.
The TAG biosynthesis pathway in microalgae is not fully known but thought to resemble that in higher plants (Chen and Smith 2012). In the model microalga Chlamydomonas reinhardtii, two sets of homologous enzymes catalyze two distinct and parallel TAG biosynthesis pathways located in plastid and endoplasmic reticulum (ER) (Fan et al. 2011; Goncalves et al. 2016b; Yamaoka et al. 2016; Nobusawa et al. 2017). TAG can be synthesized by sequential transfer of fatty acyl chains from acyl-CoA through the glycerol 3-phosphate pathway (Ohlrogge and Browse 1995; Coleman and Lee 2004); diacylglycerol acyltransferase (DGAT) catalyzing the final and committed step has been determined as the rate-limiting enzyme (Jako et al. 2001; Lung and Weselake 2006). Most microalgal species possess one DGAT type 1 (DGAT1) and multiple DGAT type 2 (DGAT2) genes (Chen and Smith 2012) localized in chloroplast and ER, respectively (Li-Beisson et al. 2015). DGAT1 and DGAT2 do not share any significant amino acid sequence similarity, although both catalyze the same enzymatic reaction. Only green alga seem to have DGAT2 in the higher plant clade, while all algal species had at least one representative DGAT2 in the animal clade (Chen and Smith 2012). DGAT2 has been shown as the potent enzyme in TAG biosynthesis (Gong et al. 2013; Hung et al. 2013; Chungjatupornchai and Watcharawipas 2015).
Identifying the regulatory mechanisms controlling DGAT2 expression has been attempted so far in few microalgal species. In C. reinhardtii, MYB-related transcription factor ROC40 and DGAT2 (DGTT1) can be induced upon −N; in the roc40 mutant strain, induction of ROC40, DGTT1, and its ability to increase TAG accumulation by −N has been shown to be impaired. In addition, several putative MYB transcription factor-binding sites have been found at the promoter region of DGTT1, suggesting that ROC40 may has a role in −N induced lipid accumulation (Goncalves et al. 2016a).
In Chromochloris (Chlorella) zofingiensis, using yeast one-hybrid assay, bZIP3 transcription factor has been shown to bind with both DGAT2 promoters (CzDGAT1A and CzDGTT5), while MYB1 transcription factor only binds with CzDGTT5 promoter. Similar to CzDGAT1A and CzDGTT5, the transcripts of bZIP3 and MYB1 are up-regulated under −N, suggesting bZIP3 and MYB1 as the transcription factors regulating CzDGAT1A and CzDGTT5 (Mao et al. 2019). However, regulation of DGAT2 expression has not been explored so far in the oleaginous microalga Neochloris oleoabundans.
Neochloris oleoabundans, a taxonomic synonym of Ettlia oleoabundans (Deason et al. 1991), produces 35–54% lipids of dry cell weight under −N condition, up to 80% of its total lipids is TAG (Tornabene et al. 1983). Similar to C. reinhardtii, N. oleoabundans has been suggested to possess two distinct and parallel TAG biosynthesis pathways localized in chloroplast and ER (Klaitong et al. 2017; Chungjatupornchai et al. 2019). However, the data regarding N. oleoabundans is very limited; no genomic sequences are available. The cDNA encoding a functional DGAT2 protein of N. oleoabundans (NeoDGAT2) has been cloned and characterized (Chungjatupornchai and Watcharawipas 2015). Overexpression of NeoDGAT2 in N. oleoabundans has been shown to dramatically increase TAG accumulation (Klaitong et al. 2017).
In this study, we explored the regulation of NeoDGAT2 expression. The upstream sequence of NeoDGAT2 gene was cloned, and the cis-regulatory elements were predicted. The functional regions of NeoDGAT2 regulatory sequence were mapped by deletion analysis using the modified cyan fluorescent protein gene as a reporter. The interaction between the −N inducible region of NeoDGAT2 regulatory sequence and DNA-binding domain of MYB-related transcription factor ROC40 was analyzed.
Materials and methods
Strain and growth conditions
Neochloris oleoabundans strain UTEX 1185 obtained from the Algal Culture Collection at University of Texas was cultured on solid (1.5% Bacto agar) or in liquid Bold’s Basal Medium (BBM) (Bischoff and Bold 1963) under constant illumination of 55–60 μmol photons m−2 s−1 at 30 °C. Cultures in liquid medium started with cells at density of ~ 1.5 × 107 cells mL−1 (OD750 = 0.3). BBM contained NaNO3 as the sole source of nitrogen. For nitrogen starvation (−N) condition, the cells grown on solid BBM were inoculated on solid BBM without NaNO3 (BBM-N) or resuspended in a 50-mL Erlenmeyer flask containing 15 mL of BBM-N shaken continuously at 100 rpm.
Cloning of upstream sequence of NeoDGAT2 gene
The upstream sequence of NeoDGAT2 gene was determined using rapid amplification of genomic ends (RAGE) (Cormack and Somssich 1997) as follows. To construct A-tailed digested genomic DNA libraries, genomic DNA extracted from N. oleoabundans as described (Draper 1988) was digested with restriction enzymes and tailed with nucleotides A at 3′ termini using terminal deoxynucleotidyl transferase (Thermo Fisher Scientific, USA). The A-tailed digested genomic DNA libraries was used as templates for genome walking by RAGE-PCR with specific primers designed based on the sequence of coding region and 5’UTR of NeoDGAT2 gene. The RAGE-PCR products were cloned into Escherichia coli DH5α using pGEM-T Easy vector system (Promega, USA) and verified by automated DNA sequencing analysis. The resulting partial upstream NeoDGAT2 sequences from each round of RAGE-PCR were assembled to obtain the predicted full-length upstream sequence of NeoDGAT2 gene. PCR of full-length upstream NeoDGAT2 sequence was performed using intact genomic DNA of N. oleoabundans as template and specific primers U-DGAT2-F1 and N-DGAT2-R3 (see primers sequences in supplement Table S1) designed based on the 5’end of full-length upstream NeoDGAT2 sequence and coding region in the first exon of NeoDGAT2 gene, respectively. The PCR product was cloned into Escherichia coli DH5α using pGEM-T Easy vector system and verified by automated DNA sequencing analysis. The resulting 1954-bp upstream sequence of NeoDGAT2 gene was submitted to GenBank database under accession number MK208997.
Analysis of NeoDGAT2 regulatory sequence
To analyze the regulatory sequence of NeoDGAT2 (RDG), various programs were used as follows: PLACE (http://www.dna.affrc.go.jp/htdocs/PLACE/) (Higo et al. 1998), PlantTFDB 4.0 (http://planttfdb.cbi.pku.edu.cn/) (Yang et al. 2016), PlantCARE (http://www.bioinformatics.psb.ugent.be/webtools/plantcare/htmL/) (Lescot et al. 2002), The Berkeley Drosophila Genome Project (BDGP, https://www.fruitfly.org/) and YAPP eukaryotic core promoter predictor (http://www.bioinformatics.org/yapp/cgi-bin/yapp.cgi).
Plasmid construction
To evaluate CFP gene mTurquoise2 (Tq) (Goedhart et al. 2012) as a reporter in N. oleoabundans, plasmids pAR-Tq, pAR-Tq1, and pAR-Tq2 harboring the gene cassettes AR-Tq-3′rbcS2, AR-Tq1–3′rbcS2, and AR-Tq2–3′rbcS2, respectively, were constructed by replacing the AR-ChGfp-3′rbcS2 fragment of pChGFP-Hyg3 (Chungjatupornchai et al. 2016) with the PCR fragments containing: (i) promoter HSP70-RBCS2 (AR) of C. reinhardtii from pCB740 (Schroda et al. 2000) (ii) Tq from plasmid pmTurquoise2-C1 (Goedhart et al. 2012). Tq1 and Tq2 containing one and two copies of C. reinhardtii rbcS2 intron 1 (Int1) from pHyg3 (Berthold et al. 2002), respectively. The Int1 located downstream of the start codon and nucleotide 343 of the Tq coding sequence and (iii) 3′UTR of 3′rbcS2 from pCrGFP (Fuhrmann et al. 1999).
To examine the functional region of RDG by deletion analysis, plasmids pRDG, pRDG-D1, pRDG-D2, pRDG-D3, pRDG-D4, pRDG-D5, and pRDG-D6 harboring various length of RGD upstream of Tq2 reporter gene were constructed by replacing the AR-ChGfp-3′rbcS2 fragment of pChGFP-Hyg3 (Chungjatupornchai et al. 2016) with the PCR fragments containing (i) various length of RDG obtained using specific primers as shown in supplement Table S1, (ii) Tq2 reporter gene, and (iii) 3′UTR of 3′rbcS2 from pCrGFP (Fuhrmann et al. 1999). All constructed plasmids harbored hygromycin B resistance gene Hyg3 (Berthold et al. 2002; Chungjatupornchai et al. 2016) used as a selectable marker.
Transformation of N. oleoabundans
To generate transformants expressing CFP, the constructed plasmids were transformed into N. oleoabundans using electroporation as described previously (Chungjatupornchai et al. 2016). Cells were electroporated using a Gene Pulser (Bio-Rad, USA) set electric field strength at 1000 V cm−1, resistance at 200 Ω, and capacitance at 25 μF. The cells were spread on BBM agar plates containing 5 μg mL−1 of hygromycin B. The resulting transformants appeared after incubation for 2 weeks.
CFP activity assay and microscopy
The CFP fluorescence intensity of the transformants was measured in a 96-well plate using a spectrofluorometer (Infinite M200PRO, TECAN, Switzerland) with excitation at 434 nm and emission at 474 nm. Values of control without CFP were subtracted. Specific fluorescence intensities were normalized by the optical density measured at 750 nm. The images of CFP fluorescence signal in the transformants were obtained using a confocal laser scanning microscope (CLSM) (ZEISS LSM800, Germany) with excitation at 433 nm and emission at 475 nm. The chlorophyll autofluorescence was detected using excitation at 652 nm and emission at 668 nm.
Recombinant protein production of ROC40 DNA–binding domain
To produce ROC40 DNA–binding domain (ROC40-DBD) of C. reinhardtii, DNA fragment encoding amino acids 43 to 103 of ROC40 (Matsuo et al. 2008) was cloned into BamHI/EcoRI sites of expression vector pGEX-4T-1 (Genscript, USA). The resulting plasmid pROC40-DBD contained the 61 amino acids of ROC40-DBD fused in-frame at the C-terminus of glutathione S-transferase (GST) was transformed into E. coli BL21 (DE3) pLysS. The expression of GST-(ROC40-DBD) was induced by 0.4 mM IPTG. To extract total proteins from E. coli harboring pROC40-DBD, the cell pellet of 200 OD600 was suspended in lysis buffer: 1X PBS, pH 7.4; 0.2 mg mL−1 lysozyme (Bio Basic Inc., Canada); 1 mM MgCl2; 1X complete EDTA-free Protease Inhibitor Cocktail (Roche, Germany); 1X RQ1 RNase-free DNase buffer and 50 units of RQ1 RNase-free DNase (Promega, USA), for 30 min at 4 °C. The cells were lysed using sonication (Vibra-Cell, Sonics & Materials, Inc., USA). The GST-(ROC40-DBD) was purified using glutathione sepharose 4B (GE Healthcare, USA) according to the manufacturer’s instructions. To remove the GST moiety from ROC40-DBD, 500 μg of GST-(ROC40-DBD) was treated with human thrombin (Merck, USA) for 16–18 h at 25 °C.
Electrophoretic mobility shift assay
To label the DNA used in electrophoretic mobility shift assay (EMSA), the 220-bp (nt. − 1486 to − 1267) of RDG amplified by PCR using primers RDG-D2-F and RDG-R (Table S1) was labeled with DIG-11-dUTP according to the manufacturer’s instructions (Roche, Germany). For protein-DNA binding reactions, recombinant protein GST-(ROC40-DBD), GST or ROC40-DBD, was added to the DIG-labeled 220-bp of RDG in a 20-μL reaction including the binding buffer (5% glycerol, 4 mM KCl, 5 mM MgCl2, 1 mM EDTA and 25 mM HEPES/KOH), 100 μg of bovine serum albumin and 1 μg poly(dI-dC) (Thermo Fisher Scientific, USA). For competition reactions, 70-fold molar excess of unlabeled 220-bp of RDG or unrelated Actin gene of N. oleoabundans (NeoActin) was added to the binding reaction carried out for 1 h at 30 °C. The protein-DNA binding reactions were subjected to 6% native polyacrylamide gel and transferred onto Amersham Hybond-N+ nylon membranes (GE Healthcare, USA) by electroblotting followed by UV crosslinking equipped with 254 nm bulb (Spectrolinker XL-1500 UV crosslinker, Spectronics Corporation, USA). The DIG-labeled 220-bp of RDG was detected with the Fab antiDIG antibody conjugated with alkaline phosphatase (Roche, Germanhy). The chemiluminescent signal developed using CDP-Star (Roche, Germany) was captured with X-ray film.
Results
The efficient CFP reporter in N. oleoabundans
We previously cloned and characterized the NeoDGAT2 cDNA (Chungjatupornchai and Watcharawipas 2015) and found that overexpression of NeoDGAT2 in N. oleoabundans dramatically increase TAG accumulation (Klaitong et al. 2017). In this study, we explored the regulation of NeoDGAT2 expression by analysis of NeoDGAT2 regulatory sequence.
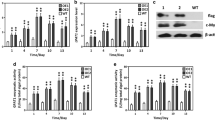
Functional analysis of NeoDGAT2 regulatory sequence requires an efficient reporter. The cyan fluorescent protein (CFP) mTurquoise2 (Tq), a variant of green fluorescent protein, has been shown to have high photostability and quantum yield that contributes to an increase of its brightness in mammalian cells of almost 20% (Goedhart et al. 2012). However, there is no report concerning Tq gene as a reporter in microalgae. To test whether the Tq gene could be used as a reporter in N. oleoabundans, plasmid pAR-Tq containing Tq gene under the control of promoter HSP70-RBCS2 (AR) from C. reinhardtii (Fig. 1a) was constructed and electroporated into N. oleoabundans to obtain transformant AR-Tq. To determine the Tq gene expression, the fluorescence intensity of transformant AR-Tq whole cell was measured using a spectrofluorometer. The CFP activity of transformant AR-Tq was detected (Fig. 1b), indicating the Tq expression in N. oleoabundans.
Development of CFP as a reporter in N. oleoabundans. a Schematic diagram of plasmids harboring CFP gene. AR, HSP70A/RBCS2 hybrid promoter of C. reinhardtii (Schroda et al. 2000); Tq, CFP gene mTurquoise2 (Goedhart et al. 2012); Tq1 and Tq2, mTurquoise2 gene including one and two copies of C. reinhardtii rbcS2 intron 1 (Int1), respectively; 3’rbcS2, 3’ UTR of C. reinhardtii rbcS2 gene (Fuhrmann et al. 1999). b The CFP activity in the transformants. Plasmids pAR-Tq, pAR-Tq1 and pAR-Tq2 were electroporated into N. oleoabundans to obtain transformants AR-Tq, AR-Tq1 and AR-Tq2, respectively. The CFP fluorescence intensity was determined using spectrofluorometer. Values of control without CFP were subtracted. Each value obtained from each transformant clone represents the mean of at least three independent experiments. Average fluorescence intensities denoted by lines. Significant difference among the transformants is indicated (*p < 0.01, t test). c CLSM images of transformant AR-Tq2 expressing CFP. DIC, differential interference contrast; CFP, CFP fluorescence (ex: 433 nm; em: 475 nm); chlorophyll, chlorophyll autofluorescence (ex: 652 nm; em: 668 nm); merged, merged image of CFP and chlorophyll. Wild type strain was used as negative control. The scale bar indicates 5 μm
Heterologous gene expression in C. reinhardtii has been shown to increase by introducing intron1 of endogenous rbcS2 gene into the coding region (Lumbreras et al. 1998; Berthold et al. 2002). To investigate the effect of intron 1 (Int1) from C. reinhardtii rbcS2 gene on Tq expression, plasmids pAR-Tq1 and pAR-Tq2 harboring Tq gene including one (Tq1) and two copies of the Int1 (Tq2), respectively, were constructed (Fig.1a) and electroporated into N. oleoabundans to obtain transformants AR-Tq1 and AR-Tq2, respectively. The CFP activities of transformants AR-Tq and AR-Tq1 were not significantly different; while transformant AR-Tq2 was found to have highest CFP activity, 2.7-fold higher, indicating the improved efficiency of Tq2 expression by the presence of two copies of Int1 (Fig. 1a, b). The Tq2 expression in transformant AR-Tq2 was confirmed by visualization under confocal laser scanning microscope (CLSM). Very bright cyan fluorescence signal was clearly visible in transformant AR-Tq2, but not in wild-type strain used as negative control (Fig. 1c). Therefore, Tq2 gene was used as a reporter for subsequent experiments.
The cis-regulatory elements of NeoDGAT2 gene
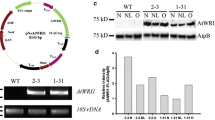
The 1954-bp regulatory sequence of NeoDGAT2 gene (RDG) successfully determined using RAGE-PCR method has been submitted to GenBank database under accession number MK208997. The transcription initiation sites of NeoDGAT2 determined by sequencing analysis of the 31 independent clones of 5’rapid amplification of cDNA ends (5’RACE)-PCR from our previous study (Chungjatupornchai and Watcharawipas 2015) were indicated as frequency (%) with the major transcription initiation site (+ 1) located at nt. 48 upstream of the start codon (Fig. 2). Core promoter region was predicted by BDGP (https://www.fruitfly.org/). TATA box was predicted by YAPP eukaryotic core promoter predictor (http://www.bioinformatics.org/yapp/cgi-bin/yapp.cgi). The CAAT box was predicted by PlantCARE (Lescot et al. 2002) and PLACE (Higo et al. 1998). The putative binding sites of basic helix-loop-helix (bHLH-bs), basic leucine zipper (bZIP-bs) and myeloblastosis (MYB) transcription factor (MYB-bs) were predicted by PlantTFDB (Yang et al. 2016) (Fig. 2).
Regulatory sequence of NeoDGAT2 gene. Locations of the putative binding sites of myeloblastosis transcription factor (MYB-bs) highlighted in red, basic helix-loop-helix (bHLH-bs) highlighted in yellow and basic leucine zipper (bZIP-bs) indicated by a box were predicted by PlantTFDB (Yang et al. 2016). The CAAT box indicated in blue was predicted by PlantCARE (Lescot et al. 2002) and PLACE (Higo et al. 1998). The TATA box indicated by a box was predicted by YAPP (http://www.bioinformatics.org/yapp/cgi-bin/yapp.cgi). The core promoter region highlighted in green was predicted by BDGP (https://www.fruitfly.org/). The boundaries of RDG truncated fragments in plasmids pRDG-D1 to pRDG-D6 used for deletion analysis are indicated. Transcription initiation sites determined by 5’RACE-PCR are indicated as frequency (%) with the major transcription initiation site marked by +1. The numbers of nucleotides refer to the transcription initiation site. Start codon (ATG) is underlined
The NeoDGAT2 regulatory sequence induced under −N condition
To investigate whether RDG could be induced under −N condition, plasmid pRDG containing the reporter gene Tq2 under the control of RDG (Fig. 3a) was introduced into N. oleoabundans via electroporation to obtain transformant RDG. The CFP activity of transformant RDG grown in BBM including various concentration of NaNO3 as the sole source of nitrogen was determined using spectrofluorometer; NaNO3 at concentrations 3, 0.5, and 0.15 mM were found to have low CFP activities not significantly different (at p < 0.01), whereas without NaNO3 (0 mM) was found to have CFP activity increased 2.7-fold (Fig. 3b), indicating the −N inducible RDG.
N-starvation inducible promoter of NeoDGAT2 gene. a Schematic diagram of plasmid pRDG. RDG, regulatory sequence of NeoDGAT2 gene; Tq2, CFP gene mTurquoise2 (Goedhart et al. 2012) including two copies of intron 1 (Int1) from C. reinhardtii rbcS2 gene; 3’rbcS2, 3’ UTR of C. reinhardtii rbcS2 gene (Fuhrmann et al. 1999). The numbers of nucleotides refer to the transcription initiation site (+ 1). b Effect of −N on CFP activity. Transformant RGD obtained by introducing plasmid pRDG into N. oleoabundans was grown in BBM containing various NaNO3 concentrations. The CFP fluorescence intensity was measured using spectrofluorometer. Values of control without CFP were subtracted. Each value obtained from each transformant clone represents the mean of at least three independent experiments. Average fluorescence intensities denoted by lines. Significant difference among the transformants is indicated (*p < 0.01, t test). c CLSM images of transformant RDG under −N condition. +N, N-replete (3 mM NaNO3) condition; −N, N-starvation (0 mM NaNO3) condition; DIC, differential interference contrast; CFP, CFP fluorescence (ex: 433 nm; em: 475 nm); chlorophyll, chlorophyll autofluorescence (ex: 652 nm; em: 668 nm); merged, merged image of CFP and chlorophyll. Wild type strain was used as negative control. The scale bar indicates 5 μm
The CFP activity of transformant RDG under −N condition was confirmed by visualization under CLSM. The cyan fluorescence signal was clearly visible in transformant RDG under −N but not +N condition, while no such signal was detected in wild-type strain both under +N and −N conditions (Fig. 3c). Therefore, RDG was induced under −N condition.
Sequences required for the function of cis-acting regulator and basal promoter of NeoDGAT2 gene
In order to analyze the regions for cis-acting regulator and basal promoter of NeoDGAT2 gene, plasmids pRDG-D1 to pRDG-D6 containing various length of RDG upstream of Tq2 reporter gene were constructed (Fig. 4a) and introduced into N. oleoabundans via electroporation to obtain transformants RDG-D1 to RDG-D6, respectively. When compared with +N condition, CFP activities of the transformants grown under −N condition were significantly increased (at p < 0.01) (Fig. 4b). Transformants RDG, RDG-D1, and RDG-D2 were observed to have CFP activities dramatically increased (up to 4-fold), whereas CFP activities of transformants RDG-D3, RDG-D4, RDG-D5, and RDG-D6 slightly increased (up to 1.9-fold), suggesting the basal promoter activities under −N condition. The results indicated that (i) the predicted binding site of MYB transcription factor (MYB-bs) was involved in −N induction, (ii) predicted binding sites of bZIP and bHLH transcription factors (bZIP-bs and bHLH-bs, respectively) were not involved in −N induction, (iii) predicted CAAT box was not involved in the basal promoter activity, and (iv) the core promoter region including TATA box possessed basal promoter activity (Fig. 2 and 4a, b).
Deletion analysis of NeoDGAT2 regulatory sequence. a Schematic diagram of pRDG-derivative plasmids. Locations of putative TATA box, CAAT box, MYB-binding site (MYB-bs), and 220-bp fragment used for electrophoretic mobility shift assay are indicated. Tq2, CFP gene mTurquoise2 (Goedhart et al. 2012) including two copies of intron 1 (Int1) from C. reinhardtii rbcS2 gene; 3’rbcS2, 3’ UTR of C. reinhardtii rbcS2 gene (Fuhrmann et al. 1999). The numbers of nucleotides refer to the transcription initiation site (+ 1). b CFP activity of RDG-derivative transformants under −N condition. Transformants obtained by introducing pRDG-derivative plasmids into N. oleoabundans were grown in +N (3 mM NaNO3) and −N (0 mM NaNO3) conditions. The CFP fluorescence intensity was measured using spectrofluorometer. Values of control without CFP were subtracted. Each value obtained from each transformant clone represents the mean of at least three independent experiments. Average fluorescence intensities denoted by lines
Specific interaction of RDG MYB-bs and ROC40-DBD
In C. reinhardtii, MYB-related transcription factor ROC40 has been shown to be upregulated by −N and suggested to play a regulatory role in −N induced expression of DGAT2 gene (CrDGTT1) and TAG accumulation (Goncalves et al. 2016a). To test whether the predicted MYB-bs of RDG was the DNA target of MYB transcription factor, EMSA was performed with the 220-bp −N inducible region harboring MYB-bs of RDG (Fig. 4a) and the recombinant protein ROC40-DBD of C. reinhardtii. The shifted bands were detected in DIG-labeled 220-bp of RDG incubated with various concentrations of recombinant fusion protein GST-(ROC40-DBD), the intensity of shifted bands increased corresponding to the increased concentration of GST-(ROC40-DBD) (Fig. 5a). While no shifted band was detected when incubated with GST (Fig. 5b). A shifted band was clearly detected in the reaction containing thrombin-cleaved ROC40-DBD, but no shifted band was detected in the presence of excess unlabeled 220-bp used as a competitor; the shifted band was also observed in the presence of unrelated competitor Actin (Fig. 5c), indicating the specific binding of MYB-bs and ROC40-DBD. Therefore, the MYB-bs of RDG was the DNA target of MYB-related transcription factor ROC40.
Specific interaction between ROC40-DBD and the 220-bp DNA fragment of RDG. The binding activity of recombinant protein ROC40-DBD to the labeled 220-bp fragment containing MYB-bs was performed using EMSA. Sixty fmol of labeled 220-bp fragment was used in all reactions. a DNA binding reactions performed with fusion protein GST-(ROC40-DBD). Lane 1, excluding GST-(ROC40-DBD) used as negative control; lanes 2–6, including 2, 4, 6, 8, and 10 μg of GST-(ROC40-DBD), respectively. b DNA binding reactions performed with protein GST. Lane 1, excluding GST used as negative control; lanes 2–6, including 2, 4, 6, 8, and 10 μg of GST, respectively. c DNA binding reactions performed with thrombin-cleaved ROC40-DBD protein. Lane 1, excluding ROC40-DBD used as negative control; lanes 2–4, including (+) and excluding (−) of 40 μg of ROC40-DBD, 70-fold excess of unlabeled 220-bp competitor and Actin gene used as an unrelated competitor are indicated. The shifted 220-bp fragments are indicated by arrows. Location of the 220-bp fragment is shown in Fig. 4a
Discussion
This study is based on identification of NeoDGAT2 regulation under −N condition that is one of the necessary steps prior to design novel solutions for development of products derived from microalgal TAG at a commercial scale.
The functional regions of NeoDGAT2 regulatory sequence were analyzed using the improved CFP gene Tq2 including two copies of the Int1 from C. reinhardtii rbcS2 gene as a reporter. Introns have been shown to have a positive effect on gene expression in eukaryotes, because their splicing improves and accelerates nuclear mRNA export (Rose and Last 1997; Reed and Hurt 2002). In C. reinhardtii heterologous gene expression can be increased by introducing Int1 of endogenous rbcS2 gene into the coding region, the presence of the int1 in selectable marker genes Aph7” and ble increases transformation frequency (Lumbreras et al. 1998; Berthold et al. 2002). Introducing the int1 of C. reinhardtii rbcS2 gene into the selectable marker genes Aph7” (Hyg3) (Berthold et al. 2002) also increases transformation frequency in N. oleoabundans (Chungjatupornchai et al. 2016). However, in this study the Tq gene without Int1 and Tq1 with one copy of the Int1 was observed to have CFP activities not significantly different, while Tq2 with two copies of the Int1 exhibited the highest CFP activity, indicating the improved efficiency of Tq2 expression (Fig. 1a, b). The Tq2 expression in transformant AR-Tq2 was confirmed as a very bright cyan fluorescence signal visualized under CLSM (Fig. 1c). Therefore, Tq2 gene was used as a reporter for functional analysis of RDG. In the transformant cells with high CFP fluorescence signal the chlorophyll fluorescence became lighter than other cells or even invisible (Figs. 1c and 3c), in agreement with the bleaching appearance of the corresponding cells observed under bright field microscope (data not shown). Whether the Tq2 expression affects chlorophyll synthesis or enhances chlorophyll breakdown remains to be investigated.
The transformant RDG grown under +N with NaNO3 at concentrations 3, 0.5, and 0.15 mM were found to have low CFP activities and not significantly different (at p < 0.01) (Fig. 3b), thus, no induction in the present of N. Similar results have been observed in transcriptome of N. oleoabundans, the gene encoding DGAT displays relatively no change in its expression under N limitation at 0.65 and 0.13 mM (Rismani-Yazdi et al. 2012). However, in this study, the CFP activity of transformant RDG grown under −N (0 mM NaNO3) increased 2.7-fold (Fig. 3b), indicating the −N inducible of NeoDGAT2 promoter. Proteomics studies of C. vulgaris also revealed overexpression of DGAT under N-free condition (Guarnieri et al. 2011).
Analysis of RDG by PlantTFDB (Yang et al. 2016) predicted potential binding sites of MYB, bZIP and bHLH (Fig. 2). Deletion analysis of RDG revealed that the DNA fragment harboring the predicted bZIP-bs and bHLH-bs were not induced upon −N, whereas, that harboring the MYB-bs was −N inducible (Fig. 4a, b). Transcription factors, a group of regulators, control their target gene expression at the transcriptional level through binding certain upstream elements. An increasing number of possible transcription factors involved in lipid synthesis have been suggested in microalgae, including MYB (Boyle et al. 2012; Hu et al. 2014; de Lomana et al. 2015; Ngan et al. 2015). MYB transcription factors regulate fundamental cellular processes and specific facets of metabolism through modulation of transcription at target genes, to which they bind in a sequence-specific manner (Prouse and Campbell 2012). In C. zofingiensis, MYB1 transcription factor upregulated under N-starvation has been shown to bind with DGAT2 (CzDGTT5) in yeast one-hybrid assay, suggesting MYB1 as the transcription factors regulating DGAT2 (Mao et al. 2019). In C. reinhardtii, the MYB-related transcription factor ROC40 upregulated upon −N condition has been suggested to play a role in −N induced lipid accumulation (Goncalves et al. 2016a). The DNA-binding domain of MYB transcription factor is conserved among different green algae, and it is phylogenetically related to ROC40-DBD of C. reinhardtii (Goncalves et al. 2016a). So far, neither MYB transcription factors nor their sequences have been identified in N. oleoabundans. Therefore, ROC40-DBD from C. reinhardtii was used to verify the predicted MYB-bs of RDG. The specific binding of MYB-bs of RDG and ROC40-DBD was observed in EMSA (Fig. 5c), indicating MYB-bs as DNA target of MYB-related transcription factor ROC40. Thus, the corresponding MYB transcription factor in of N. oleoabundans is probably the transcription factor regulating NeoDGAT2. The interaction between MYB transcription factor and the MYB-bs may play a role in regulating −N induced expression of NeoDGAT2, affecting TAG accumulation.
Conclusions
We characterized the cis-regulatory elements of NeoDGAT2 gene and successfully identified the NeoDGAT2 regulation under −N condition using the improved efficient Tq2 with high CFP activity as a reporter. The RDG region harboring the MYB-bs was −N inducible. The MYB-bs bound specifically to ROC40-DBD from C. reinhardtii, indicating MYB-bs as DNA target of MYB-related transcription factor ROC40. Therefore, the corresponding MYB transcription factor of N. oleoabundans is probably the transcription factor regulating −N induced expression of NeoDGAT2, affecting TAG accumulation. MYB transcription factors can be the potential targets for engineering to increase TAG content. Increasing TAG content is essential for products derived from microalgal TAG to achieve economic viability.
References
Berthold P, Schmitt R, Mages W (2002) An engineered Streptomyces hygroscopicus aph 7″ gene mediates dominant resistance against Hygromycin B in Chlamydomonas reinhardtii. Protist 153:401–412
Bischoff HW, Bold HC (1963) Some soil algae from Enchanted Rock and related algal species. Phycological Studies, vol IV. University of Texas, Austin, pp 1–95
Boyle NR, Page MD, Liu B, Blaby IK, Casero D, Kropat J, Cokus SJ, Hong-Hermesdorf A, Shaw J, Karpowicz SJ, Gallaher SD, Johnson S, Benning C, Pellegrini M, Grossman A, Merchant SS (2012) Three acyltransferases and nitrogen-responsive regulator are implicated in nitrogen starvation-induced triacylglycerol accumulation in Chlamydomonas. J Biol Chem 287:15811–15825
Chen JE, Smith AG (2012) A look at diacylglycerol acyltransferases (DGATs) in algae. J Biotechnol 162:28–39
Chisti Y (2007) Biodiesel from microalgae. Biotechnol Adv 25:294–306
Chungjatupornchai W, Watcharawipas A (2015) Diacylglycerol acyltransferase type 2 cDNA from the oleaginous microalga Neochloris oleoabundans: cloning and functional characterization. J Appl Phycol 27:1499–1507
Chungjatupornchai W, Kitraksa P, Fa-aroonsawat S (2016) Stable nuclear transformation of the oleaginous microalga Neochloris oleoabundans by electroporation. J Appl Phycol 28:191–199
Chungjatupornchai W, Areerat K, Fa-Aroonsawat S (2019) Increased triacylglycerol production in oleaginous microalga Neochloris oleoabundans by overexpression of plastidial lysophosphatidic acid acyltransferase. Microb Cell Factories 18:53
Coleman RA, Lee DP (2004) Enzymes of triacylglycerol synthesis and their regulation. Prog Lipid Res 43:134–176
Cormack RS, Somssich IE (1997) Rapid amplification of genomic ends (RAGE) as a simple method to clone flanking genomic DNA. Gene 194:273–276
de Lomana ALG, Schäuble S, Valenzuela J, Imam S, Carter W, Bilgin DD, Yohn CB, Turkarslan S, Reiss DJ, Orellana MV (2015) Transcriptional program for nitrogen starvation-induced lipid accumulation in Chlamydomonas reinhardtii. Biotechnol Biofuels 8:207
Deason TR, Silva PC, Watanabe S, Floyd GL (1991) Taxonomic status of the species of the green algal genus Neochloris. Plant Systemat Evol 177:213–219
Draaisma RB, Wijffels RH, Slegers PE, Brentner LB, Roy A, Barbosa MJ (2013) Food commodities from microalgae. Curr Opin Biotech 24:169–177
Draper J (1988) Plant genetic transformation and gene expression : a laboratory manual. Blackwell Scientific Publications, Oxford
Fan J, Andre C, Xu C (2011) A chloroplast pathway for the de novo biosynthesis of triacylglycerol in Chlamydomonas reinhardtii. FEBS Lett 585:1985–1991
Fuhrmann M, Oertel W, Hegemann P (1999) A synthetic gene coding for the green fluorescent protein (GFP) is a versatile reporter in Chlamydomonas reinhardtii. Plant J 19:353–361
Goedhart J, von Stetten D, Noirclerc-Savoye M, Lelimousin M, Joosen L, Hink MA, van Weeren L, Gadella TWJ Jr, Royant A (2012) Structure-guided evolution of cyan fluorescent proteins towards a quantum yield of 93%. Nat Commun 3:751
Goncalves EC, Koh J, Zhu N, Yoo M-J, Chen S, Matsuo T, Johnson JV, Rathinasabapathi B (2016a) Nitrogen starvation-induced accumulation of triacylglycerol in the green algae: evidence for a role for ROC40, a transcription factor involved in circadian rhythm. Plant J 85:743–757
Goncalves EC, Wilkie AC, Kirst M, Rathinasabapathi B (2016b) Metabolic regulation of triacylglycerol accumulation in the green algae: identification of potential targets for engineering to improve oil yield. Plant Biotech J 14:1649–1660
Gong Y, Zhang J, Guo X, Wan X, Liang Z, Hu CJ, Jiang M (2013) Identification and characterization of PtDGAT2B, an acyltransferase of the DGAT2 acyl-coenzyme a: diacylglycerol acyltransferase family in the diatom Phaeodactylum tricornutum. FEBS Lett 587:481–487
Guarnieri MT, Nag A, Smolinski SL, Darzins A, Seibert M, Pienkos PT (2011) Examination of triacylglycerol biosynthetic pathways via de novo transcriptomic and proteomic analyses in an unsequenced microalga. PLoS One 6:e25851
Higo H, Iwamoto M, Ugawa Y, Higo K (1998) PLACE: a database of plant cis -acting regulatory DNA elements. Nucl Acids Res 26:358–359
Hu Q, Sommerfeld M, Jarvis E, Ghirardi M, Posewitz M, Seibert M, Darzins A (2008) Microalgal triacylglycerols as feedstocks for biofuel production: perspectives and advances. Plant J 54:621–639
Hu J, Wang D, Li J, Jing G, Ning K, Xu J (2014) Genome-wide identification of transcription factors and transcription-factor binding sites in oleaginous microalgae Nannochloropsis. Sci Rep 4:5454
Hung C-H, Ho M-Y, Kanehara K, Nakamura Y (2013) Functional study of diacylglycerol acyltransferase type 2 family in Chlamydomonas reinhardtii. FEBS Lett 587:2364–2370
Jako C, Kumar A, Wei Y, Zou J, Barton DL, Giblin EM, Covello PS, Taylor DC (2001) Seed-specific over-expression of an Arabidopsis cDNA encoding a diacylglycerol acyltransferase enhances seed oil content and seed weight. Plant Physiol 126:861–874
Klaitong P, Fa-Aroonsawat S, Chungjatupornchai W (2017) Accelerated triacylglycerol production and altered fatty acid composition in oleaginous microalga Neochloris oleoabundans by overexpression of diacylglycerol acyltransferase 2. Microb Cell Factories 16:61–61
Klok A, Lamers P, Martens D, Draaisma R, Wijffels R (2014) Edible oils from microalgae: insights in TAG accumulation. Trends Biotechnol 32:521–528
Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouzé P, Rombauts S (2002) PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res 30:325–327
Li-Beisson Y, Beisson F, Riekhof W (2015) Metabolism of acyl-lipids in Chlamydomonas reinhardtii. Plant J 82:504–522
Lumbreras V, Stevens DR, Purton S (1998) Efficient foreign gene expression in Chlamydomonas reinhardtii mediated by an endogenous intron. Plant J 14:441–447
Lung S-C, Weselake RJ (2006) Diacylglycerol acyltransferase: a key mediator of plant triacylglycerol synthesis. Lipids 41:1073–1088
Mao X, Wu T, Kou Y, Shi Y, Zhang Y, Liu J (2019) Characterization of type I and type II diacylglycerol acyltransferases from the emerging model alga Chlorella zofingiensis reveals their functional complementarity and engineering potential. Biotechnol Biofuels 12:28
Mata TM, Martins AA, Caetano NS (2010) Microalgae for biodiesel production and other applications: a review. Renew Sust Energ Rev 14:217–232
Matsuo T, Okamoto K, Onai K, Niwa Y, Shimogawara K, Ishiura M (2008) A systematic forward genetic analysis identified components of the Chlamydomonas circadian system. Genes Dev 22:918–930
Ngan CY, Wong C-H, Choi C, Yoshinaga Y, Louie K, Jia J, Chen C, Bowen B, Cheng H, Leonelli L, Kuo R, Baran R, García-Cerdán JG, Pratap A, Wang M, Lim J, Tice H, Daum C, Xu J, Northen T, Visel A, Bristow J, Niyogi KK, Wei C-L (2015) Lineage-specific chromatin signatures reveal a regulator of lipid metabolism in microalgae. Nature Plants 1:15107
Nobusawa T, Hori K, Mori H, Kurokawa K, Ohta H (2017) Differently localized lysophosphatidic acid acyltransferases crucial for triacylglycerol biosynthesis in the oleaginous alga Nannochloropsis. Plant J 90:547–559
Ohlrogge J, Browse J (1995) Lipid biosynthesis. Plant Cell 7:957–970
Prouse MB, Campbell MM (2012) The interaction between MYB proteins and their target DNA binding sites. Biochim Biophys Acta -Gene Regul Mech 1819:67–77
Reed R, Hurt E (2002) A conserved mRNA export machinery coupled to pre-mRNA splicing. Cell 108:523–531
Rismani-Yazdi H, Haznedaroglu BZ, Hsin C, Peccia J (2012) Transcriptomic analysis of the oleaginous microalga Neochloris oleoabundans reveals metabolic insights into triacylglyceride accumulation. Biotechnol Biofuels 5:74
Rose AB, Last RL (1997) Introns act post-transcriptionally to increase expression of the Arabidopsis thaliana tryptophan pathway gene PAT1. Plant J 11:455–464
Schroda M, Blöcker D, Beck CF (2000) The HSP70A promoter as a tool for the improved expression of transgenes in Chlamydomonas. Plant J 21:121–131
Scragg A, Illman A, Carden A, Shales S, Bioenergy (2002) Growth of microalgae with increased calorific values in a tubular bioreactor. Biomass Bioenergy 23:67–73
Song M, Pei H, Hu W, Ma G (2013) Evaluation of the potential of 10 microalgal strains for biodiesel production. Bioresour Technol 141:245–251
Tornabene TG, Holzer G, Lien S, Burris N (1983) Lipid composition of the nitrogen starved green alga Neochloris oleoabundans. Enz Microb Technol 5:435–440
Yamaoka Y, Achard D, Jang S, Legéret B, Kamisuki S, Ko D, Schulz-Raffelt M, Kim Y, Song W-Y, Nishida I, Li-Beisson Y, Lee Y (2016) Identification of a Chlamydomonas plastidial 2-lysophosphatidic acid acyltransferase and its use to engineer microalgae with increased oil content. Plant Biotech J 14:2158–2167
Yang D-C, Jin J, Kong L, Meng Y-Q, Gao G, Luo J, Tian F (2016) PlantTFDB 4.0: toward a central hub for transcription factors and regulatory interactions in plants. Nucl Acids Res 45:D1040–D1045
Acknowledgments
We thank Dr. Ir. Joachim Goedhart, University of Amsterdam, The Netherlands, for providing plasmid pmTurquoise2-C1. Paeka Klaitong was supported by The Royal Golden Jubilee PhD Scholarship from The Thailand Research Fund (TRF).
Funding
This work was supported by Mahidol University and TRF (grant number: BRG5780005) to Wipa Chungjatupornchai.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
ESM 1
(DOCX 41 kb)
Rights and permissions
About this article
Cite this article
Klaitong, P., Watcharawipas, A., Fa-aroonsawat, S. et al. Nitrogen starvation-inducible promoter of microalga Neochloris oleoabundans lipogenic gene encoding diacylglycerol acyltransferase 2. J Appl Phycol 33, 331–341 (2021). https://doi.org/10.1007/s10811-020-02307-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10811-020-02307-w