Abstract
To clarify the presence of algal-susceptible indigenous bacteria (SIB) in the gut, ICR mice were fed high-sucrose (50% w/w) diets containing either no fibre (NF), 5% brown alga “arame”, Eisenia bicyclis; 5% red alga “tsunomata”, Chondrus ocellatus; 5% (w/w) green alga “hitoegusa”, Monostroma nitidum; or 5% (w/w) cyanobacterium “blue-green alga”, Aphanizomenon flos-aquae for 14 days. Faecal frequency and weight were the highest in mice fed M. nitidum. Plasma cholesterol was the lowest in the mice fed C. ocellatus. The caecal microbiome was examined by 16S rDNA (V4) amplicon sequencing. Principal component analysis of operational taxonomical units (OTUs) revealed that the edible algae altered the microbiome. An increase in abundance levels of OTUs by E. bicyclis (Bacteroides acidifaciens-, Bacteroides intestinalis-, Bifidobacterium pseudolongum-like), C. ocellatus (Bacteroides vulgatus- and Escherichia coli-like), M. nitidum (Faecalibaculum rodentium- and Muribaculum sp.-like), and A. flos-aquae (Muribaculum sp.) was detected. Abundance of Lactobacillus johnsonii was the lowest in mice fed the algal diets. Bacteria that increased in numbers were identified as algal SIBs. SIBs might have different effects on host health depending on the food material consumed. From the algal SIBs, B. pseudolongum, B. vulgatus, F. rodentium, and L. johnsonii were isolated using blood-liver agar and identified with the 16S rDNA BLAST search. Future studies should be focused on isolation of other SIBs.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In East Asian countries such as China, South Korea, and Japan, life expectancy of the aging population has increased with improved medical, sanitary, hygiene, and nutritional conditions (Kido 2015; Lee et al. 2016; Bao et al. 2017). However, the incidence of lifestyle diseases such as obesity, diabetes, hyperlipidaemia, and hypertension have increased with increasing adoption of the westernized diet that contains high-fat and low–dietary fibre (Park et al. 2016; Htun et al. 2017; Winglee et al. 2017). For example, lifestyle diseases in Japan, including malignant neoplasms, which are mainly the result of aging and poor diet, consume a third of total medical expenses, and are responsible for deaths of more than half population (Cabinet Office Japan 2018). Although exercise and diet therapy are generally effective for prevention of lifestyle diseases, the demand for functional food materials has increased recently, including probiotics and prebiotics (Santeramo et al. 2018).
In healthy normal humans and other mammals, 11–12 log cells g−1 and 500–1000 bacterial species are present in the large intestine and the gut microbiome affects host health (Marchesi et al. 2016). In general, Bifidobacterium and Lactobacillus spp. are recognised as beneficial gut bacteria present in humans and laboratory rodents (Kato et al. 2017). Therefore, studies on various food ingredients that promote gut Bifidobacterium and Lactobacillus have been reported (Delgado and Tamashiro 2018). However, it is speculated that functional ingredient-susceptible indigenous bacteria (SIB) species in the intestine vary with host species and/or individuals. It is also important to investigate whether an increase or decrease in SIB abundance actually affects the health of the host.
In a previous study, Lactobacillus plantarum AN1, screened for its in vitro antioxidant and anti-inflammatory properties, ameliorated the activity of dextran sodium sulphate-induced inflammatory bowel disease with increased gut indigenous Lactobacillus reuteri (Yokota et al. 2018a) in the ICR mouse model. Indigenous L. reuteri abundance also increased in normal ICR mice in the presence of L. plantarum AN1, but decreased in ICR mice fed a high-fat, high-sucrose, low–dietary fibre diet (Kuda et al. 2017; Yokota et al. 2018b). In the case of normal BALB/c mice, L. plantarum AN1 increased the abundance of gut indigenous L. plantarum (Kuda et al. 2019).
The traditional Japanese diet is believed to have potential health benefits (Sugawara et al. 2018). Edible algae are a major component of traditional Japanese cuisine and have various beneficial functions, such as hypocholesterolemia, antioxidation, and antiglycation, due to their richness in dietary fibres, minerals, polyphenols (phlorotannins), and flavonoids (Kuda and Ikemori 2009; Eda et al. 2016; Takei et al. 2017; Zhao et al. 2018). Although the effects of their functional compounds on the gut microbiome using the culture-dependent method have been reported (Nakata et al. 2016; Shang et al. 2018; Yang et al. 2019), the detailed effects of the whole algal body as a food stuff on the microbiome and algal SIBs have not been yet been revealed.
In this study, to verify the presence and influence of algal SIB on the host, the gut microbiomes of ICR mice fed diets containing the brown alga “arame”, Eisenia bicyclis; red alga “tsunomata”, Chondrus ocellatus; green alga “hitoegusa”, Monostroma nitidum; or cyanobacterium “blue-green alga”, Aphanizomenon flos-aquae were determined by 16S rDNA amplicon sequencing method using a next-generation sequencer MiSeq system (Jovel et al. 2016). The three marine algae have been used in Japanese cuisine since ancient times (Takei et al. 2017) and the dried powder of the cyanobacteria has more recently been used as a health food (Taniguchi et al. 2019). Furthermore, in order to explore the algal SIB, typical gut bacterial colonies were isolated on nonselective and highly discriminating media (blood and liver (BL) agar plates) (Mitsuoka 2014).
Materials and methods
Samples
Dried powder products of E. bicyclis (EB), C. ocellatus (CO), and M. nitidum (MN) harvested in the Boso Peninsula, Japan, were obtained from Suzuki Nori Co. (Choshi, Japan). Powder of A. flos-aquae (AFA), harvested in Upper Klamath Lake, Oregon, was obtained from Dr’s Choice Co. (Tokyo, Japan). The mineral composition, carbohydrate, polysaccharide and phenolic compound content, and antioxidant properties of EB, CO and MN were shown in a previous study (Takei et al. 2017). The compositions of mineral, sugars, organic acids, and free amino acids in AFA were also previously reported (Taniguchi et al. 2019).
Animal care
Animal experiments were performed in compliance with the “Fundamental Guidelines for Proper Conduct of Animal Experiment and Related Activities in Academic Research Institutions” under the jurisdiction of the Ministry of Education, Culture, Sports, Science, and Technology, and approved by the animal experiment committee of the Tokyo University of Marine Science and Technology (approval No. H30-4).
Thirty 5-week-old male ICR mice were purchased from Tokyo Laboratory Animal Science (Tokyo, Japan) and were housed in metal wire cages (three mice per cage), at 22 ± 2 °C. The mice were acclimated to a high-sucrose-low–dietary fibre diet (NF), as shown in Table 1, with distilled water for drinking ad libitum. After 7 days, mice were divided into five groups (n = 6) and fed diets containing either no fibre (NF), 5% (w/w) EB, 5% CO, 5% MN, or 5% AFA for 14 days. During the 11–13 feeding days, defecation frequency and faecal weight were measured.
After feeding, the mice were anaesthetised with isoflurane (Fujifilm Wako Pure Chemical Corporation, Japan) and bled from the abdominal aorta. Following lethal exsanguination, the liver, spleen, kidneys, and epididymal fat pads were removed and weighed. The cecum was ligated before being excised and cooled on ice until further use for microbial analysis.
Plasma lipid and glucose levels
Plasma triacylglyceride (TG), total cholesterol (TC), and glucose (Glu) levels were determined using commercial kits (Triglyceride E-Test Wako, Total Cholesterol E-Test Wako, Glucose CII-Test Wako, respectively; Fujifilm Wako Pure Chemical Corporation).
Viable counts and identification of caecal microbiota using blood-liver agar and BLAST search
The culture-independent method was carried out as described in a previous study (Kuda et al. 2019). The content was immediately prepared as a 1:100 dilution with phosphate-buffered saline (PBS; Nissui Pharmaceuticals, Japan), containing 0.1% (w/v) agar. The diluted samples (0.03 mL) were plated on blood-liver (BL) agar (Nissui Pharmaceuticals) plates containing 5% (v/v) defibrinated horse blood (Nippon Bio-Supp. Center, Tokyo, Japan), and incubated at 37 °C for 48 h under anaerobic conditions, using an AnaeroPack (Mitsubishi Gas Chemical, Japan). According to the method described by Mitsuoka (2014), the detected colonies were enumerated for each morphology. Three to five of the typical colonies were isolated and purified using BL agar and AnaeroPack. Amplification of the 16S rRNA gene, using PCR primers 27F and 1492R, and sequencing of the amplicons, was performed by Macrogen Japan Corp. (Kyoto, Japan). The homology search was performed using BLASTn of the DNA Data Bank of Japan (http://ddbj.nig.ac.jp/blast/blastn).
Direct bacterial cell count
The caecal direct bacterial cell count was performed following the procedure previously described (Shikano et al. 2019). Briefly, bacterial cells were prepared as a 1:100 dilution with PBS, and counted by dielectrophoretic impedance measurement (DEPIM) (Hirota et al. 2014) using a Bacteria Counter (PHC, Tokyo, Japan). Both live and dead cells were counted using DEPIM.
Analysis of caecal microbiota using 16S rDNA amplicon sequencing
Amplicon sequencing of the 16S rDNA was conducted by Fasmac Co., Ltd. (Atsugi, Japan). DNA was extracted from the samples using an MPure bacterial DNA extraction kit (MP Bio Japan, Japan). The DNA library was prepared using a two-step polymerase chain reaction (PCR) (Sinclair et al. 2015). The V4 region was amplified in a 23-cycle PCR using a forward primer 515f: 5′-ACACTCTTTCCCTACACGACGCTCTTCCGATCTGTGCCAGCMGCCGCGGTAA-3′ and reverse primer 806r: 5′-GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTGGACTCHVGGGTWTCTAAT-3′. Thereafter, individual DNA fragments were tagged in an eight-cycle PCR using forward primer: 5′-AATGATACGGCGACCACCGAGATCTACAC-[sequence for individual mouse]-ACACTCTTTCCCTACACCGACGC-3′ and reverse primer: 5′-CAAGCAGAAGACGGCATACGAGAT-[sequence for individual mouse]-CTGACTGGAGTTCAGACGTGTG-3′. DNA libraries were multiplexed and loaded in an Illumina MiSeq instrument (Illumina, USA). Readings with a mismatched sequence at the start region were filtered using the FASTX Toolkit (http://hannonlab.cshl.edu/fastx_toolkit/); reads with quality below 20 and shorter than 40 base pairs were omitted using Sickle (https://github.com/ucdavis-bioinformatics/sickle). The shortlisted reads were merged using the pair-end merge script FLASH (http://ccb.jhu.edu/software/FLASH/), and 240–260 base length reads were selected. From the selected reads, chimeras were identified using a bioinformatics pipeline QIIME (http://qiime.org/tutorials/chimera_checking.html) and omitted. Sequences were clustered into operational taxonomic units (OTUs) according to a 97% identity cut-off using the QIIME workflow script and Greengenes database (http://greengenes.lbl.gov/).
Diversity of the microbiota
The alpha and beta diversity of the gut microbiome in mice was expressed using the Shannon-Wiener index (H′) and principal component analysis (PCA), respectively (Gafan et al. 2005; Shibayama et al. 2018). PCA was calculated based on the number of OTUs in dominant bacterial genes (filtered by 0.1% counts of whole OTU), and analysed using Easy PCA software (http://hoxom-hist.appspot.com/pca.html).
Statistical analysis
The results of body and organ weights and alpha diversity indices were expressed as the mean value ± standard error of the mean. Data were subjected to analysis of variance and Tukey’s post hoc tests, using a statistical software package (Excel Statistic Ver. 6, Japan). p < 0.05 was considered statistically significant.
Results
Body, organ, and faecal weights, and plasma lipid and Glu levels
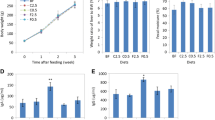
During the feeding period, no symptoms or abnormalities were observed in any of the mice. Body, faecal, and organ weights of the test mice are summarised in Table 2. No significant differences in body and organ weights were observed among the diet groups, though they tended to be lower in mice fed CO. Defecation frequency and faecal weight were significantly increased by dietary intake of three marine algae EB, CO, and MN, and were the highest in mice fed MN. The plasma TG level tended to be low in the CO group, though the difference was not statistically significant. The plasma TC level in the CO group was 30% lower than that in the NF group. Plasma Glu levels tended to be lower in MN and AFA groups.
Direct cell count and diversity of caecal microbiome
The direct bacterial cell count using DEPIM was approximately 11.2 log cells g−1 caecal matter in mice fed the NF diet and the count was not affected significantly by the addition of algae to the diet (Table 3). The total read number ranged from 101,000–136,000 and tended to be low in mice fed AFA and EB. Compared with the NF group, the number of OTU types was 32% lower in the AFA group. On the other hand, there were no significant differences in Shannon-Wiener H′ as an α-diversity index.
Figure 1 shows PCA of the major OTUs (> 0.1% abundance) of caecal bacteria. PC1 vs. PC2 and PC2 vs. PC3 show that microbiome diversity varied with the intake of different algae, particularly for EB and AFA. Among the five groups, individual differences in the microbiome were small for the MN group.
Abundances of caecal microbiome at phylum and genus levels
Caecal microbiome at the phylum and genus levels are summarized in Fig. 2. Firmicutes was the most dominant phylum in the NF group, comprising approximately 66% of the microbiome, but it was lowered significantly to 41% in mice fed EB. The second-most dominant phylum in the NF group was Actinobacteria (25%), the abundance of which was decreased (to 13%) in the CO group. The third-most dominant phylum in the NF group was Bacteroidetes (8%) and its abundance was high (32%) in the EB group. Proteobacteria was also detected as a dominant phylum in the NF group (1%) and its abundance was high (11%) in the CO group.
Composition of the caecal microbiome at the phylum and genus levels in mice fed high-sucrose diets containing either no fibre (NF), 5% (w/w) Eisenia bicyclis (EB), 5% Chondrus ocellatus (CO), 5% Monostroma nitidum (MN), or 5% Aphanizomenon flos-aquae (AFA). * Values are mean of six mice. a-c Values with different letters are significantly different with p < 0.05
At the genus level within the Firmicutes, Allobaculum was dominant (33%) in the NF group, followed by Lactobacillus (10%), Clostridiaceae g. (8%), and Turicibacter (5%). Among the dominant genera, abundance of Clostridiaceae g. was low in mice fed marine algae EB, CO, and MN. Furthermore, Turicibacter was hardly detected in the EB group. The dominant member of Actinobacteria genus in all mouse groups was Bifidobacterium, followed by Coriobacteriaceae g. Compared with the NF group, Bifidobacterium abundance was low in CO and AFA groups. Bacteroidetes in the NF group were divided mainly into Bacteroidales, f_S24-7 g., and Bacteroides. The abundance of Bacteroides was significantly high (25%) in the EB group. Among the phyla, the abundance of Prevotella was high in the CO and MN groups. Bacteroidales, f_S24-7 g., and Rikenellaceae g. abundances were high in the AFA group. Almost all the Proteobacteria in the CO group were Enterobacteriaceae g.
Dominant OTUs and SIBs
Figure 3 shows a heat map of the relative abundance of identified OTUs (filtered by 0.25% counts of whole OTUs) and estimated species names of typical OTUs defined by BLAST analysis of de novo sequences. The dominant OTU taxonomies were Allobaculum stercoricanis- (similarity = 92%), Faecalibaculum rodentium- (100%), and/or Bifidobacterium pseudolongum-like (100%) bacteria. Most Clostridiaceae g. decreased with the intake of the three marine algae (Fig. 2), as estimated by Clostridium disporicum-like (99%) bacteria.
Heat map showing the relative abundance of identified OTUs filtered by 0.25% counts of whole OTUs of the caecal microbiome in mice fed with high-sucrose diet containing either no fibre (NF), 5% (w/w) Eisenia bicyclis (EB), 5% Chondrus ocellatus (CO), 5% Monostroma nitidum (MN), or 5% Aphanizomenon flos-aquae (AFA). P: Proteobacteria, F: Firmicutes, B: Bacteroidetes, A: Actinobacteria. *, a-c Values with different letters are significantly different with p < 0.05
Compared with the NF group, abundances of two Bacteroides OTUs, B. acidifaciens- (100%) and B. intestinalis-like (100%) bacteria, were high in the EB group. Each OTU of Coriobacteriaceae, which was defined as B. pseudolongum- (92%), Longibaculum muris- (97%), and Coprobacillus sp.-like (100%) bacteria, was also highly abundant in the EB group.
In the mice fed CO, the two Bacteroides B. vulgatus- (100%) and Escherichia coli-like (100%) bacteria were typically abundant OTUs. The abundances of B. acidifacience/rodentium- (100%), Prevotellaceae- (93%), and Alistipes finegoldii-like (96%) bacteria were also high. On the other hand, the abundances of B. pseudolongum- and Lactobacillus johnsonii-like (100%) bacteria were low. With MN feeding, F. rodentium-like bacteria became more abundant. Three Bacteroidales, f_S24-7 g, s OTUs defined as Muribaculum sp.-like (92–95%) bacteria were also highly abundant in the MN group. On the other hand, the abundance of L. johnsonii was low in this group. In mice fed with AFA, the abundances of two OTUs, Muribaculum sp.- (90, 93%) and Alistipes inops-like (100%) bacteria, were high. However, B. pseudomonas abundance was low in this group.
Isolation and identification of SIB
The counts of each colony type on BL agar plates are shown in Table 4. The agar plate showed six typical colony morphologies. Among them, three (Algae-1, Algae-3, Algae-7) were defined as B. pseudolongum using 16S rDNA-BLAST search. Algae-1 was high in mice fed EB, and as shown in Fig. 3, B. pseudolongum Algae-1 can be considered as one of the EB-SIBs. However, B. acidifaciens and B. intestinalis, which were also considered EB-SIBs, could not be isolated. Cloudy white colonies of one typical morphology were divided into two groups using Gram staining. The Gram-negative (Algae-4) and Gram-positive (Algae-5) were identified as B. vulgatus and F. rodentium, respectively. These strains can be considered CO-SIB and MN-SIB. Muribaculum, identified as an AFA-SIB, shown in Fig. 3, could not be isolated in this experiment. Viable counts of L. johnsonii (Algae-2) were low in mice fed marine algae, as shown in Fig. 3.
Discussion
The present study aimed to clarify the presence of, and isolate, algal-susceptible indigenous bacteria (SIB) in the gut. It is well known that edible algae are rich in water-soluble polysaccharides, a form of dietary fibre. Among various polysaccharides, brown algae have alginic acid (mannuronic and guluronic acid polymer), fucoidan (mainly sulphated fucan), and laminaran (mainly β-1,3-glucan), and E. bicyclis is rich in alginic acid and laminaran (Kuda et al. 1998). Red algae have various water-soluble polysaccharides and C. ocellatus contains high levels of carrageenans (Van de Verde 2008). Typical sulphated polysaccharides in the green alga M. nitidum have been reported (Mao et al. 2008). Various amelioration capacities of marine algal dietary fibres, polyphenols and lipids, have been reported in model animals for lifestyle-related diseases (Wells et al. 2017). In this experiment, some samples did not cause significant differences in body and fat tissue weights and blood lipid levels (Table 2) because of the short feeding period. However, it has been revealed that the gut microbiome fluctuates quickly within a few days of a diet change (David et al. 2014).
It has been reported that gut microbiome α-diversity is correlated with vegetable (dietary fibre) intake, and may be responsible for some of the microbiome’s health benefits (Deehan and Walter 2016). However, there was no significant difference in the Shannon H′ index (Table 3), even though differences in the microbiota were shown in beta diversity analysis (Fig. 1). Alpha diversity is lowered with ingestion of some fermentable dietary fibres such as laminaran, oligosaccharides such as soybean oligosaccharides, prebiotics, and selected lactic acid bacteria (Nakata et al. 2016; Nakata et al. 2017).
The positive correlation between the Firmicutes/Bacteroidetes (F/B) ratio and obesity and/or diabetes has been reported several times (Tilg and Kaser 2011; Houghton et al. 2018). The F/B ratio was low in mice fed EB, as shown in Fig. 2. Alginic acid and laminaran, which are brown algal polysaccharides, can be degraded and fermented by several species of human intestinal Bacteroides, such as B. ovatus, B. thetaiotaomicron, and B. distasonis (Salyers et al. 1977; Fujii et al. 1992). In the case of laboratory rats, degrading bacteria in the gut increased significantly following polysaccharide feeding (An et al. 2013a). The fermentation of alginate and laminaran in the gut produces short chain fatty acids that may ameliorate the intestinal environment and lipid metabolism, and inhibit generation of some putative carcinogen-related compounds, such as ammonia, H2S, and phenol (Kuda et al. 2005; An et al. 2013b).
Although Bifidobacterium, the abundance of which was low in the CO and AFA groups, is regarded as beneficial, it is highly present in mice fed a high-calorie and low–dietary fibre diet (Kuda et al. 2017). The abundance of Proteobacteria Enterobacteriaceae, which was highly abundant only in the CO group, was shown to increase during gut inflammation (Shibayama et al. 2018).
The predominant OTUs in the NF group, estimated as A. stercoricanis-, F. rodentium-, B. pseudolongum-, and C. disporicum-like bacteria (Fig. 3), have been reported as dominant bacteria in the gut of mammals, including human and laboratory rodents (Sasajima et al. 2010; Lim et al. 2016; Shikano et al. 2019). B. acidifaciens, one of the estimated bacterial species that increased in the EB group (EB-SIB), reportedly has a preventative effect on obesity in mice (Yang et al. 2017). B. intestinalis abundance is increased by ingestion of several water-soluble dietary fibres similar to B. ovatus and B. thetaiotaomicron (Chung et al. 2016). B. pseudolongum combined with oligosaccharides ameliorated gut inflammation (Sasajima et al. 2010). These EB-SIBs may be affected by the fermentable water-soluble polysaccharides alginic acid and laminaran, and an immune modulable polysaccharide, fucoidan, present in E. bicyclis (Kuda et al. 1998).
Among the CO-SIB-estimated species, B. vulgatus and E. coli are in general gut indigenous bacteria that may act as symbionts in a healthy host, though their abundance increases with intestinal inflammation (Salyers et al. 1977; Chung et al. 2016; Shibayama et al. 2018). It has been reported that Bifidobacterium and L. johnsonii were dominant in ICR mice fed AIN-76 without cellulose (same as the NF diet in this study) and normal chow diet, respectively, and thus, these are regarded as beneficial symbionts (Kuda et al. 2017; Yokota et al. 2018b). F. rodentium, a MN-SIB-estimated species, is detected as a lactic acid–producing bacteria in the faeces of laboratory mice (Chang et al. 2015). Upregulation of Faecalibaculum in calorie-restricted mice has been reported (Wang et al. 2018).
Among the AFA-SIB-estimated species, Muribaculum is predominant in the gut of mammals, including human and laboratory mice, and it is considered the dominant host-affecting species; however, this hypothesis has not yet been validated (Lagkouvardos et al. 2019). Some gut Alistipes OTUs in diet-induced obese mice are lowered with consumption of probiotics and prebiotics demonstrating improvement of metabolic disturbances (Ke et al. 2019). As mentioned above, Bifidobacterium is abundant in mice fed a high-calorie and low–dietary fibre diet (Kuda et al. 2017).
Antibacterial activity of algal phytochemicals, such as phlorotannins in brown algae and sulphated polysaccharides, has been reported particularly against Gram-positive bacteria (Kuda et al. 2007; Berri et al. 2016). These algal phytochemicals might suppress Gram-positive bacteria, such as C. disporicum in mice fed EB, B. pseudomonas in mice fed CO, and AFA and L. johnsonii in mice fed CO and MN.
BL agar, containing 5% horse blood, under anaerobic conditions, can maintain the growth of various anaerobes, such as Bacteroides, Clostridium, Eubacterium, Fusobacterium, Veillonella, Peptostreptococcus, and Bifidobacterium (Mitsuoka 2014). In the present study, the caecal content was prepared and plated on BL agar plates to isolate the SIBs under not strictly anaerobic conditions, although the AnaeroPack system was used. As shown in Table 4, although several algal SIBs could be detected in the present study, the abundances of strict obligate anaerobes, including dominants shown in Fig. 3, such as Bacteroides sp. Allobaculum sp., Muribaculum sp., and Turicibacter sp., might have been drastically decreased. In future studies, B. acidifaciens, B. intestinalis, and Muribaculum, estimated as EB-, MN-, and AFA-SIBs, respectively, should be isolated.
Significant differences were demonstrated in the gut microbiome among ICR mice fed NF-, EB-, CO-, MN-, or AFA-containing diets for 14 days in the present study. Typical EB-SIBs (B. acidifaciens-, B. intestinalis-, and B. pseudolongum-like), CO-SIBs (B. vulgates- and E. coli-like), MN-SIBs (F. rodentium- and Muribaculum sp.-like), and AFA-SIBs (Muribaculum sp.) were detected by the 16S rDNA amplicon sequencing. These SIBs may affect host health depending on the food material ingested. From the SIBs, B. pseudolongum, B. vulgatus, and F. rodentium could be isolated and identified employing BL agar and the 16S rDNA BLAST search. Some parts of the algal SIBs have beneficial synergistic effects when the host consumes edible algae. In the future, new probiotics may be created from human algal SIBs detected and isolated with the in vitro faecal culture method (Charoensiddhi et al. 2017).
Functional properties, such as immunomodulation, intestinal epithelial cell protection activity, inhibitory activity on invasion of pathogens, and bile acid lowering (Hirano et al. 2017; Shibayama et al. 2018; Yokota et al. 2018a), of these isolated SIBs in vitro are currently being studied. The in vivo functional study and isolation of SIBs from human faeces is needed for future studies.
Conclusion
Significant differences in the gut microbiome were demonstrated among ICR mice fed NF-, EB-, CO-, MN-, or AFA-containing diets for 14 days. Typical EB-SIBs (B. acidifaciens-, B. intestinalis-, and B. pseudolongum-like), CO-SIBs (B. vulgates- and E. coli-like), MN-SIBs (F. rodentium- and Muribaculum sp.-like), and AFA-SIBs (Muribaculum sp.) were detected by the 16S rDNA amplicon sequencing. These SIBs may affect host health depending on the food material consumed. From the SIBs, B. pseudolongum, B. vulgatus, and F. rodentium could be isolated and identified employing BL agar and the 16S rDNA BLAST search. Other SIBs are expected to be isolated in future studies.
References
An C, Yazaki T, Takahashi H, Kuda T, Kimura B (2013a) Diet-induced changes in alginate- and laminaran-fermenting bacterial levels in the caecal contents of rats. J Funct Foods 5:389–394
An C, Kuda T, Yazaki T, Takahashi H, Kimura B (2013b) FLX pyrosequencing analysis of the effects of the brown-algal fermentable polysaccharides alginate and laminaran on rat caecal microbiotas. Appl Environ Microbiol 79:860–866
Bao J, Tang Q, Chen Y (2017) Individual nursing care for the elderly among China's aging population. BioSci Trends 11:694–696
Berri M, Slugocki C, Olivier M, Helloin E, Jacques I, Salmon H, Demais H, Le Goff M, Collen PN (2016) Marine-sulfated polysaccharides extract of Ulva armoricana green algae exhibits an antimicrobial activity and stimulates cytokine expression by intestinal epithelial cells. J Appl Phycol 28:2999–3008
Cabinet Office Japan, Annual Report on the Aging Society. (2018) https://www8.cao.go.jp/kourei/english/annualreport/2018/2018pdf_e.html
Chang D, Rhee M, Ahn S, Bang B, Oh JE, Lee HK, Kim BC (2015) Faecalibaculum rodentium gen. nov., sp. nov., isolated from the faeces of a laboratory mouse. Antonie Van Leeuwenhoek 108:1309–1318
Charoensiddhi S, Conlon MA, Vuaran MS, Franco CMM, Zhang W (2017) Polysaccharide and phlorotannin-enriched extracts of the brown seaweed Ecklonia radiata influence human gut microbiota and fermentation in vitro. J Appl Phycol 29:2407–2416
Chung WS, Walker AW, Louis P, Parkhill J, Vermeiren J, Bosscher D, Duncan SH, Flint HJ (2016) Modulation of the human gut microbiota by dietary fibres occurs at the species level. BMC Biol 14:3
David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, Biddinger SB, Dutton RJ, Turnbaugh PJ (2014) Diet rapidly and reproducibly alters the human gut microbiome. Nature 505:559–563
Deehan EC, Walter J (2016) The fiber gap and the disappearing gut microbiome: implications for human nutrition. Trends Endocrinol Metab 27:239–242
Delgado GTCD, Tamashiro WMSC (2018) Role of prebiotics in regulation of microbiota and prevention of obesity. Food Res Int 113:183–188
Eda M, Kuda T, Kataoka M, Takahashi H, Kimura B (2016) Anti-glycation properties of the aqueous extract solutions of dried algae products harvested and made in the Miura Peninsula, Japan, and effect of lactic acid fermentation on the properties. J Appl Phycol 28:3617–3624
Fujii T, Kuda T, Saheki K, Okuzumi M (1992) Fermentation of water-soluble polysaccharides of brown algae by human intestinal bacteria in vitro. Nippon Suisan Gakkaishi 58:147–152
Gafan GP, Lucas VS, Roberts GJ, Petrie A, Wilson M, Spratt DA (2005) Statistical analyses of complex denaturing gradient gel electrophoresis profiles. J Clin Microbiol 43:3971–3978
Hirano S, Yokota S, Eda M, Kuda T, Shikano A, Tkahashi H, Kimura B (2017) Effect of Lactobacillus plantarum Tennozu-SU2 on Salmonella typhimurium infection in human enterocyte-like HT-29-Luc cells and BALB/c mice. Probiotics Antimicro Prot 9:64–70
Hirota K, Inagaki S, Hamada R, Ishihara K, Miyake Y (2014) Evaluation of a rapid oral bacteria quantification system using dielectrophoresis and the impedance measurement. Biocontrol Sci 19:45–49
Houghton D, Hardy T, Stewaart C, Errington L, Day CP, Trenell ML, Avery L (2018) Systematic review assessing the effectiveness of dietary intervention on gut microbiota in adults with type 2 diabetes. Diabetol 61:1700–1711
Htun NC, Suga H, Imai S, Shimizu A, Takimoto H (2017) Food intake patterns and cardiovascular risk factors in Japanese adults: analyses from the 2012 National Health and nutrition survey, Japan. Nutr J 16:61
Jovel J, Patterson J, Wang W, Hotte N, O’Keefe S, Mitchel T, Perry T, Kao D, Mason AL, Madsen KL, Wong GK (2016) Characterization of the gut microbiome using 16S or shotgun metagenomics. Front Microbiol 7:459
Kato K, Odamaki T, Mitsuyama E, Sugahara H, Xiao J, Osawa R (2017) Age-related changes in the composition of gut Bifidobacterium species. Curr Microbiol 74:987–995
Ke X, Walker A, Haange S, Lagkouvardos I, Liu Y, Schmitt-Kopplin P, von Bergen M, Jehmlich N, He X, Clavel X, Cheung PCK (2019) Synbiotic-driven improvement of metabolic disturbances is associated with changes in the gut microbiome in diet-induced obese mice. Mol Metab 22:96–109
Kido Y (2015) The issue of nutrition in an aging society. J Nutr Sci Vitaminol 61:S176–S177
Kuda T, Goto H, Yokoyama M, Fujii T (1998) Fermentable dietary fiber in dried products of brown algae and their effects on cecal microflora and levels of plasma lipid in rats. Fisheries Sci 64:582–588
Kuda T, Ikemori T (2009) Minerals, polysaccharides and antioxidant properties of aqueous solutions obtained from macroalgal beach-casts in the Noto Peninsula, Ishikawa, Japan. Food Chem 112:575–581
Kuda T, Kunii T, Goto H, Suzuki T, Yano T (2007) Varieties of antioxidant and antibacterial properties of Ecklonia stolonifera and Ecklonia kurome products harvested and processed in the Noto peninsula, Japan. Food Chem 103:900–905
Kuda T, Yano T, Matsuda N, Nishizawa M (2005) Inhibitory effects of laminaran and low molecular alginate against the putrefactive compounds produced by intestinal microflora in vitro and in rats. Food Chem 91:745–749
Kuda T, Yokota Y, Haraguchi Y, Takahashi H, Kimura B (2019) Susceptibility of gut indigenous lactic acid bacteria in BALB/c mice to oral administered Lactobacillus plantarum. Int J Food Sci Nutr 70:53–62
Kuda T, Yokota Y, Shikano A, Takei M, Takahashi H, Kimura B (2017) Dietary and lifestyle disease indices and caecal microbiota in high fat diet, dietary fibre free diet, or DSS induced IBD models in ICR mice. J Funct Foods 35:605–614
Lagkouvardos I, Lesker TR, Hitch TC, Gálvez EJ, Smit N, Neuhaus K, Wang J, Baines JF, Abt B, Stecher B, Overmann J, Strowig T, Clavel T (2019) Sequence and cultivation study of Muribaculaceae reveals novel species, host preference, and functional potential of this yet undescribed family. Microbiome 7:28
Lee H, Oh S, Cho H, Cho H, Kang Y (2016) Prevalence and socio-economic burden of heart failure in an aging society of South Korea. BMC Cardiovasc Disord 16:215
Lim S, Chang D, Ahn S, Kim B (2016) Whole genome sequencing of “Faecalibaculum rodentium” ALO17, isolated from C57BL/6J laboratory mouse feces. Gut Pathogen 8:3
Mao W, Fang F, Li H, Qi X, Sun H, Chen Y, Guo S (2008) Heparinoid-active two sulfated polysaccharides isolated from marine green algae Monostroma nitidum. Carbohydr Polym 74:834–839
Marchesi JR, Adams DH, Fava F, Hermes GDA, Hirschfield GM, Hold G, Quraishi MN, Kinross J, Smidt H, Tuohy KM, Thomas LV, Zoetendal EG, Hart A (2016) The gut microbiota and host health: a new clinical frontier. Gut 65:330–339
Mitsuoka T (2014) Establishment of intestinal bacteriology. Biosci Microb Food Health 33:99–116
Nakata T, Kyoui D, Takahashi H, Kimura B, Kuda T (2016) Inhibitory effects of laminaran and alginate on production of putrefactive compounds from soy protein by intestinal microbiota in vitro and in rats. Carbohydr Polym 143:61–69
Nakata T, Kyoui D, Takahashi H, Kimura B, Kuda T (2017) Inhibitory effects of soybean oligosaccharides and water-soluble soybean fibre on formation of putrefactive compounds from soy protein by gut microbiota. Int J Biol Macromol 97:173–180
Park Y, Lee J, Oh JH, Shin A, Kim J (2016) Dietary patterns and colorectal cancer risk in a Korean population. Medicine 95:25
Salyers AA, Vercellotti JR, West SEH, Wilkins TD (1977) Fermentation of mucin and plant polysaccharides by strains of Bacteroides from the human colon. Appl Environ Microbiol 33:319–322
Santeramo FG, Carlucci D, De Devitiis B, Seccia A, Stasi A, Viscecchia R, Nardone G (2018) Emerging trends in European food, diets and food industry. Food Res Int 104:39–47
Sasajima N, Ogasawara T, Takemura N, Fujiwara R, Watanabe J, Sonoyama K (2010) Role of intestinal Bifidobacterial pseudolongum in dietary fructo-oligosaccharide inhibition of 2,4-dinitrofluorobenzene-induced contact hypersensitivity in mice. Br J Nutr 103:539–548
Shang Q, Jiang H, Cai C, Hao J, Li G, Yu G (2018) Gut microbiota fermentation of marine polysaccharides and its effects on intestinal ecology: an overview. Carbohydr Polym 179:173–185
Shibayama J, Kuda T, Shikano A, Fukunaga M, Takahashi H, Kimura B, Ishizaki S (2018) Effects of rice bran and fermented rice bran suspensions on caecal microbiota in dextran sodium sulphate-induced inflammatory bowel disease model mice. Food Biosci 25:8–14
Shikano A, Kuda T, Shinayama J, Toyama A, Yuka I, Takahashi H, Kimura B (2019) Effects of Lactobacillus plantarum Uruma-SU4 fermented green loofah on plasma lipid levels and gut microbiome of high-fat diet fed mice. Food Res Int 121:817–824
Sinclair L, Osman OA, Bertilsson S, Eiler A (2015) Microbial community composition and diversity via 16S rRNA gene amplicons: evaluating the Illumina platform. PLoS One 10:e0116955
Sugawara S, Kushida M, Iwagaki Y, Asano M, Yamamoto K, Tomata Y, Tsuji I, Tsuduki T (2018) The 1975 type Japanese diet improves lipid metabolic parameters in younger adults: a randomized controlled trial. J Oleo Sci 67:599–607
Takei M, Kuda T, Eda M, Shikano A, Takahashi H, Kimura B (2017) Antioxidant and fermentation properties of aqueous solutions of dried algal products from the Boso Peninsula, Japan. Food Biosci 19:85–91
Taniguchi M, Kuda T, Shibayama J, Sasaki T, Michihata T, Takahashi H, Kimura B (2019) In vitro antioxidant, anti-glycation and immunomodulation activities of fermented blue-green algae Aphanizomenon flos-aquae. Mol Biol Rep 46:1775–1786
Tilg H, Kaser A (2011) Gut microbiome, obesity, and metabolic dysfunction. J Clin Invest 121:2126–2132
Van de Verde F (2008) Structure and function of hybrid carrageenans. Food Hydrocoll 22:727–734
Wang S, Huang M, You X, Zhao J, Chen L, Wang L (2018) Gut microbiota mediates the antiobesity effect of calorie restriction in mice. Sci Rep 8:13037
Wells ML, Potin P, Craigie JS, Raven JA, Merchant SS, Helliwell KE, Smith AG, Camire ME, Brawley SH (2017) Algae as nutritional and functional food sources: revisiting our understanding. J Appl Phycol 29:949–982
Winglee K, Howard AG, Sha W, Gharaibeh RZ, Liu J, Jin D, Fodor AA, Gordon-Larsen P (2017) Recent urbanization in China is correlated with a Westernized microbiome encoding increased virulence and antibiotic resistance genes. Microbiome 5:121
Yang C, Lai S, Chen Y, Liu D, Liu B, Ai C, Wan X, Gao L, Chen X, Zhao C (2019) Anti-diabetic effect of oligosaccharides from seaweed Sargassum confusum via JNK-IRS1/PI3K signalling pathways and regulation of gut microbiota. Food Chem Toxicol 131:110562
Yang JY, Lee YS, Lee SH, Ryu S, Fukuda S, Hase K, Yang CS, Lim HS, Kim MS, Kim HM, Ahn SH, Kwon BE, Ko HJ, Kweon MN (2017) Gut commensal Bacteroides acidifaciens prevents obesity and improves insulin sensitivity in mice. Mucosal Immunol 10:104–116
Yokota Y, Shikano A, Kuda T, Takei M, Takahashi H, Kimura B (2018a) Lactobacillus plantarum AN1 cells increase caecal L. reuteri in an ICR mouse model of dextran sodium sulphate-induced inflammatory bowel disease. Int Immunopharmacol 56:119–127
Yokota Y, Haraguchi Y, Shikano A, Kuda T, Takahashi H, Kimura B (2018b) Induction of gut Lactobacillus reuteri in normal ICR mice by oral administration of L. plantarum AN1. J Food Biochem 42:e12589
Zhao C, Yang C, Liu B, Lin L, Sarker SD, Nahar S, Yu H, Cao H, Xiao J (2018) Bioactive compounds from marine macroalgae and their hypoglycemic benefits. Trends Food Sci Technol 72:1–12
Acknowledgements
This work was partially supported by the Towa Foundation for Food Science & Research, Tokyo, Japan.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Statement of animal rights
Animal experiments were performed in compliance with the Fundamental Guidelines for Proper Conduct of Animal Experiment and Related Activities in Academic Research Institutions, under the jurisdiction of the Ministry of Education, Culture, Sports, Science, and Technology, and approved by the animal experiment committee of the Tokyo University of Marine Science and Technology (approval No. H30-4).
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(XLSX 15277 kb).
Rights and permissions
About this article
Cite this article
Takei, M., Kuda, T., Fukunaga, M. et al. Effects of edible algae on caecal microbiomes of ICR mice fed a high-sucrose and low–dietary fibre diet. J Appl Phycol 31, 3969–3978 (2019). https://doi.org/10.1007/s10811-019-01866-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10811-019-01866-x