Abstract
Objective Recent studies have revealed a link between toll-like receptors (TLRs), Kruppel-like factors (KLFs), and the adipose tissue inflammation associated with obesity. TLR4 is associated with chronic inflammation in obesity. KLF7 is known to play an important role in the differentiation of adipocytes, but its role in visceral adipose tissue inflammation has not yet been investigated. Thus, the objective of this study was to determine the correlation of TLR4 and KLF7 in inflammation induced by obesity. Methods A total of 32 Wistar male rat subjects were fed in the center for experimental animals of Shihezi University. The rats were divided into normal control (NC) and high-fat diet (HFD) group. Surgical instruments were used to collect rats’ visceral adipose tissue samples in the 10th week after HFD feeding. Ninety-five Uygur subjects between 20 and 90 years old were enrolled in the present study. The subjects were divided into two groups: the normal control group (NC, 18.0 kg/m2 ≤ BMI ≤ 23.9 kg/m2, n = 50) and the obesity group (OB, BMI ≥ 28 kg/m2, n = 45), and visceral adipose tissue was collected from the subjects. Anthropometric and clinical parameters were measured using standard procedures; biochemical indices were detected using the glucose oxidase-peroxidase method and a standardized automatic biochemistry analyzer; the plasma levels of inflammatory factors and adipocytokines were measured by enzyme-linked immunosorbent assay (ELISA); the mRNA and protein expression levels of key genes involved in the inflammatory signaling pathway were measured by real-time PCR and Western blot. Results In rats, compared with the NC group, the weight, Lee’s index, waist circumference, visceral fat mass, and the plasma level of Glu, TG, FFA, and TNF-α were higher in the HFD group, while the plasma levels of LPT and APN were significantly lower in the HFD group in the 10th week. Furthermore, compared with the NC group, visceral adipose tissue’s mRNA expression levels of TLR4, KLF7, and SRC were higher in the HFD group, and KLF7 was significantly positively correlated with LDL, TLR4, SRC, and IL-6 (P < 0.05). Meanwhile, in the Uygur population, the plasma levels of TG, LDL, and TNF-α in the OB group were significantly higher than those in the NC group (P < 0.05). Moreover, compared with the NC group, visceral adipose tissue’s mRNA expression levels of TLR4, KLF7, and SRC were significantly higher in the OB group (P < 0.05), and KLF7 was significantly positively correlated with TC, TLR4, MYD88, SRC, and IL-6 (P < 0.05); the protein expression levels of TLR4 and KLF7 were significantly higher than those in the NC group (P < 0.05). Conclusion Higher expression of TLR4 and KLF7 may play a vital role in the process of inflammation induced by obesity in visceral adipose tissue.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
INTRODUCTION
Obesity, particularly abdominal obesity, has become a worldwide health problem [1]. Accumulating evidence suggests that obesity-induced inflammation plays an important role in the development of a number of diseases, such as metabolic syndrome and fatty liver disease [2]. Abdominal obesity, as one of the principal locations of the systemic inflammation response, is a risk factor for this cluster of metabolic dysfunctions that is independent of the risk conferred by overall obesity [3]. The mechanism for the initiation or acceleration of the inflammatory response of visceral adipose tissue has not been completely elucidated. However, there is evidence implicating partial involvement of innate pattern recognition receptors in such a process.
Toll-like receptors (TLRs) are pattern recognition molecules, and their signaling pathways are associated with chronic inflammation in obesity [4]. It was found out that free fatty acids (FFA) can serve as an agonist of the toll-like receptor 4 (TLR4) complex. Stimulation of TLR4 activates the pro-inflammatory pathway and induces cytokine expression in various cells [5]. Kruppel-like factors (KLFs), as a transcription factor family, are composed of 17 members with a zinc finger structure and are widely involved in cell proliferation, differentiation, and embryonic developmental regulation [6]. In the mature adipocyte, KLF7 can regulate the expression and release of cytokines, and it is involved in the process of lipid metabolism [7]. In the inflammation state induced by abdominal obesity, it remains unclear whether or not KLF7 plays an important role and whether or not TLR4 correlates with KLF7.
Uygur and Kazak are two of the main minorities of Xinjiang region. Our group has previously reported that at the same body mass index (BMI) level, both male and female Uygur subjects had significantly greater WHR and visceral fat compared with Kazak subjects [8]. Thus, this study aimed to evaluate the correlation of TLR4 and KLF7 with obesity-induced inflammation of visceral adipose tissue in both an obesity rat model and the Uygur population.
METHODS
Obesity High-Fat Diet Rats
A total of 32 Wistar male rats provided by the Centers for Disease Control in Xinjiang Uygur Autonomous region were fed in the center for experimental animals of Shihezi University. After a week of adaptive feeding, rats were randomly divided into two groups: the control group (normal control, NC, n = 16) and the experimental group (high-fat diet, HFD, n = 16). The NC group was fed on a standard diet (Beijing Keaoxieli cooperation Feed Co., Ltd.), with the following composition: corn, soybean meal, fish meal, flour, bran, salt, phosphate calcium ammonia, powder, multiple vitamins, trace elements, amino acids, and others, including 13.4 % fat ratio. The nutritional and calorific percentages of the two groups are shown in Tables S1 and S2. The HFD group was fed with 40 % fat ratio high-fat diet. On a weekly basis, the water was changed, the feces were removed, and the animal room was disinfected. Rat weight was measured every 2 weeks, and the Lee’s index was calculated using the following equation: Lee’s index = [body weight × 103/body length (cm)]1/3.
Subjects
From January to December 2014, 95 Uygur subjects were enrolled with ages between 20 and 90 years old in the People’s Hospital of Xinjiang Uygur Autonomous Region for physical examination and evaluation of dyslipidemia. In the process of operating with general anesthetic, we collected visceral adipose tissue and analyzed the mRNA and protein expression levels of key genes related to inflammation. The subjects were divided into two groups: the normal control group (NC, 18.0 kg/m2 ≤ BMI ≤ 23.9 kg/m2, n = 50) and the obesity group (OB, BMI ≥ 28 kg/m2, n = 45). Exclusion criteria included the following: type 1 diabetes (T1DM) (fasting insulin lower than 5 μ IU/mL); type 2 diabetes (T2DM) (FPG ≥ 7.0 mmol/L, 2 h PG ≥ 11.1 mmol/L, and the subjects who take any medicine and do exercise/diet control to treat T2DM in the days or weeks before surgery); cancer; acute inflammation (diagnosed clinically and by measuring granulocyte levels); kidney disease (diagnosed by measuring proteinuria, hypoproteinemia, and hyperproteinemia); and use of medications that modulate glucose and lipid metabolism such as biguanides, sulphonylureas, and statins.
Anthropometric and Clinical Parameters
We measured the following clinical parameters using standard procedures: height, weight, BMI, waist circumference (WC), hip circumference (HC), waist-to-hip ratio (WHR), systolic blood pressure (SBP), and diastolic blood pressure (DBP). BMI was calculated by dividing weight (in kilograms) by height (in meters) squared. WC and HC were measured using a flexible tape with tension calipers at the extremity (Gulick-creative Health Product, Inc., Plymouth, MI), midway between the xiphoid and umbilicus during the mid-expiratory phase, and at the maximum circumference in the hip area, respectively. WHR was calculated by dividing WC by HC.
Plasma Sample Collection
After a period of fasting, animals were anesthetized, 3 mL of retinal venous blood was collected from the angular vein, and plasma was anti-coagulated by immediate placement in a tube containing sodium heparin. Human venous blood was collected after overnight fasting into EDTA-K2 collection tubes and allowed to stand for 30 min, and plasma was collected after centrifugation (4000 rpm × 10 min). Plasma was stored at −80 °C.
Measurement of Plasma Glucose, Lipids, and Cytokines
Frozen plasma samples were thawed at 37 °C. Fasting plasma glucose was detected using the glucose oxidase-peroxidase method [9]. Plasma levels of glucose (Glu), triglyceride (TG), total cholesterol (TC), low density lipoprotein cholesterol (LDL-C), high density lipoprotein cholesterol (HDL-C), and FFA were measured using an Olympus AU2700 biochemical analyzer. The levels of tumor necrosis factor-α (TNF-α), leptin (LEP), and adiponectin (APN) were measured by ELISA according to the instruction kit. All the biochemical indicators (including glucose, lipids, and cytokines) were tested in the first affiliated hospital clinical laboratory of medicine of Shihezi University.
Tissue Samples
Surgical instruments were used to collect rat visceral adipose tissue samples after high-fat diet feeding. Approximately 3 cm × 3 cm of tissue was placed in 5 mL of freeze-stored tube, and the time, rat identifier, and group information were recorded. For the Uygur population, 3 cm × 3 cm visceral adipose tissue samples were removed on the day of abdominal surgery. After the samples were removed, they were washed three times with PBS buffer solution, and they were then placed into cryopreserved tubes and tagged with identifier, gender, medical record number, and group. All samples were snap-frozen in liquid nitrogen until RNA extraction was performed. Total RNA was extracted from the tissue within a week, and then stored at −80 °C.
RNA Isolation and Real-Time PCR
Total RNA was isolated from visceral adipose tissue using TRIZOL reagent (Cat#15596-026, Life technologies, Carlsbad, CA, USA) and purified using an RNeasy mini kit (Cat#74106, QIAGEN, GmBH, Germany). RNA purity was evaluated using an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). Reverse transcription was performed as follows: 25 °C for 5 min, 42 °C for 60 min, and 70 °C for 15 min. Real-time PCR was performed using SYBR Premix Ex Taq (Takara) on a 7500 Real-Time PCR System (Applied Biosystems, Foster City, CA). Primer sequences are listed in Table S3, using glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as an internal control. One microliter (1 μL) of each RT reaction product was amplified in a 20-μL PCR reaction using an ABI Prism 7500 sequence detection system (Applied Biosystems). The PCR protocol was performed as follows: 95 °C for 30s and 40 cycles consisting of 5 s at 95 °C, 34 s at 60 °C. Dissociation curves were analyzed using the Dissociation Curve 1.0 software (Applied Biosystems) to detect and eliminate possible primer-dimer artifacts. All reactions were performed in triplicate. The relative amounts of target gene transcripts were calculated using the comparative cycle time method.
Western Blot
The total protein collected was isolated from visceral adipose tissue using PMSF and 1 % RIPA buffer. Thirty to fifty micrograms of total protein was mixed with 6× Laemmli sample buffer containing 6 % (v/v) β-mercaptoethanol. Samples were heated for 10 min at 95 °C, analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and electro-blotted to hybond nitrocellulose membrane (GE Healthcare, USA). Membranes were blocked in Tris-buffered saline containing 0.5 % Tween-20 (TBST) in 5 % skim milk for 2–3 h. Membranes were then incubated at 4 °C overnight with antibodies against TLR4 1:250 97 kDa (Abcam, USA) or KLF7 1:1000 38 kDa (Abcam, USA), then re-probed with β-actin 1:1000 42 kDa (ZSGB-BIO, CHINA). Membranes were washed using TBST and incubated with Peroxidase-Conjugated AffiniPure Goat Anti-Mouse IgG (H + L) secondary antibody for 2 h at room temperature. Proteins were visualized using the enhanced chemiluminescence (ECL) detection system (FluorChem HD2, USA).
Subject’s Consent and Ethics Statement
All subjects provided informed and voluntary consent prior to enrollment in the study. This consent included understanding that clinical information and biological samples would be used for research. The consent form and ethical approval were provided by the Medical Ethics Committee at First Affiliated Hospital, Shihezi University School of Medicine (reference number 2014LL22).
Statistical Analysis
SPSS statistical package (version 13.0, SPSS Inc, Chicago, IL, USA) was used for data analysis. Clinical characteristics and biochemical data are expressed as mean ± standard deviation (SD); mRNA expression levels of rats are expressed as median and interquartile ranges [M (QL·QU)]; the mRNA expression levels of the Uygur population were expressed as mean ± standard error of the mean (SEM). Student’s t test and Wilcoxon rank sum test were used to compare groups. Correlation analysis was performed using the Pearson method, and a P value <0.05 was defined as statistically significant.
Quality Control of Laboratory Testing
This study implemented strict quality control methods for the collection of subjects’ general information and visceral adipose tissue samples. A team of researchers designed questionnaires, and two independent operators performed molecular biology techniques. Attempts were made to choose subjects from different groups who were well matched in terms of age and gender.
RESULTS
Construction of the Obesity Animal Model
At the beginning of the study (weeks 0th, 2nd), there was no statistical difference in weight between the NC group and the HFD group (P > 0.05). In the 4th, 6th, 8th, and 10th weeks after high-fat diet feeding, the weight of the HFD group was significantly higher than that of the NC group (P < 0.05). In the 10th week, the weight of the HFD group did not appear to increase, but it was still higher than that of the NC group (P < 0.01) (Fig. 1a). Moreover, compared with the weight of the NC group (318.50 ± 38.07), the HFD group (403.00 ± 50.38) was 20 % higher. The Lee’s index (337.10 ± 4.72) of the HFD group was significantly higher than that of the NC group (323.42 ± 4.72) (P < 0.01). In addition, the waist circumference and visceral fat mass were higher in the HFD mice (Fig. 1b/c), and HFD mice had fatty liver (Fig. 1c).
Construction of obese animal model. The rats were monitored for 10 weeks while being fed normal diet and high-fat diet. a Body weight (n = 8 rat per group). b The appearance of the rat. Scale bar 1 cm. c The visceral fat mass and liver appearance. Scale bar 1 cm. t test, values are given as the mean ± SD. * P < 0.05, ** P < 0.01 compared with NC group.
Biochemical Indices and Inflammatory Factor Levels in Rats
In the 4th week after high-fat diet feeding, the levels of FFA, TG, TC, and LDL in the HFD group were significantly higher than those in the NC group (P < 0.05). In the 10th week, the plasma levels of Glu, TG, FFA, and TNF-α in the HFD group were significantly higher than those in the NC group (P < 0.05), and the levels of LPT and APN were significantly lower than those in the NC group (P < 0.05) (Table 1).
The mRNA Expression Level of TLR4, KLF7, and Inflammation-Related Factors of Visceral Adipose Tissue in Rats
The mRNA expression level of MYD88 in the HFD group was lower than that in the NC group, whereas the mRNA expression levels of TLR4, KLF7, SRC, TNF-α, and IL-6 were higher than those in the NC group (Fig. 2).
Correlation of TLR4, KLF7, and Other Factors
In the HFD group, the mRNA expression level of KLF7 was significantly positively correlated with LDL, TLR4, SRC, and IL-6 (P < 0.05). In addition, KLF7 was slightly positively correlated with MYD88, and slightly negatively correlated with APN (Fig. 3).
Clinical Characteristics of Uygur Subjects in the NC and OB Groups
The SBP and DBP in the OB group were higher than in the NC group, but were not statistically significant. The weight, WC, HC, WHR, and BMI of individuals in the OB group were significantly higher than those in the NC group (P < 0.05) (Table 2).
The Biochemical Indices and Inflammatory Factor Levels in the Uygur Population
The plasma levels of TC and HDL in the OB group were higher than in the NC group, but were not statistically significant. The levels of TG, LDL, and TNF-α in the OB group were significantly higher than those in the NC group (P < 0.05), and the APN in the OB group was slightly lower than that in the NC group (Table 3).
The mRNA and Protein Expression Levels of LTR4, KLF7, and Inflammation-Related Factors in the Visceral Adipose Tissue in the Uygur Population
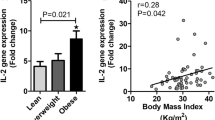
Compared with the NC group, the mRNA expression levels of TLR4, KLF7, MYD88, SRC, TNF-α, and IL-6 were significantly higher in the OB group (P < 0.05) (Fig. 4a). Moreover, the protein levels of TLR4 and KLF7 were significantly higher in the OB group (P < 0.05) (Fig. 4b).
The mRNA and protein expression level of critical gene in inflammation signaling pathways of visceral adipose tissue in Uygur population. a The mRNA expression level of TLR4, MYD88, SRC, KLF7, APN, MCP-1, NF-kB, TNF-α, and IL-6. Values are normalized to GAPDH mRNA expression. b The protein expression level of LTR4 and KLF7. Values are normalized to β-actin protein expression. t test,*P < 0.05, **P < 0.01. The difference between the two groups has statistical significance.
Correlation of TLR4, KLF7, and Inflammation Indices of the OB Group in the Uygur Population.
In the OB group, the mRNA expression level of KLF7 was significantly positively correlated with TC, TLR4, MYD88, SRC, and IL-6 (P < 0.05) (Fig. 5).
DISCUSSION
Obesity, particularly abdominal obesity, has become a worldwide health problem, which is closely related to chronic inflammation state [10], and increased lipolyis and blood FFA levels lead to a chronic inflammatory state [11]. In this study, the results showed that in obesity animal models and human tissue, along with weight and plasma lipid level increases, the levels of inflammatory factor TNF-α and IL-6 increased, while the level of anti-inflammatory factor APN decreased. The above results showed that the obesity animal models and the obesity Uygur individuals in this study were all in the state of inflammation.
The study selected male Wistar rats to eliminate the effects of the estrogen level. Previous studies have shown that weight difference (the weight of HFD group being more than 20 % greater than that of the NC group) and blood lipid level difference can be used to determine the successful execution of the obesity animal model design [12]. Results showed that in the 10th week, the weight of the HFD group was over 20 % greater than that of the NC group; the Lee’s index of the HFD group was significantly higher than that of the NC group; the waist circumference and visceral fat mass were higher in the HFD mice; HFD mice had the formation of fatty liver; the levels of Glu, TG, FFA, and TNF-α in the HFD group were significantly higher than those in the NC group; and the levels of LPT and APN were significantly lower compared to the NC group. The above results suggested that the obesity animal model was constructed successfully. In the 10th week, the level of TC and LDL returned to normal, which may correlate with physical signs in HFD group (limb ulcer, eating less that may cause the mobilization of lipids in vivo) and the sample size. More importantly, the weight of the HFD group did not appear to increase, but it was still higher than the NC group. This is the reason why rats in the 10th week were chosen in evaluating the mRNA expression levels of TLR4, KLF7, and other inflammation-related factors in the visceral adipose tissue.
TLRs are important molecules involved in non-specific immunity, and they play an important role in inflammatory signaling pathways of congenital immune responses [13]. Studies on different strains of mice showed that expression and activation of TLR4 were involved in aortic inflammation and atherogenesis [14]. In particular, TLR4 activation caused the upregulation of IL-6 and the release of MCP-1 via activation of both the ERK and NF-kB pathways [15]. Recent discoveries underscore that LPS engagement of TLR4 caused Src activation, which stabilized the LPS-TLR4-MyD88 complex and activated the downstream signaling pathway, resulting in pro-inflammatory cytokine release [16]. Therefore, this study was able to determine the expression levels of TLR4, MYD88, SRC, and key inflammatory factors. Results showed that in the HFD group of rats, the mRNA expression levels of TLR4, SRC, TNF-α, and IL-6 were higher than in the NC group. Conversely, the mRNA expression level of MYD88 was lower than that in the NC group, which may be related to the sample size; similarly, in the OB group of the Uygur population, the mRNA expression levels of TLR4, MYD88, SRC, TNF-α, and IL-6 and the protein expression level of TLR4 were higher than those in the NC group. The above results show that in obese rats and the Uygur population, TLR4, SRC, and MYD88 might play an important role in the process of inflammation caused by obesity. However, the specific molecular mechanism underlying how these factors regulate the expression of inflammatory factors remains unclear.
KLF7 is expressed in adipose tissue, pancreas, liver, and bone tissues, and it was initially found to be involved in cell differentiation [17, 18]. However, there are only a few literature talking about its correlation with inflammation. Recent studies have shown that KLF7 can inhibit the synthesis of APN and LPT and promote the release of inflammatory factors, such as IL-6, in fat cells [18]. It was also recently reported that KLF7 functioned as a key mediator in low-level TGF-β signaling. Notably, the expression of KLF7 was regulated by TGF-β, and the knockdown of KLF7 diminished low-level TGF-β [19]. At the same time, TGF-β could significantly increase the pro-inflammatory factors’ secretion, such as MIF and MCP-1 [20]. The results demonstrated that in the HFD group of rats, the mRNA expression level of KLF7 was slightly higher than that in the NC group, and KLF7 was significantly positively correlated with LDL, TLR4, SRC, and IL-6. Furthermore, in the OB group of the Uygur population, the mRNA and protein expression levels of KLF7 were significantly higher than those in the NC group, and KLF7 was significantly positively correlated with TC, TLR4, MYD88, SRC, and IL-6. The above results showed that the difference in KLF7 expression and the correlation of TLR4, KLF7, and other inflammatory indices may play an important role in the process of obesity-induced inflammation.
In conclusion, it was found out that in the HFD group of rats, the mRNA expression levels of TLR4 and KLF7 were higher than those in the NC group, and in the OB group of the Uygur population, the mRNA and protein levels of TLR4 and KLF7 were significantly higher than those in the NC group. It was also found out that TLR4 is significantly positively correlated with KLF7 in rats and in the Uygur population. Thus, TLR4 and KLF7 are likely to play a key role in regulating adipose tissue inflammation, although additional investigations are required to elucidate the underlying mechanisms. Probing the cell type-specific mechanism of the correlation of TLR4 and KLF7 may provide further useful therapeutic approaches for the treatment of inflammation-related diseases.
References
Kotas, M.E., and R. Medzhitov. 2015. Homeostasis, inflammation, and disease susceptibility. Cell 160(5): 816–827.
Ding, J., L.M. Reynolds, T. Zeller, et al. 2015. Alterations of a cellular cholesterol metabolism network are a molecular feature of obesity-related Type 2 diabetes and cardiovascular disease. Diabetes 64(10): 3464–3474.
Chalmem, J., and M.E. Cooper. 2008. UKPDS and the legacy effect. New England Journal of Medicine 359: 1618–1620.
Park, Mi-Young, and Seong Taek Mun. 2014. Carnosic acid inhibits TLR4-MyD88 signaling pathway in LPS-stimulated 3T3-L1 adipocytes. Nutrition Research and Practice 8(5): 516–520.
Cullberg, K.B., J.O. Larsen, S.B. Pedersen, et al. 2014. Effects of LPS and dietary free fatty acids on MCP-1 in 3T3-L1 adipocytes and macrophages in vitro. Nutrition and Diabetes 4(3): e113.
Bieker, J.J. 2001. Krüppel-like factors: three fingers in many pies. Journal of Biological Chemistry 276(37): 34355–34358.
McConnell, Beth B., and Vincent W. Yang. 2010. Mammalian Krüppel-Like Factors in Health and Diseases. Physiological Reviews 90(4): 1337–1381.
Zhang, J., Z. Zhang, Y. Ding, et al. 2015. Adipose tissues characteristics of normal, obesity, and Type 2 diabetes in uygurs population. Journal of Diabetes Research 2015: 905042.
Gupta, S., A.K. Gupta, M. Verma, et al. 2016. A study to compare the plasma glucose level obtained in sodium fluoride and citrate buffer tubes at a tertiary care hospital in Punjab. International Journal of Applied and Basic Medical Research 6(1): 50–53.
Jagannadham, J., H.K. Jaiswal, S. Agrawal, et al. 2016. Comprehensive map of molecules implicated in obesity. PLoS One 11(2), e0146759.
Chathyan, P., J.B. Clifford, and R.G. Helen. 2015. Aging, adipose tissue, fatty acids and inflammation. Biogerontology 16(2): 235–248.
Gaur, A., G.K. Pal, P.H. Ananthanarayanan, et al. 2014. Role of Ventromedial hypothalamus in high fat diet induced obesity in male rats: association with lipid profile, thyroid profile and insulin resistance. Annals of Neurosciences 21(3): 104–107.
Li, Q., and I.M. Verma. 2002. NF-kB regulation in the immune system. Nature Reviews Immunology 2(10): 725–734.
Poulain, L., V. Richard, P. Lévy, et al. 2015. Toll-like receptor-4 mediated inflammation is involved in the cardiometabolic alterations induced by intermittent hypoxia. Mediators of Inflammation 2015: 620258.
Zhang, J., W. Dou, E. Zhang, et al. 2014. Paeoniflorin abrogates DSS-induced colitis via a TLR4-dependent pathway. American Journal of Physiology. Gastrointestinal and Liver Physiology 306(1): G27–36.
Jiao, Hao, Zhang Yang, Zhibo Yan, et al. 2013. Caveolin-1 Tyr14 phosphorylation induces interaction with TLR4 in endothelial cells and mediates MyD88-dependent signaling and sepsis-induced lung inflammation. Journal of Immunology 191(12): 619–9. doi:10.4049/jimmunol.1300873.
Chen, L., Y.M. Dai, C.B. Ji, et al. 2014. MiR-146b is a regulator of human visceral preadipocyte proliferation and differentiation and its expression is altered in human obesity. Molecular and Cellular Endocrinology 393(1–2): 65–74.
Kawamura, Y., Y. Tanaka, R. Kawamori, et al. 2006. Overexpression of Kruppel-like factor 7 regulates adipocytokine gene expressions in human adipocytes and inhibits glucose-induced insulin secretion in pancreatic beta-cell line. Molecular Endocrinology 20(4): 844–856.
Wang, X., Q.W. Shen, J. Wang, et al. 2016. KLF7 regulates satellite cell quiescence in response to extracellular signaling. Stem Cells. doi:10.1002/stem.2346.
Mreich, E., X.M. Chen, A. Zaky, et al. 2015. The role of Krüppel-like factor 4 in transforming growth factor-β-induced inflammatory and fibrotic responses inhuman proximal tubule cells. Clinical and Experimental Pharmacology and Physiology 42(6): 680–686.
Acknowledgments
This work is supported by the Natural Science Foundation of China (no. 81360142), the Program for High-Level Personnel Start at Shihezi University (nos. RCZX201230, gxjs2012-zdgg02), and the Applied basic research project of Xinjiang corps (2015AG016).
Authors’ Contribution
Cuizhe Wang and Xiaodan Ha contributed equally to the paper.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of Interests
The authors declare that there is no conflict of interests regarding the publication of this paper.
Additional information
Cuizhe Wang and Xiaodan Ha contributed equally to this work.
Rights and permissions
About this article
Cite this article
Wang, C., Ha, X., Li, W. et al. Correlation of TLR4 and KLF7 in Inflammation Induced by Obesity. Inflammation 40, 42–51 (2017). https://doi.org/10.1007/s10753-016-0450-z
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10753-016-0450-z