Abstract
The cleavage and secretion of pro-inflammatory cytokines IL-1β and IL-18 is regulated by NLRP3 (NACHT, LRR, and PYD domain-containing protein 3) inflammasome activation. Kupffer cells (KCs) are implicated in the pathogenesis of various liver diseases, such as non-alcoholic fatty liver disease (NAFLD), alcoholic liver disease, and liver fibrosis. However, the role of NLRP3 played in the non-alcoholic steatohepatitis (NASH) has yet to be evaluated. In the present study, methionine–choline-deficient (MCD) diet was used to establish the mice NASH model. The expression levels of F4/80 and NLRP3 in liver tissues were evaluated, and the IL-1β and IL-18 in serum were also evaluated. KCs were isolated from wild-type (WT) mice and NLRP3 knockout (NLRP3−/−) mice and then randomly divided into two groups: the control and palmitic acid (PA) groups. The expression levels of NLRP3, ASC, and caspase-1 in KCs were determined by RT-PCR, western blotting, and immunofluorescence. The levels of IL-1β and IL-18 in the supernatant (SN) of KCs were evaluated by enzyme-linked immunosorbent assay (ELISA). We found that KCs and NLRP3 play pro-inflammatory roles in the progression of NASH, probably through secretions of IL-1β and IL-18 by KCs induced by PA. PA could act as a kind of damage-associated molecular patterns to elevate the messenger RNA and protein expression levels of NLRP3, ASC, and caspase-1 in KCs from WT mice. In the contrast, NLRP3 deletion could inhibit the NLRP3 inflammasome upregulation and activation in KCs induced by PA. Furthermore, the levels of pro-inflammatory cytokines IL-1β and IL-18 in the SN of KCs from WT mice were all elevated with the stimulation of PA, and the increase of these cytokines in the SN was blocked by NLRP3 deletion. In conclusion, our novel findings demonstrate that NLRP3 plays a pivotal role in NASH development and pro-inflammatory cytokines IL-1β and IL-18 secretion induced by PA stimulation, and NLRP3 might be an effective potential target for the treatment of liver inflammatory diseases associated with NLRP3 inflammasome activation.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
INTRODUCTION
Non-alcoholic fatty liver disease (NAFLD) is currently the most common cause of chronic liver disease in many developed countries worldwide [1, 2]. NAFLD is characterized by excessive hepatic fat accumulation, associated with insulin resistance (IR), and defined by the presence of steatosis in >5% of hepatocytes according to histological analysis. NAFLD includes two pathologically distinct conditions with different prognoses: non-alcoholic fatty liver (NAFL) and non-alcoholic steatohepatitis (NASH), which can eventually progress to end-stage liver diseases such as cirrhosis and hepatocellular carcinoma [1, 3]. The pathogenesis of NAFLD is commonly based on the double-hit hypothesis, which was proposed by Day and James in 1998 [4]. According to this theory, the first hit is the initial hepatocellular lipid accumulation, followed by a second hit, in which pro-inflammatory mediators induce inflammation and hepatocellular injury, thus promoting the progression to NASH. Recently, the progression from NAFL to NASH has been deemed as the crucial step in the development of NAFLD [5]. Metaflammation, a low-grade form of chronic inflammation, plays an important role in the genesis and development of NASH [6]. However, the exact mechanism underlying the development of NASH remains to be clearly defined.
NLRP3 (NACHT, LRR, and PYD domain-containing protein 3) is the most well-characterized member of the inflammasome family, which consists of the NOD-like receptor NLRP3, the adaptor ASC, and the effector pro-caspase-1 [7]. It is considered that the activation of NLRP3 inflammasome contains two steps. The first step consists of cell priming with an NF-κB activator, leading to the upregulation of messenger RNA (mRNA) levels of NLRP3 and pro-IL-1β, and the second step is triggered by various activators such as ATP and free fatty acid (FFA) [8,9,10]. It has been proved that FFA acts as a kind of damage-associated molecular patterns (DAMPs) to activate NLRP3 inflammasome in the NAFLD patients and mice models [11, 12]. However, the effect of FFA has on the NLRP3 inflammasome activation and IL-1β secretion in vitro is still unclear.
Kupffer cells (KCs), which constitute 80–90% of the tissue macrophages in the body, are increasingly deemed as an important immune cell in liver homeostasis and inflammatory diseases [13, 14]. Upon activation, KCs release various products, including cytokines, nitric oxide, and reactive oxygen species (ROS), and KCs have been implicated in the pathogenesis of various liver diseases, such as viral hepatitis, steatohepatitis, alcoholic liver disease, and activation or rejection of the liver after liver transplantation [15, 16]. Activated KCs play a central role in pathogenesis and the progression of liver disease by contributing to parenchymal inflammation, hepatocyte injury, and initiation of fibrosis via cytokine secretion (TNF-α, TGF-β) [17]. It has also been proved that the KCs are the predominant source of pro-inflammatory cytokine IL-1β, when compared with other cell types in the liver such as hepatocytes and hepatic stellate cells (HSCs) [18, 19]. Nevertheless, the exact mechanism of the IL-1β secretion in KCs induced by palmitic acid (PA) and the function of NLRP3 in this process are still needed to be evaluated.
In the current study, we explore the role of NLRP3 in NASH development and secretions of IL-1β and IL-18 in KCs induced by PA. We assume that NLRP3 deletion could prevent the development of mice NASH, and PA induces IL-1β and IL-18 secretion through activating NLRP3 inflammasome in KCs, which could be suppressed by NLRP3 deletion.
MATERIALS AND METHODS
Animals
NLRP3 knockout (NLRP3−/−) male mice aged 8 weeks which were purchased from Jackson Laboratories (Bar Harbor, ME, USA) were bred in the Laboratory Animal Research Center of Chongqing Medical University (Chongqing, China). Wild-type (WT) C57BL/6 mice were obtained from the Laboratory Animal Research Center of Chongqing Medical University. All animals were housed under specific pathogen-free condition and allowed free access to sterile water and food. The animals received humane care in compliance with the institution’s guidelines, as outlined in the Guide for the Care and Use of Laboratory Animals prepared by the National Academy of Sciences. All experimental protocols described in this study complied with the ethics review committee for animal experimentation of Chongqing Medical University.
Histological Analysis
Sections of formalin-fixed livers were stained with hematoxylin-eosin (HE), Oil Red O, and immunohistochemical staining for F4/80 (ab16911; Abcam, UK) and NLRP3 (sc-66846; Santa Cruz, USA). The quantitative immunohistochemical staining values (QISVs) were calculated as the integrated OD divided by the total area occupied by the brown cells in each slide.
Serum Analysis
Serum aminotransferase levels were determined according to the rate method. Serum levels of FFA were measured using an enzymatic method (Clinimate NEFA kit, Sekisui Medical Company, Japan). Serum IL-1β and IL-18 levels were determined by chemiluminescence (Bio-Rad, USA).
In vitro Experiments
Primary KCs were isolated from mouse livers according to a previously described procedure [20]. Briefly, KCs were cultured in six-well plates at a density of 1 × 106 cells/well in Dulbecco’s modified Eagle’s medium (DMEM) (HyClone, USA) supplemented with 10% FBS (HyClone, USA) and antibiotics (100 U/ml of penicillin G and 100 mg/ml of streptomycin sulfate) at 37 °C in the presence of 5% CO2. The viability of KCs (>90%) was determined by trypan blue exclusion experiment. KCs were randomly divided into two groups: the control (CON) and PA (0.32 mM) groups. Cell culture supernatant (SN), morphology, gene, and protein expression levels of NLRP3 inflammasome were further analyzed.
RNA Analysis
The mRNA expression levels of NLRP3, ASC, and caspase-1 in KCs were analyzed by RT-qPCR. Briefly, total RNA was extracted from cell samples using TRIzol® reagent (Invitrogen Life Technologies, Carlsbad, USA) according to the manufacturer’s instructions and quantified via the 260:280 nm absorption ratio of RNA samples (model 722; Bio-Rad Laboratories, Hercules, USA). The amplification of sample cDNA was monitored using the SsoFast EvaGreen Supermix (Bio-Rad Laboratories, Hercules, USA) and the DNA fluorescent dye SYBR Green (Molecular Probes, Eugene, USA).
Sequences of specific NLRP3, ASC, caspase-1, and β-actin primers were as follows: NLRP3, forward: 5′-ACTTGCAGAAGCTGGGGTTG-3′, reverse: 5′-AGTTTACAGTCCGGGTGCAG-3′; ASC, forward: 5′-ACAGAAGTGGACGGAGTGCT-3′, reverse: 5′-TCATCTTGTCTTGGCTGGTG-3′; caspase-1, forward: 5′-TGCCCAGAGCACAAGACTTC-3′, reverse: 5′-TCCTTGTTTCTCTCCACGGC-3′; and β-actin, forward: 5′-TGACGTGGACATCCGCAAAG-3′, reverse: 5′-CTGGAAGGTGGACAGCGAGG-3′.
Western Blotting Analysis
The protein expression levels of NLRP3, ASC, and caspase-1 in KCs were detected by western blotting. Protein extracts were obtained by homogenizing samples in a cell lysis buffer (RIPA buffer, R0278; Sigma, UK). The protein concentration was determined with a BCA Protein Assay Kit (23227; Thermo, UK). An equal amount of protein samples was each separated on 10% Tris–HCl gels (Bio-Rad, USA) by electrophoresis and transferred onto a polyvinylidene fluoride membrane. Then, the membrane was blocked for 1 h with 5% non-fat dry milk and incubated with primary antibodies specific for NLRP3 (sc-66846; Santa Cruz, USA), ASC (sc-22514-R; Santa Cruz, USA), and caspase-1 (sc-56036; Santa Cruz, USA) at 4 °C overnight. The membrane was washed and incubated for 1 h at room temperature with the secondary antibodies. Finally, the membrane was developed using an Enhanced Chemiluminescence Detection Kit (Pierce, USA) and exposed to an auto-radiographic film (Kodak, USA). The relative amount of the proteins was quantified by the relative density of protein bands using the image analysis system (Bio-Rad Gel Doc 2000, USA).
Caspase-1 Activity Assay
Caspase-1 activity in KCs was determined with a colorimetric assay (R&D Systems, USA).
Immunofluorescence Analysis
The protein expression levels of NLRP3, ASC, and caspase-1 in KCs were also detected by immunofluorescence (IF). IF was performed according to the instruction. In detail, the cells were fixed by 4% paraformaldehyde (PFA) for 15 min and then permeated in 0.2% Triton X-100 in PBS for 10 min, followed by blocking with 10% serum in 0.2% Triton X-100 in PBS for 1 h. The cells were then incubated with the primary antibody introduced above (1:200), overnight at 4 °C. Then, the cells were incubated with the secondary antibody (1:200) (Invitrogen, UK) for 1 h. After washing with PBS, cells were covered with the mounting medium and the slides were viewed by confocal microscopy (Nikon, Japan).
Enzyme-Linked Immunosorbent Assay Analysis
Enzyme-linked immunosorbent assay (ELISA) was used to measure the levels of IL-1β and IL-18 in the SN according to the manufacturer’s instructions.
Statistical Analysis
Data were expressed as mean ± SD, and statistical calculations were determined by analysis of variance (ANOVA) using the statistical package SPSS, version 18.0. The comparison of the means was analyzed by t test. A value of P < 0.05 was considered to be statistically significant.
RESULTS
KCs and NLRP3 and Are Involved in the Development of NASH in Mice
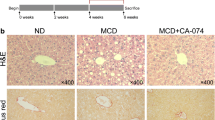
We used diet-induced model of NASH to detect the roles of KCs and NLRP3 in NASH development. Immunohistochemical staining revealed a greater number of F4/80- and NLRP3-positive cells in the livers of WT mice in the NASH group (arrows) than in the normal group (Fig. 1a), which indicated that KCs and NLRP3 might be involved in the progression of NASH. Next, we used NLRP3−/− mice to determine the role of NLRP3 during NASH development by using methionine–choline-deficient (MCD) diet-induced NASH models. There is no significant difference in liver pathological characteristics among the two types of mice fed normal diet (ND) (Fig. 2). Hepatocyte ballooning and inflammatory cell infiltration were obviously increased in MCD diet-fed group of WT mice, which were markedly attenuated in the MCD diet-fed group of NLRP3−/− mice (Fig. 2, arrows; hepatocyte ballooning and inflammatory cells).
KC infiltration and NLRP3 are involved in the NASH progression of WT mice. Immunohistochemical staining for F4/80 (a) and NLRP3 (b) in liver sections (arrows, positive expression levels for KCs and NLRP3). The quantitative immunohistochemical staining values (QISVs) were analyzed for F4/80 and NLRP3 protein expression levels. Data are expressed as the mean ± SD. *P < 0.05, vs normal group. The original magnification is labeled in each picture.
The levels of ALT and AST in the MCD diet-fed group of WT mice were significantly increased than in the ND-fed group of WT mice, whereas NLRP3 deletion obviously suppressed the levels of ALT and AST in the MCD diet-fed group (Fig. 3a, b). The FFA levels were all significantly elevated in these MCD diet-fed groups, and there is no significant difference among these MCD diet-fed groups (Fig. 3c). In these MCD diet-fed groups, IL-1β and IL-18 levels of NLRP3−/− mice were significantly decreased than in the WT mice (Fig. 3d, e). Taken together, these results suggest that KCs and NLRP3 play pro-inflammatory roles in the progression of NASH, probably through secretions of IL-1β and IL-18 by KCs induced by FFA.
NLRP3 deletion ameliorated liver function and reduced pro-inflammatory cytokines in serum. a, b Serum ALT and AST levels. c–e Serum FFA, IL-1β, and IL-18 concentrations. Data are expressed as the mean ± SD. *P < 0.05, vs ND group; # P < 0.05, vs MCD diet-fed group of WT mice; ***P < 0.001, vs ND group.
PA Increases the mRNA and Protein Expression Levels of NLRP3 Inflammasome in KCs
As a kind of DAMPs and activators of NLRP3 inflammasome, PA could increase the mRNA and protein expression levels of NLRP3, ASC, and caspase-1 in KCs, and there were significant differences in the expression levels of components of NLRP3 inflammasome between CON and PA groups (P < 0.05) (Fig. 4a–c). NLRP3 deletion could inhibit the upregulation of NLRP3, ASC, and caspase-1 in KCs induced by PA, and there was no significant difference in the expression levels of components of NLRP3 inflammasome between CON and PA groups (Fig. 4a–c), which suggested that PA was not able to increase the mRNA and protein expression levels of NLRP3, ASC, and caspase-1 in KCs from NLRP3−/− mice. Furthermore, we found that caspase-1 activity was significantly increased in the KCs from WT mice stimulated by PA, whereas PA was not able to upregulate the caspase-1 activity in the KCs from NLRP3−/− mice (Fig. 4d). Taken together, these results demonstrated that PA could effectively induce the upregulation of NLRP3 inflammasome in KCs, which is mainly controlled by NLRP3. Deletion of NLRP3 could remarkable inhibit the upregulation of NLRP3 inflammasome induced by PA.
NLRP3 deletion inhibits the upregulation of mRNA and protein expression levels of NLRP3, ASC, and caspase-1 in KCs induced by PA. The relative NLRP3, ASC, and caspase-1 protein expression levels in KCs and caspase-1 activity were determined as described in the “MATERIALS AND METHODS” and summarized in the graphs. Values are expressed as mean ± standard deviation. *P < 0.05, vs CON group; ***P < 0.001, vs CON group. Data are representative of four individual experiments.
PA Induces the Co-localization of NLRP3, ASC, and Caspase-1 in KCs
The results of IF showed that PA increased expression levels and co-localization of NLRP3 inflammasome in KCs from WT mice in comparison with the CON group (Fig. 5a, arrows; co-localization of NLRP3, ASC, and caspase-1). However, no clear influences on expression levels of NLRP3 inflammasome in KCs from NLRP3−/− mice were detected (Fig. 5b). These data demonstrate that PA could mechanistically act as a kind of DAMPs, increasing the expression levels and co-localization of NLRP3 inflammasome in KCs. Moreover, NLRP3 was basically indispensable for NLRP3 inflammasome activation induced by PA.
PA induces the upregulation and co-localization of NLRP3, ASC, and caspase-1 in KCs. The expression levels and co-localization of NLRP3, ASC, and caspase-1 were observed under a confocal microscopy. a PA increased expression levels and co-localization of NLRP3 inflammasome in KCs from WT mice (×600). b NLRP3 deletion inhibits the expression levels and co-localization of NLRP3, ASC, and caspase-1 in KCs (×600).
NLRP3 Deletion Decreases the Levels of IL-1β and IL-18 in the SN Induced by PA
To further investigate the inflammation induced by PA and the role of NLRP3 in this process, we measured the levels of pro-inflammatory cytokines IL-1β and IL-18 in the SN of each KC group. Our results showed that PA could increase the levels of IL-1β and IL-18 in the SN of KCs from WT mice, the levels of IL-1β and IL-18 in the PA group were significantly higher than those in the CON group (Fig. 6). However, as a consequence of NLRP3 deletion, there was no significant difference in the levels of IL-1β and IL-18 between CON and PA groups in the SN of KCs from NLRP3−/− mice (Fig. 6). Taken together, these results demonstrate that PA could induce the NLRP3 inflammasome activation and, subsequently, IL-1β and IL-18 secretion. Moreover, NLRP3 deletion could suppress the IL-1β and IL-18 secretion after PA stimulation, which suggests that NLRP3 plays a fundamental role in the NLRP3 inflammasome activation and in the secretion of pro-inflammatory cytokines IL-1β and IL-18, and NLRP3 might be an effective potential target for the treatment of inflammatory diseases associated with NLRP3 inflammasome activation and with secretion of pro-inflammatory cytokines IL-1β and IL-18.
NLRP3 deletion decreases the levels of IL-1β and IL-18 in the SN stimulated by PA. The levels of pro-inflammatory cytokines IL-1β and IL-18 in the SN were determined by ELISA as described in the “MATERIALS AND METHODS” and are shown in the graphs. Values are expressed as mean ± standard deviation. ***P < 0.001, vs CON group. Data are representative of four individual experiments.
DISCUSSION
Inflammation contributes to the pathogenesis of most acute and chronic liver diseases, such as alcoholic fatty liver disease, NAFLD, viral hepatitis, and liver transplantation [21]. NLRP3 inflammasome is a multiprotein complex that can sense various danger signals from damaged cells and pathogens and assemble to mediate caspase-1 activation, which proteolytically activates the cytokines IL-1β and IL-18 [22]. NLRP3 inflammasome activation has been widely studied in different human and experimental liver diseases and has been identified as a major contributor to hepatocyte damage, immune cell activation, and amplification of liver inflammation [23]. KCs, an important kind of innate immune cell, play an essential role in the pathogenetic mechanisms of various liver diseases, and NLRP3 inflammasome activation in KCs has been received extensively focused and studied [24,25,26].
In the present study, we demonstrated that KCs and NLRP3 inflammasome activation are involved in the NASH progression, mainly by secretions of pro-inflammatory cytokines IL-1β and IL-18. Furthermore, NLRP3 deletion could suppress the mice NASH development induced by MCD diet. In in vitro experiments, we found that FFA can act as a mechanistic activator of NLRP3 inflammasome in KCs, triggering the NLRP3 inflammasome activation and IL-1β and IL-18 secretion. The results of the IF showed the upregulation and co-localization of NLRP3, ASC, and caspase-1 in KCs induced by PA. Our results showed that the caspase-1 protein levels are not consistent with the caspase-1 mRNA levels. However, the results of western blotting (Fig. 4B) and immunofluorescence (Fig. 5) show the same pattern, which indicates that NLRP3 inflammasome expression and activation are probably controlled at the transcriptional and translational levels to prevent unwanted excessive inflammation, and possibly, NLRP3 deletion has an inhibitory effect on the protein expression of caspase-1. Moreover, NLRP3 inflammasome activation and IL-1β and IL-18 secretion in KCs were blocked in the absence of NLRP3, which indicated that NLRP3 was fundamental for the NLRP3 inflammasome activation and subsequent IL-1β and IL-18 secretion.
For the relationship between hepatocytes and KCs, it is reported that hepatocytes exposed to FFA release danger signals that trigger NLRP3 inflammasome activation in KCs, and hepatocytes play a key role in orchestrating tissue responses to danger signals [11]. Nevertheless, we deemed that FFA could activate NLRP3 inflammasome and IL-1β and IL-18 secretion in KCs, which would be harmful to hepatocytes. There are also supporting data for a major role for macrophages, which comes from the fact that baseline levels of the components of NLRP3 inflammasome were 20-fold higher in mononuclear cells than hepatocytes, LPS only induced an increase in the mononuclear fraction, and KCs are the primary site of the NLRP3 inflammasome activation and primary source of IL-1β in the liver, which further highlighted the essential role of KCs for NLRP3 inflammasome activation and IL-1β and IL-18 secretion [19].
Notably, the mechanism mentioned above is called canonical NLRP3 inflammasome. In addition, NLRP3 needs additional co-factors for the processing of IL-1β in response to Gram-negative bacteria, and this pathway has been termed the non-canonical NLRP3 inflammasome [27]. The activation of caspase-1 following the recognition of live Gram-negative bacteria depends on inflammatory pro-caspase-11 in mice (human orthologues are pro-caspase-4 and pro-caspase-5), which results in non-canonical NLRP3 inflammasome activation. The non-canonical pathway initiates the activation of caspase-11 in mice and caspase-4 and caspase-5 in human in response to LPS (a component of the Gram-negative bacterial cell wall) in the host cell cytosol. Upon activation, caspase-4, caspase-5, and caspase-11 initiate pyroptosis similar to caspase-1, but they do not cleave pro-IL-1β or pro-IL-18 [28]. However, this mechanism is different from the mechanism of PA-induced NLRP3 inflammasome activation, and it still remains to be shown whether additional factors are required to promote cytosolic LPS recognition.
In summary, our novel findings demonstrate that NLRP3 plays a pivotal role in NASH development and pro-inflammatory cytokines IL-1β and IL-18 secretion induced by PA stimulation, and NLRP3 might be an effective potential target for the treatment of liver inflammatory diseases associated with NLRP3 inflammasome activation. Future studies are needed to identify the co-factors which bridge the PA and NLRP3 and clarify the mechanism of NLRP3 inflammasome activation.
References
European Association for the Study of the Liver (EASL).; European Association for the Study of Diabetes (EASD).; European Association for the Study of Obesity (EASO). 2016. EASL-EASD-EASO Clinical Practice Guidelines for the management of non-alcoholic fatty liver disease. Journal of Hepatology 64 (6): 1388–1402.
Wree, A., L. Broderick, A. Canbay, H.M. Hoffman, and A.E. Feldstein. 2013. From NAFLD to NASH to cirrhosis-new insights into disease mechanisms. Nature Reviews. Gastroenterology & Hepatology 10: 627–636.
Wong, R.J., R. Cheung, and A. Ahmed. 2014. Nonalcoholic steatohepatitis is the most rapidly growing indication for liver transplantation in patients with hepatocellular carcinoma in the U.S. Hepatology 59: 2188–2195.
Day, C.P., and O.F. James. 1998. Steatohepatitis: A tale of two “hits”? Gastroenterology 114: 842–845.
Seki, E., and R.F. Schwabe. 2015. Hepatic inflammation and fibrosis: Functional links and key pathways. Hepatology 61: 1066–1079.
Pais, R., F. Charlotte, L. Fedchuk, P. Bedossa, P. Lebray, T. Poynard, et al. 2013. A systematic review of follow-up biopsies reveals disease progression in patients with non-alcoholic fatty liver. Journal of Hepatology 59: 550–556.
Latz, E., T.S. Xiao, and A. Stutz. 2013. Activation and regulation of the inflammasomes. Nature Reviews. Immunology 13 (6): 397–411.
Lamkanfi, M., and V.M. Dixit. 2012. Inflammasomes and their roles in health and disease. Annual Review of Cell and Developmental Biology 28: 137–161.
Strowig, T., J. Henao-Mejia, E. Elinav, and R. Flavell. 2012. Inflammasomes in health and disease. Nature 481 (7381): 278–286.
Elliott, E.I., and F.S. Sutterwala. 2015. Initiation and perpetuation of NLRP3 inflammasome activation and assembly. Immunological Reviews 265 (1): 35–52.
Csak, T., M. Ganz, J. Pespisa, et al. 2011. Fatty acid and endotoxin activate inflammasomes in mouse hepatocytes that release danger signals to stimulate immune cells. Hepatology 54 (1): 133–144.
Miura, K., L. Yang, N. van Rooijen, et al. 2013. Toll-like receptor 2 and palmitic acid cooperatively contribute to the development of nonalcoholic steatohepatitis through inflammasome activation in mice. Hepatology 57 (2): 577–589.
Jenne, C.N., and P. Kubes. 2013. Immune surveillance by the liver. Nature Immunology 14 (10): 996–1006.
Wynn, T.A., A. Chawla, and J.W. Pollard. 2013. Macrophage biology in development, homeostasis and disease. Nature 496 (7446): 445–455.
Li, P., K. He, J. Li, et al. 2017. The role of Kupffer cells in hepatic diseases. Molecular Immunology 85: 222–229.
Heymann, F., and F. Tacke. 2016. Immunology in the liver—from homeostasis to disease. Nature Reviews. Gastroenterology & Hepatology 13 (2): 88–110.
Tiniakos, D.G., M.B. Vos, and E.M. Brunt. 2010. Nonalcoholic fatty liver disease: Pathology and pathogenesis. Annual Review of Pathology 5: 145–171.
Szabo, G., and T. Csak. 2012. Inflammasomes in liver diseases. Journal of Hepatology 57 (3): 642–654.
Hoque, R., Y. Vodovotz, and W. Mehal. 2013. Therapeutic strategies in inflammasome mediated diseases of the liver. Journal of Hepatology 58 (5): 1047–1052.
Li, P.Z., J.Z. Li, M. Li, et al. 2014. An efficient method to isolate and culture mouse Kupffer cells. Immunology Letters 158 (1–2): 52–56.
Racanelli, V., and B. Rehermann. 2006. The liver as an immunological organ. Hepatology 43 (2 Suppl 1): S54–S62.
Lamkanfi, M., and V.M. Dixit. 2014. Mechanisms and functions of inflammasomes. Cell 157 (5): 1013–1022.
Jin, C., J. Henao-Mejia, and R.A. Flavell. 2013. Innate immune receptors: Key regulators of metabolic disease progression. Cell Metabolism 17 (6): 873–882.
Baffy, G. 2009. Kupffer cells in non-alcoholic fatty liver disease: The emerging view. Journal of Hepatology 51 (1): 212–223.
Stienstra, R., F. Saudale, C. Duval, et al. 2010. Kupffer cells promote hepatic steatosis via interleukin-1beta-dependent suppression of peroxisome proliferator-activated receptor alpha activity. Hepatology 51 (2): 511–522.
Bilzer, M., F. Roggel, and A.L. Gerbes. 2006. Role of Kupffer cells in host defense and liver disease. Liver International 26 (10): 1175–1186.
Broz, P., and V.M. Dixit. 2016. Inflammasomes: Mechanism of assembly, regulation and signalling. Nature Reviews. Immunology 16 (7): 407–420.
Guo, H., J.B. Callaway, and J.P. Ting. 2015. Inflammasomes: Mechanism of action, role in disease, and therapeutics. Nature Medicine 21 (7): 677–687.
Acknowledgments
The work was supported by the National Nature Science Foundation of China (No. 81400614 and No. 31370753).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The animals received humane care in compliance with the institution’s guidelines, as outlined in the Guide for the Care and Use of Laboratory Animals prepared by the National Academy of Sciences. All experimental protocols described in this study complied with the ethics review committee for animal experimentation of Chongqing Medical University.
Conflict of Interest
The authors declare that they have no competing interests.
Rights and permissions
About this article
Cite this article
Cai, C., Zhu, X., Li, P. et al. NLRP3 Deletion Inhibits the Non-alcoholic Steatohepatitis Development and Inflammation in Kupffer Cells Induced by Palmitic Acid. Inflammation 40, 1875–1883 (2017). https://doi.org/10.1007/s10753-017-0628-z
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10753-017-0628-z