Abstract
Environmental DNA (eDNA) is useful to detect the presence of aquatic organisms from water samples, especially for rare and cryptic species. Two freshwater unionid mussels¸ Pronodularia japanensis and Nodularia douglasiae, have rapidly declined over the last three decades and are now threatened with extinction on the Matsuyama Plain, south-western Japan. We designed a species-specific eDNA marker targeting the COI region of P. japanensis. The distribution of this species in the Kunichi River system on the Matsuyama Plain was investigated using both quantitative PCR with this eDNA marker and conventional surveying. We show that the distribution area of P. japanensis did not change between 2013–2014 and 2020–2021, but its density decreased by 99%. eDNA of P. japanensis was detected, with 100% success, from sites where this species was collected by hand. Furthermore, eDNA was detected at nine sites where P. japanensis was not collected but was expected to occur. This study has established a species-specific eDNA marker targeting the COI region of P. japanensis, and this eDNA marker has been validated as effective for surveying the distribution of this species. Using this eDNA marker, extensive investigation of remaining populations and the monitoring of them should improve conservation practices.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Freshwater bivalves are one of the most species-diverse and widely distributed groups of organisms in freshwater habitats (Lopes-Lima et al., 2021). The freshwater bivalves of the family Unionidae are described in 157 genera and 753 species, and are widespread in rivers, lakes, and marshes throughout the world (Graf & Cummings, 2021). Freshwater bivalves play key roles in freshwater ecosystems by contributing to water quality, primary production, and nutrient cycling (Spooner & Vaughn, 2006; Hoellein et al., 2017), providing hard shells as habitats for other organisms (Vaughn & Hakenkamp, 2001) and in supplying a breeding substratum for bitterling fishes (Acheilognathinae, Cyprinidae; Wiepkema, 1961; Smith et al., 2004). On the other hand, aquatic ecosystems are threatened by human impacts, such as habitat degradation, non-native species introduction, overexploitation, flow modification, and water pollution (Brookes, 1988; Allan & Flecker, 1993; Dudgeon, 2006). As a consequence of such threats, one third of these unionid species are in danger of extinction (Böhm et al., 2021). In Japan, there are 26 species of Unionidae in 13 genera, including 20 endemic species, as well as two alien species (Sinanodonta cf. woodiana and Sinohyriopsis cf. cumingii; Lopes-Lima et al., 2020). Of these species, 13 are listed on the Japanese Red List of Threatened Species (Ministry of the Environment, Japan, 2020).
Japan is a mountainous, wet, and forested country with a rich freshwater fauna and a high proportion of endemic species (Yoshimura et al., 2005). People are concentrated in densely populated urban areas along the coast and on alluvial plains, and recent urbanisation and intensive agriculture have destroyed and degraded natural freshwater habitats and traditional agricultural habitats, and now most unionid species are endangered (Onikura et al., 2006, 2016; Kondo, 2008; Negishi et al., 2008). On Matsuyama Plain, Ehime Prefecture, western Japan, there are three species of the family Unionidae—Pronodularia japanensis (Lea), Nodularia douglasiae (Griffith & Pidgeon), and Sinanodonta lauta (Martens). Populations of the fluvial unionids P. japanensis and N. douglasiae have decreased rapidly between 1988–1991 and 2013–2014 in this area through habitat fragmentation by weirs (Figs. 1, 2; Kuwahara et al., 2017). Glochidium larvae of these unionids mostly parasitise an amphidromous goby, Rhinogobius nagoyae Jordan and Seale, but weirs prevent these glochidium larvae from moving upstream and restrict unionid dispersion to downstream of the weirs. S. lauta has also decreased in streams, but this species prefers lentic water to streams, and populations remain in several ponds in Ehime (Ishikawa & Chiba, 1999). On Matsuyama Plain, the rapid decline of unionids is the cause behind a decrease of the native bitterling fish Tanakia lanceolata (Temminck and Schlegel). Bitterling fishes spawn on the gills of live freshwater mussels of the families Unionidae and Margaritiferidae and obligately depend on the mussels for reproduction (Wiepkema, 1961; Smith et al., 2004). On Matsuyama Plain, T. lanceolata spawn mainly on P. japanensis, and no species of the family Margaritiferidae lives here (Kuwahara et al., 2017; Hata et al., 2021b). The decrease in P. japanensis and the other two unionid species (N. dougrasiae and S. lauta) causes the rapid decrease in the T. lanceolata population through a loss of breeding substrate. Furthermore, a congeneric bittering, T. limbata (Temminck and Schlegel), was artificially introduced from the island of Kyushu in the 1970s, and the decline of unionid populations enhances hybridisation and genetic introgression between T. lanceolata and T. limbata through an increase in competition for breeding substrate between these two species (Matsuba et al., 2014; Uemura et al., 2018; Hata et al., 2019, 2021b).
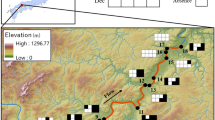
Map of study sites in the Kunichi River system (a) on the Matsuyama Plain, Ehime. Thick vertical lines on the streams indicate weirs over 1 m high. Bars with diagonal lines show floodgates. (b) inset showing location of the study site in Japan. These maps are based on those provided under CC BY 4.0 by the Geospatial Information Authority of Japan (https://maps.gsi.go.jp)
Distribution of unionid mussels found in previous studies [left column, 1988–1991 (Matsukawa et al. 1993); middle column, 2013–2014 (Kuwahara et al. 2017)], and the present study [right column, 2020–2021]. Top row, P. japanensis; middle row, N. douglasiae; bottom row, S. lauta. Black circles indicate the presence of mussels with density represented by circle size; white circles indicate their absence. Thick vertical lines on streams indicate weirs over 1 m high
The environmental DNA (eDNA) approach is an effective tool to detect the presence of target species in aquatic environments (Minamoto et al., 2012; Taberlet et al., 2012; Thomsen et al., 2012). Knowing species distributions is a primary step for the conservation and management of endangered species, and eDNA assays are especially useful for rare and cryptic species (Belle et al., 2019; Beng & Corlett, 2020). The distribution of freshwater bivalves has been successfully analysed by species-specific eDNA assays in many parts of the world, for example, for 13 species of Unionidae: Lampsilis fasciola Rafinesque, Ligumia nasuta (Say), Ptychobranchus fasciolaris (Rafinesque), Quadrula quadrula (Rafinesque)(Currier et al., 2018), Lampsilis siliquoidea (Barnes)(Sansom & Sassoubre, 2017), Lasmigona decorata (Lea)(Schmidt et al., 2021), Lampsilis fasciola Rafinesque (Gasparini et al., 2020), Gonidea angulata Lea, Anodonta nuttalliana Lea, and Anodonta oregonensis Lea (Rodgers et al., 2020; Preece et al., 2021), Alasmidonta varicosa (Lamarck)(LeBlanc et al., 2021), Alasmidonta heterodon (Lea)(Schill & Galbraith, 2019), Unio tumidus Philipsson (Deiner & Altermatt, 2014), U. crassus Philipsson (Stoeckle et al., 2021); three species of Margaritiferidae: Margaritifera margaritifera (Linnaeus)(Stoeckle et al., 2016; Carlsson et al., 2017; Wacker et al., 2019), M. falcata (Gould) (Dysthe et al., 2018; Preece et al., 2021), and M. monodonta (Say) (Lor et al., 2020); and one species of Dreissenidae: Dreisseina polymorpha (Pallas) (Shogren et al., 2019). Regarding eDNA marker targeting for unionid species in Japan, only one eDNA marker for the 12S region of Sinanodonta spp. has been developed, and this has been used to reveal the distribution of these species in farm ponds (Togaki et al., 2019). No eDNA marker has been available for P. japanensis.
This study aims to establish an eDNA marker specific for the Japanese endemic unionid P. japanensis and to monitor the population of P. japanensis on Matsuyama Plain via both conventional survey and eDNA assays; the outcomes of these two approaches are compared to ascertain the status of the P. japanensis population and to establish an effective method for further monitoring of endangered freshwater bivalves.
Materials and methods
Study sites and field survey
The study was conducted in the Kunichi and Koyori streams (both in the Kunichi River system; main stem length 5.7 km) on the Matsuyama Plain, which is on the island of Shikoku in south-western Japan (coordinate range 33° 47′ N–33° 48′ N, 132° 41′ E–132° 45′ E; Figs. 1, 2). The region is part of an alluvial fan formed by the Shigenobu River (watershed area 445 km2; main stem length 36 km). The Kunichi River system is an irrigation canal with some weirs for water intake into nearby paddy fields. The riverbanks are lined with concrete, and a floodgate (width 30 m) is installed at the mouth of the river. We set 23 study sites approximately 300 m apart from each other in the Kunichi River system, following Kuwahara et al. (2017). Study sites were set at varying stream widths between 2.0 and 20.0 m and between 25 and 100 m in reach length, and survey areas were between 100 and 1400 m2. The substrate at each site was excavated to a depth of 10 cm using a hand dredge with a 10 mm mesh sieve, and then Pronodularia japanensis, Nodularia douglasiae, and Sinanodonta lauta were gathered by hand from both the sieve and from the newly exposed substrate that had just been revealed; this was done from September to December 2020, and in September 2021. Note that sites 1 and 2 were too deep to access for this conventional survey (Fig. 3). The number of individuals for each species was counted for each site and shell length of all individuals recorded. Collected individuals were released back to the site where collected. The density of each species at each study site was calculated by dividing the number of collected individuals by the study site area (m2). Empty shells were also collected and counted if they were complete and unbroken. Mussels and empty mussel shells were identified at species level following Kondo (2008).
a eDNA concentration (number of copies per litre of filtered water) and b the density of P. japanensis (number of individuals/m2) at each site. White circles indicate their absence. The value for eDNA concentration is the mean of four samples in a river cross section at each site. Site number is shown for each site. Thick vertical lines on streams indicate weirs over 1 m high
Design of species-specific primers and a probe for Pronodularia japanensis eDNA assays
From June to November 2013, we collected 29 individuals of P. japanensis from the Kunichi River system on Matsuyama Plain. A small foot-tissue sample was collected from each individual using the nonlethal method of Naimo et al. (1998). Samples were preserved in 99% ethanol, and genomic DNA was extracted from the samples using a Wizard Genomic DNA Purification Kit (Promega, Madison) following the manufacturer's instructions. Doubly uniparental inheritance of mitochondrial DNA has been known in unionids, and males inherit both the female-lineage mitogenome from their mother and the male-lineage mitogenome from their father (Breton et al., 2007). Recent molecular phylogenetic studies of Japanese unionids focus only on the female-lineage mitogenome (Sano et al., 2020; Lopes-Lima et al., 2021). Therefore, we designed an eDNA marker for the female-lineage mitogenome of P. japanensis (Table S1). This is appropriate because the male-lineage mitogenome generally occurs only in the gametic tissue of males (Venetis et al., 2006), and both female and male individuals have a female-lineage mitogenome. The partial COII and COI gene regions of the female-lineage mitogenome were amplified using the primers CO2.2 (Curole & Kocher, 2002) and HCO2198 (Folmer et al., 1994) as forward and reverse primers, respectively, via polymerase chain reaction (PCR). Each PCR was performed in a 10 μl volume containing 3.35 μl of ultra-pure water, 5.0 μl Ampdirect Plus (Shimadzu, Kyoto), 0.3 μl of each primer (10 μM solutions), 0.05 μl Taq DNA polymerase (BIOTAQ HS DNA Polymerase, Bioline, London), and 1 μl template. The thermal cycle profile was as follows: initial denaturation at 95 °C for 10 min; 25 cycles of 95 °C for 45 s, 59 °C for 1 min and 72 °C for 90 s; and a final extension at 72 °C for 10 min. PCR products were analysed by electrophoresis using a 1% agarose gel, and the focal band was cut and purified using a Fast Gene Gel/PCR Extraction Kit (Nippon Genetics, Tokyo) according to the manufacturer's instructions. Purified products were subjected to direct cycle sequencing employing BigDye Terminator version 3.1 (Applied Biosystems, Foster City) using the above‐mentioned PCR primers. These sequences are stored at the DNA Databank of Japan (DDBJ), accession nos. LC651381–LC651409. Note that P. japanensis consists of three clades (P. cf. japanensis 1, P. cf. japanensis 2, P. cf. japanensis 3 sensu Lopes-Lima et al. 2020). Our focal species/clade is P. cf. japanensis 1 sensu Lopes-Lima et al. (2020), which is distributed solely in western Japan, including our site and the Sea of Japan coast of eastern Japan. The next step was to download available COI sequences of female lineages of the family Unionidae. We aligned these sequences and designed a primer set using Geneious 7.7.9 (https://www.geneious.com)—forward, P_japa_COI_221F 5'-CCTGATATGGCTTTCCCTCGT-3'; reverse, P_japa_COI_339R 5'-TAAACAGTTCACCCCGTCCC-3')—and a probe—P_japa_COI_301P 5'-GTTGGTGGAGAGCGGTGTG-3')—to amplify regions of 119 bp, which matched with all the sequences of P. japanensis, including P. cf. japanensis 1, P. cf. japanensis 2, P. cf. japanensis 3, and had at least seven non-matching nucleotides compared with all the other species of Unionidae (Table S1). The forward and reverse primers and the probe are located at positions 221–241, 320–339, and 301–319, respectively, of our aligned sequences (Supplementary file 1). Note that P. japanensis (P. cf. japanensis 1) collected on Kyushu Is. has one nucleotide mismatch in the reverse primer region, and care must be taken in using this primer in this area. We then tested this primer set using Primer-BLAST and found that the primer set did not match any other unionid species, even with two mismatches allowed (Ye et al., 2012). In total, 437 sequences were obtained by Primer-BLAST, but no sequence matched our probe sequence (see positions 79–99 in Supplementary file 2). We tested qPCR with this primer set and probe on DNA templates extracted from two individuals of P. japanensis (P. cf. japanensis 1) from Koyori stream collected in 2014, and five individuals of P. cf. japanensis 2 collected from Mie in 2021; amplifications were detected for all samples (cycle threshold Cq < 19 for all samples), and the amplification was confirmed to be the focal region of COI by subsequent Sanger sequencing. On the other hand, no amplification was detected for five individuals of P. cf. japanensis 3 collected from Iwate in 2021. We amplified a partial COI region of these individuals by PCR using a primer set—LCO1490 and HCO2198 (Folmer et al., 1994)—and the PCR products were sequenced in the same manner as described above; one nucleotide mismatch was found in each of our forward primer, reverse primer, and probe regions (Table S1). Finally, we tested the qPCR on two sympatric species of Unionidae—eight individuals of Nodularia douglasiae and one individual of Sinanodonta lauta—as for P. japanensis and found no amplification. Therefore, this primer set and probe are confirmed as species-specific for P. japanensis, except for P. cf. japanensis 1 on Kyushu Is. and P. cf. japanensis 3 in Iwate Prefecture.
Quantification standard for qPCR
We designed a 219 bp artificial double-stranded DNA fragment containing the focal region of the primer set—P_japa_COI_221F and P_japa_COI_339R—based on a COI sequence of P. japanensis from Ehime (LC651386, Supplementary file 3). This DNA fragment was supplied by INTEGRATED DNA TECHNOLOGIES (Coralville, Iowa). The concentration of the solution was diluted in tenfold steps (1, 10, 100, 1000 copies/μl) for quantification standards. Note that the eDNA concentrations of our samples were less than 400 copies/reaction (i.e. 50 copies/μl) and were confirmed to be inside our standard curve.
Water sampling and on-site filtration
Throughout eDNA sampling and analyses, we avoided contamination and degradation of DNA and other materials by strictly following the eDNA Society (2019) protocols. All equipment (bottles for water sampling, forceps, filter holders) were soaked in a 1% sodium hypochlorite solution for ten minutes, thoroughly rinsed with tap water, and then rinsed with ultra-pure water before use. Water sampling was conducted in November 2020. The breeding season of Pronodularia japanensis in the study area is May to August (Kuwahara et al., 2017), and therefore November is outside the breeding season. At each site, we divided the stream cross section into four parts, and in each of these parts we collected 1 L of water from the surface in a clean bottle. Then we filtered the water immediately using a glass fibre filter (GF/F, 0.7 μm pore size, Whatman, Maidstone). We filtered each 1 L sample of water; this was done 500 ml at a time with a filter using a pump powered by a battery. After filtration, the filters were stored in an in-vehicle freezer at −20 °C. At each site, the stream width was measured and was equally divided into 10 parts; for each of these 10 parts, water depth (cm), and flow velocity (m/s) at 60% depth were measured. Flow velocity was measured five times and the mean value was used. We filtered 1 L of pure water at both the beginning and end of the survey as field blanks at field sites to evaluate contamination over the course of the field survey.
DNA extraction
Filter samples were moved to a laboratory and stored in a freezer at −20 °C until just before DNA extraction. DNA was extracted from each filter using a DNeasy Blood & Tissue Kit (QIAGEN, Hilden) following the eDNA Society (2019) protocols. For each 1 L water sample, the two lysis solutions from the two filters were put into the same spin column and mixed. Purified nucleic acids were eluted in 200 μl of elusion buffer (Buffer AE).
Real-time PCR experiments
To avoid PCR inhibition, we used TaqMan Environmental Master Mix 2.0 (Thermo Fisher Scientific, Waltham, Massachusetts), which is known to be effective for Japanese freshwater (Uchii et al., 2019; Minegishi et al., 2019). The reaction mixture contained 10.0 μl of TaqMan Environmental Master Mix, 0.1 μl of AmpErase Uracil N-Glycosylase (Thermo Fisher Scientific), 0.1 μl of both 100 μM forward and reverse primers, 0.4 μl of 10 μM TaqMan probe (Thermo Fisher Scientific), 5.0 μl of DNA template, and 4.3 μl of ultra-pure water. All PCR reactions were performed in triplicate. The four quantification standard steps were also carried out in triplicate on each 96-well reaction plate (i.e. 5, 50, 500, 5000 copies/reaction). Additionally, we put triplicates of ultra-pure water plus qPCR mixture on each plate as a negative control. The reaction protocol consisted of an initial step at 50 °C for 2 min and then at 95 °C for 10 min; this was followed by 55 cycles of 95 °C for 15 s and 60 °C for 1 min. Reactions and analyses were conducted using the StepOnePlus Real-Time PCR System (Thermo Fisher Scientific). We quantified the amount of eDNA using the quantification standard described above. The quantification mean of triplet samples was used for subsequent analyses. Each eDNA concentration was calculated as the number of copies per 1 L of environmental water multiplied by 40 (extracted final volume, 200 μl)/(qPCR template volume, 5 μl). Sanger sequencing of 18 positive results (at least one sample at each site) confirmed the specificity of the qPCR assays (Table S2). No single field blank (triplicates of four samples) and none of the nine qPCR negative controls were amplified by the qPCR marker.
qPCR limits of detection
The limit of detection (LOD) and limit of quantification (LOQ) were calculated following Merkes et al. (2019). The target DNA concentration and cycle threshold (Cq) values for all replicates of all serial dilutions were correlated (r2 = 0.97, efficiency = 95.14%, slope = −3.44, intercept = 42.04; Fig S1). The lower limit of DNA detection was 11.3 copies/reaction, and 3.8 copies/reaction when using triplets (Fig. S1). The LOQ was calculated as 30 copies/reaction. 84 samples were analysed as triplicates (yielding a total of 252 data points), and 86 of 252 were undetected without any amplification. Three of the 166 amplified samples had low values—0.01, 0.03, 3.56 copies/reaction, 47.9, 46.2, 41.1 Cq, respectively—and therefore, we treated these three samples as undetermined. A further, additional 66 data points had quantity values less than the LOQ (30 copies/reaction); therefore, to analyse eDNA concentration, we deleted these 66 and used the quantity data for the remaining 97.
Statistical analysis
The density of Pronodularia japanensis in this study was analysed by the density observed in the period 2013–2014 at each site using a generalised linear model (GLM) with Gaussian family. The response variable was the density in the period 2020–2021 at each site, and an explanatory variable was the density in the period 2013–2014 at the same site. We compared shell length of P. japanensis between individuals collected in this study and those individuals collected in the period 2013–2014 (Kuwahara et al., 2017), and between live individuals and empty shells collected in this study. We checked that these shell length distributions followed a normal distribution, and comparisons were made using a t test. To test the hypothesis that the concentration of eDNA is higher when the density of P. japanensis is higher, we constructed a GLM with Gaussian family, eDNA concentration as the response variable, density of P. japanensis, flow velocity, and their interaction factor as explanatory variables. To test whether eDNA concentrations were biased either towards the stream banks or the centre of the stream, we constructed a generalised linear mixed model (GLMM) with Gaussian family, eDNA concentration as the response variable, part of the stream cross section (the stream bank side or centre of the stream) as a fixed factor, site as a random factor. All statistical analyses were performed using R 4.0.3 (R Core Team, 2020).
Results
Conventional survey
In total, 53 individuals of Pronodularia japanensis were collected and the density was low, ranging from 0 to 0.019 individuals/m2, with an average of 0.009 individuals/m2 among sites where P. japanensis occurred (Fig. 2). The density of P. japanensis is shown to have decreased by around 99% at all sites between 2013–2014 and 2020 (Fig. 4). The density in the 2020–2021 period was not related to the density at each site in the 2013–2014 period (GLM, p = 0.73, Table S3). No individuals of Nodularia douglasiae or Sinanodonta lauta were collected; only seven incomplete empty shells of N. douglasiae were collected. The shell length of live P. japanensis was recorded as 67.8 ± 5.4 mm (average ± SD; Fig. 5) and was no different in this respect from P. japanensis in the 2013–2014 period (t test, t = 1.88, p = 0.06). Regarding empty, complete shells of P. japanensis, 249 were collected, and the shell length of these was 66.0 ± 6.2 mm, which was smaller than live individuals (t test, t = −2.76, p = 0.006).
Densities of P. japanensis collected at each study site in the Kunichi River system in the periods 2013–2014 (Kuwahara et al., 2017) and 2020–2021 (this study). Note that the scale of the x-axis is 100 times the scale of the y-axis
Histograms of shell length of (a) all the live P. japanensis collected in the period 2020–2021, (b) all the empty shells of P. japanensis collected in the period 2020–2021, and (c) all the live P. japanensis collected in the period 2013–2014 (Kuwahara et al. 2017)
Environmental DNA detection
eDNA of the unionid Pronodularia japanensis was successfully detected from water collected from streams that this species inhabits (Fig. 3). At all 10 sites where P. japanensis was collected, eDNA of P. japanensis was also detected (Table 1). On the other hand, at nine of eleven sites where P. japanensis was not collected, eDNA was still detected. Three sites (sites 1, 2, 17) were downstream from sites where P. japanensis was collected, but six sites (sites 9, 11, 12, 19, 20, 21) were upstream. eDNA concentration did not relate to the density of P. japanensis at sites in our survey (Fig. 6). The interactive effect between the observed density and flow velocity did not relate to eDNA concentration either (Fig. S2; GLM, both P > 0.05, Table S4). eDNA concentration varied among samples collected from different parts of the stream cross section at some sites, but the concentrations were not biased either towards the stream banks or the centre of the stream (Fig. S3; Tables S2, S5; GLMM, p = 0.29).
Relationship between eDNA concentration (number of copies/L filtered water) and the density of P. japanensis (number of individuals/m2) for four stream cross section samples per site for a total of 19 sites. Numbers of stream cross section parts are shown in Table S2. Error bars indicate standard deviations of triplicates of PCR reactions
Discussion
Specific eDNA marker for Pronodularia japanensis and its efficiency
This is the first study to report new species-specific primers and a new probe for eDNA assays of the endangered freshwater bivalve P. japanensis. eDNA of P. japanensis was detected, with 100% success, from sites where this species was collected by hand, even when the density was as low as 0.003 individuals/m2. Furthermore, eDNA was detected at nine sites where P. japanensis was not collected in this study. Currier et al. (2018) also reported eDNA detection of two unionids—Lampsilis fasciola and Quadrula quadrula—at multiple sites in rivers in Ontario, Canada where no mussels were collected and the minimum density observed was 0.03 individuals/m2. At our study sites, the unionid density has rapidly declined and is now quite low: 0.003–0.019 individuals/m2 (minimum–maximum). Three of nine sites where only eDNA was detected were less than 1 km downstream from sites where P. japanensis was collected, and the detected DNA fragments were possibly supplied by the local population and/or transported from an upstream population via flowing water. In fact, eDNA of Unio crassus has been reportedly transported 3 km in streams in Germany (Stoeckle et al., 2021), that of U. tumidus transported 9 km in a river in Switzerland (Deiner & Altermatt, 2014), and that of Lampsilis siliquoidea detected up to 36.7 km downstream in a creek in western New York (Sansom & Sassoubre, 2017). On the other hand, eDNA was detected but no individual collected at six sites upstream of all the sites where P. japanensis was collected in this study. This suggests that the presence of P. japanensis at these sites is highly plausible and further intensive conventional surveying in these focal areas should reveal the precise distribution of this species. In the breeding season, the eDNA concentration of freshwater mussels increases in rivers (Wacker et al., 2019), and upstream migration via host fish seems to be possible, but this would not have been a factor in this study because we collected water outside of the breeding season (Kuwahara et al., 2017). In this way, our results show that the eDNA approach is an efficient method to detect the distribution of this species in streams. The focal streams are small: main stem length 5.7 km, 30 m wide at river mouth. Still, conventional sampling using a hand dredge for shell gathering is difficult, especially when water becomes deep and turbid due to agricultural run-off in this area throughout spring and summer. Therefore, a combination of both conventional sampling and eDNA assays is more effective for monitoring. The distribution of freshwater bivalves has been successfully analysed by species-specific eDNA assays in many parts of the world. Regarding evaluation of local stocks of P. japanensis by eDNA concentration, neither a clear effect of eDNA concentration, nor an interaction effect between eDNA concentration and flow velocity on P. japanensis density was detected. This is firstly because of the quite low density of P. japanensis in this area—0.019 individuals/m2 at most—and because of the inaccuracy of unionid density surveys when done by hand in natural streams (Carlsson et al., 2017; Sanchez & Schwalb, 2019; Prié et al., 2020). In a previous study, eDNA copies of three unionid species were not correlated with mussel density in a low-density range (< 0.02 mussels/m2) but were positively correlated when all density ranges were considered together (Currier et al., 2018). Additionally, in our study, eDNA concentrations varied among samples in the river cross section at each site. Thalinger et al. (2021) demonstrated that eDNA concentration varied in river cross sections when the source of eDNA was close by and distributed heterogeneously in rivers. Therefore, the variation in eDNA concentration in river cross sections may correlate with the microhabitat use of the focal species at each site.
Conservation of Pronodularia japanensis
Populations of the unionid P. japanensis have decreased rapidly over the past 30 years in this area. The rapid decrease in density of P. japanensis in the last six to seven years seems to have been caused by mortality of individuals and absence of recruitment. In fact, more fresh empty shells (in total 249) were collected than live individuals (in total 53, Fig. 5). Matsukawa et al. (1993) reported the number of live individuals and number of empty shells of P. japanensis to be similar in the period 1990–1991 in this area. Therefore, the high ratio of empty shells compared with live individuals (nearly five times higher) in the 2020–2021 period might indicate a high mortality relative to recruitment rate. Shell size distribution also indicates that no recruitment has occurred within at least the last ten years. The conservation of this endangered unionid now needs more intensive management, such as habitat restoration and reintroduction of P. japanensis, to increase survival rates and encourage reproduction. For such a reintroduction, upstream sites from weirs are suitable because there are the areas from which P. japanensis has disappeared as a result of recruitment restriction by weirs. Monitoring using this eDNA marker would be effective as part of these management approaches. Recently, a new, small, existing population comprising more than 1000 individuals was found on Dozen Plain, Ehime (Hata et al., 2021a). This population is only 40 km away from the Matsuyama population and could be a donor population; glochidium larvae attaching to host fishes and/or young adult mussels could be artificially transported to the Matsuyama population.
Further study
To conduct conventional surveys on unionid species requires experienced and labour-intensive work because unionids are cryptic, usually buried under substratum, and the water they live in is often too deep and turbid for easy study. Still, observing the natural setting at field sites is necessary to understand the ecosystem and to practise adaptive management. Thus, use of the eDNA marker we have developed is useful as a first step to determine the extent of the occurrence and distribution of P. japanensis. Subsequently, conventional survey practices can be used in a more suitable targeted manner in those places where this species is detected by eDNA.
Regarding Nodularia douglasiae, the last records of live individuals collected in Kunichi stream are of two specimens collected in July and August 2013 (Kuwahara et al., 2017; Hata et al., 2021b). Kunichi stream was the main habitat of this species in Ehime Prefecture, but a few individuals were previously recorded from a few farm ponds (Ishikawa & Chiba, 1999). Our next aim is to construct an eDNA marker specific for this species in order to search for the remaining population in these ponds.
Data availability
All relevant data are available as supplementary tables and supplementary files.
References
Allan, J. D. & A. S. Flecker, 1993. Biodiversity conservation in running waters. Bioscience 43: 32–43.
Belle, C. C., B. C. Stoeckle & J. Geist, 2019. Taxonomic and geographical representation of freshwater environmental DNA research in aquatic conservation. Aquatic Conservation: Marine and Freshwater Ecosystems 29: 1996–2009.
Beng, K. C. & R. T. Corlett, 2020. Applications of environmental DNA (eDNA) in ecology and conservation: opportunities, challenges and prospects. Biodiversity and Conservation 29: 2089–2121.
Böhm, M., N. I. Dewhurst-Richman, M. Seddon, S. E. H. Ledger, C. Albrecht, D. Allen, A. E. Bogan, J. Cordeiro, K. S. Cummings, A. Cuttelod, G. Darrigran, W. Darwall, Z. Fehér, C. Gibson, D. L. Graf, F. Köhler, M. Lopes-Lima, G. Pastorino, K. E. Perez, K. Smith, D. van Damme, M. V. Vinarski, T. von Proschwitz, T. von Rintelen, D. C. Aldridge, N. A. Aravind, P. B. Budha, C. Clavijo, D. Van Tu, O. Gargominy, M. Ghamizi, M. Haase, C. Hilton-Taylor, P. D. Johnson, Ü. Kebapçı, J. Lajtner, C. N. Lange, D. A. W. Lepitzki, A. Martínez-Ortí, E. A. Moorkens, E. Neubert, C. M. Pollock, V. Prié, C. Radea, R. Ramirez, M. A. Ramos, S. B. Santos, R. Slapnik, M. O. Son, A.-S. Stensgaard & B. Collen, 2021. The conservation status of the world’s freshwater molluscs. Hydrobiologia 848: 3231–3254.
Breton, S., H. D. Beaupré, D. T. Stewart, W. R. Hoeh & P. U. Blier, 2007. The unusual system of doubly uniparental inheritance of mtDNA: isn’t one enough? Trends in Genetics 23: 465–474.
Brookes, A., 1988. Channelized Rivers: Perspectives For Environmental Management, A Wiley-Interscience publication, Chichester.
Carlsson, J. E. L., D. Egan, P. C. Collins, E. D. Farrell, F. Igoe & J. Carlsson, 2017. A qPCR MGB probe based eDNA assay for European freshwater pearl mussel (Margaritifera margaritifera L.). Aquatic Conservation: Marine and Freshwater Ecosystems 27: 1341–1344.
Curole, J. P. & T. D. Kocher, 2002. Ancient sex-specific extension of the cytochrome c oxidase II gene in bivalves and the fidelity of doubly-uniparental inheritance. Molecular Biology and Evolution 19: 1323–1328.
Currier, C. A., T. J. Morris, C. C. Wilson & J. R. Freeland, 2018. Validation of environmental DNA (eDNA) as a detection tool for at-risk freshwater pearly mussel species (Bivalvia: Unionidae). Aquatic Conservation: Marine and Freshwater Ecosystems 28: 545–558.
Deiner, K. & F. Altermatt, 2014. Transport distance of invertebrate environmental DNA in a natural river. PLoS ONE 9: e88786.
Dudgeon, D., 2006. The impacts of human disturbance on stream benthic invertebrates and their drift in North Sulawesi, Indonesia. Freshwater Biology 51: 1710–1729.
Dysthe, J. C., T. Rodgers, T. W. Franklin, K. J. Carim, M. K. Young, K. S. McKelvey, K. E. Mock & M. K. Schwartz, 2018. Repurposing environmental DNA samples—detecting the western pearlshell (Margaritifera falcata) as a proof of concept. Ecology and Evolution 8: 2659–2670.
Folmer, O., M. Black, W. Hoeh, R. Lutz & R. Vrijenhoek, 1994. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Molecular Marine Biology and Biotechnology 3: 294–299.
Gasparini, L., S. Crookes, R. S. Prosser & R. Hanner, 2020. Detection of freshwater mussels (Unionidae) using environmental DNA in riverine systems. Environmental DNA 2: 321–329
Graf, D. L. & K. S. Cummings, 2021. A ‘big data’ approach to global freshwater mussel diversity (Bivalvia: Unionoida), with an updated checklist of genera and species. Journal of Molluscan Studies 87: eyaa034.
Hata, H., Y. Uemura, K. Ouchi & H. Matsuba, 2019. Hybridization between an endangered freshwater fish and an introduced congeneric species and consequent genetic introgression. PLoS ONE 14: e0212452.
Hata, H., D. Togaki, K. Ogasawara, K. Matsumoto, T. Yamamoto, H. Murakami, J. Nakajima & M. Inoue, 2021a. A remaining population of an endangered freshwater unionid, Pronodularia japanensis (Lea, 1958), in a fragmented agricultural ditch. Japanese Journal of Conservation Ecology 26: 315–322.
Hata, H., Y. Uemura & K. Ouchi, 2021b. Decline of unionid mussels enhances hybridisation of native and introduced bitterling fish species through competition for breeding substrate. Freshwater Biology 66: 189–201.
Hoellein, T. J., C. B. Zarnoch, D. A. Bruesewitz & J. DeMartini, 2017. Contributions of freshwater mussels (Unionidae) to nutrient cycling in an urban river: filtration, recycling, storage, and removal. Biogeochemistry 135: 307–324.
Ishikawa, H. & N. Chiba, 1999. Catalogue of the freshwater molluscs of Ehime Prefecture. Bulletin of Ehime Prefectural Science Museum 14: 1–50.
Kondo, T., 2008. Monograph of Unionoida in Japan (Mollusca: Bivalvia), Vol. 3. Malacological Society of Japan, Japan.
Kuwahara, A., H. Matsuba, M. Inoue & H. Hata, 2017. Population decline in unionid mussels in the Matsuyama Plain, Ehime Prefecture. Japanese Journal of Conservation Ecology 22: 91–103.
LeBlanc, F., R. Steeves, V. Belliveau, F. Akaishi & N. Gagné, 2021. Detecting the brook floater, a freshwater mussel species at risk, using environmental DNA. Aquatic Conservation: Marine and Freshwater Ecosystems 31: 1233–1244.
Lopes-Lima, M., A. Hattori, T. Kondo, J. Hee Lee, S. Ki Kim, A. Shirai, H. Hayashi, T. Usui, K. Sakuma, T. Toriya, Y. Sunamura, H. Ishikawa, N. Hoshino, Y. Kusano, H. Kumaki, Y. Utsugi, S. Yabe, Y. Yoshinari, H. Hiruma, A. Tanaka, K. Sao, T. Ueda, I. Sano, J.-I. Miyazaki, D. V. Gonçalves, O. K. Klishko, E. S. Konopleva, I. V. Vikhrev, A. V. Kondakov, MYu. Gofarov, I. N. Bolotov, E. M. Sayenko, M. Soroka, A. Zieritz, A. E. Bogan & E. Froufe, 2020. Freshwater mussels (Bivalvia: Unionidae) from the rising sun (Far East Asia): phylogeny, systematics, and distribution. Molecular Phylogenetics and Evolution 146: 106755.
Lopes-Lima, M., N. Riccardi, M. Urbanska, F. Köhler, M. Vinarski, A. E. Bogan & R. Sousa, 2021. Major shortfalls impairing knowledge and conservation of freshwater molluscs. Hydrobiologia 848: 2831–2867.
Lor, Y., T. M. Schreier, D. L. Waller & C. M. Merkes, 2020. Using environmental DNA (eDNA) to detect the endangered Spectaclecase Mussel (Margaritifera monodonta). Freshwater Science 39: 837–847.
Matsuba, H., S. Yoshimi, M. Inoue & H. Hata, 2014. Origin of Tanakia limbata in Ehime prefecture indicated by phylogeographic analysis of mitochondrial cytochrome b gene sequences. Japanese Journal of Ichthyology 61: 89–96.
Matsukawa, M., M. Higuchi, T. Yamamoto & K. Ido, 1993. Primary information on death assemblages in fluvial environments—comparative analysis between living and dead fresh-water molluscan assemblages in the Kunichigawa River, southern Matsuyama, Japan. Journal of Geographic Society of Japan 99: 643–657.
Merkes, C. M., K. E. Klymus, M. J. Allison, C. Goldberg, C. C. Helbing, M. E. Hunter, C. A. Jackson, R. F. Lance, A. M. Mangan, E. M. Monroe, A. J. Piaggio, J. P. Stokdyk, C. C. Wilson & C. Richter, 2019. Generic qPCR Limit of Detection (LOD) / Limit of Quantification (LOQ) calculator. R Script. Available at: https://github.com/cmerkes/qPCR_LOD_Calc. https://doi.org/10.5066/P9GT00GB. Accessed: 1 September 2021
Minamoto, T., H. Yamanaka, T. Takahara, M. N. Honjo & Z. Kawabata, 2012. Surveillance of fish species composition using environmental DNA. Limnology 13: 193–197.
Minegishi, Y., M.K.-S. Wong, T. Kanbe, H. Araki, T. Kashiwabara, M. Ijichi, K. Kogure & S. Hyodo, 2019. Spatiotemporal distribution of juvenile chum salmon in Otsuchi Bay, Iwate, Japan, inferred from environmental DNA. PLoS ONE 14: e0222052.
Ministry of the Environment, Japan, 2020. The Japanese Red Lists 2020 [Internet]. Ministry of the Environment, Japan. Available at: http://www.env.go.jp/press/files/jp/114457.pdf.
Naimo, T. J., E. D. Damschen, R. G. Rada & E. M. Monroe, 1998. Nonlethal evaluation of the physiological health of unionid mussels: methods for biopsy and glycogen analysis. Journal of the North American Benthological Society 17: 121–128.
Negishi, J. N., Y. Kayaba, K. Tsukahara & Y. Miwa, 2008. Ecological studies on Unionoida: current status and future challenges. Japanese Journal of Ecology 58: 37–50.
Onikura, N., J. Nakajima, K. Eguchi, R. Inui, E. Higa, T. Miyake, K. Kawamura, S. Matsui & S. Oikawa, 2006. Change in distribution of bitterlings, and effects of urbanization on populations of bitterlings and unionid mussels in Tatara River System, Kyushu, Japan. Journal of Japan Society on Water Environment 29: 837–842.
Onikura, N., J. Nakajima, R. Inui & J. Kaneto, 2016. Priority maps for protecting the habitats of threatened freshwater fishes in urban areas: a case study of five rivers in the Fukuoka Plain, northern Kyushu Island, Japan. Ichthyological Research 63: 347–355.
Preece, E. P., M. Bryan, S. M. Mapes, C. Wademan & R. Dorazio, 2021. Monitoring for freshwater mussel presence in rivers using environmental DNA. Environmental DNA 3: 591–604.
Prié, V., A. Valentini, M. Lopes-Lima, E. Froufe, M. Rocle, N. Poulet, P. Taberlet & T. Dejean, 2020. Environmental DNA metabarcoding for freshwater bivalves biodiversity assessment: methods and results for the Western Palearctic (European sub-region). Hydrobiologia 848: 2931–2950.
R Core Team., 2020. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. Available at: http://www.R-project.org/
Rodgers, T. W., J. C. Dysthe, C. Tait, T. W. Franklin, M. K. Schwartz & K. E. Mock, 2020. Detection of 4 imperiled western North American freshwater mussel species from environmental DNA with multiplex qPCR assays. Freshwater Science 39: 762–772.
Sanchez, B. & A. N. Schwalb, 2019. Detectability affects the performance of survey methods: a comparison of sampling methods of freshwater mussels in Central Texas. Hydrobiologia 848: 2919–2929.
Sano, I., T. Saito, J.-I. Miyazaki, A. Shirai, T. Uechi, T. Kondo & S. Chiba, 2020. Evolutionary history and diversity of unionoid mussels (Mollusca: Bivalvia) in the Japanese Archipelago. Plankton and Benthos Research 15: 97–111.
Sansom, B. J. & L. M. Sassoubre, 2017. Environmental DNA (eDNA) shedding and decay rates to model freshwater mussel eDNA transport in a river. Environmental Science & Technology 51: 14244–14253.
Schill, W. B. & H. S. Galbraith, 2019. Detecting the undetectable: Characterization, optimization, and validation of an eDNA detection assay for the federally endangered dwarf wedgemussel, Alasmidonta heterodon (Bivalvia: Unionoida). Aquatic Conservation: Marine and Freshwater Ecosystems 29: 603–611.
Schmidt, B. C., S. F. Spear, A. Tomi & C. M. B. Jachowski, 2021. Evaluating the efficacy of environmental DNA (eDNA) to detect an endangered freshwater mussel Lasmigona decorata (Bivalvia:Unionidae). Freshwater Science 40: 354–367.
Shogren, A. J., J. L. Tank, S. P. Egan, D. Bolster & T. Riis, 2019. Riverine distribution of mussel environmental DNA reflects a balance among density, transport, and removal processes. Freshwater Biology 64: 1467–1479.
Smith, C., M. Reichard, P. Jurajda & M. Przybylski, 2004. The reproductive ecology of the European bitterling (Rhodeus sericeus). Journal of Zoology 262: 107–124.
Spooner, D. E. & C. C. Vaughn, 2006. Context-dependent effects of freshwater mussels on stream benthic communities. Freshwater Biology 51: 1016–1024.
Stoeckle, B. C., R. Kuehn & J. Geist, 2016. Environmental DNA as a monitoring tool for the endangered freshwater pearl mussel (Margaritifera margaritifera L.): a substitute for classical monitoring approaches? Aquatic Conservation: Marine and Freshwater Ecosystems 26: 1120–1129.
Stoeckle, B. C., S. Beggel, R. Kuehn & J. Geist, 2021. Influence of stream characteristics and population size on downstream transport of freshwater mollusk environmental DNA. Freshwater Science 40: 191–201.
Taberlet, P., E. Coissac, M. Hajibabaei & L. H. Rieseberg, 2012. Environmental DNA. Molecular Ecology 21: 1789–1793.
Thalinger, B., D. Kirschner, Y. Pütz, C. Moritz, R. Schwarzenberger, J. Wanzenböck & M. Traugott, 2021. Lateral and longitudinal fish environmental DNA distribution in dynamic riverine habitats. Environmental DNA 3: 305–318.
The eDNA Society, 2019. Environmental DNA Sampling and Experiment Manual Version 2.1. The eDNA Society, Otsu, Japan
Thomsen, P. F., J. Kielgast, L. L. Iversen, C. Wiuf, M. Rasmussen, M. T. P. Gilbert, L. Orlando & E. Willerslev, 2012. Monitoring endangered freshwater biodiversity using environmental DNA. Molecular Ecology 21: 2565–2573.
Togaki, D., H. Doi & I. Katano, 2019. Detection of freshwater mussels (Sinanodonta spp.) in artificial ponds through environmental DNA: a comparison with traditional hand collection methods. Limnology 21: 59–65.
Uchii, K., H. Doi, T. Okahashi, I. Katano, H. Yamanaka, M. K. Sakata & T. Minamoto, 2019. Comparison of inhibition resistance among PCR reagents for detection and quantification of environmental DNA. Environmental DNA 1: 359–367.
Uemura, Y., S. Yoshimi & H. Hata, 2018. Hybridization between two bitterling fish species in their sympatric range and a river where one species is native and the other is introduced. PLoS ONE 13: e0203423.
Vaughn, C. C. & C. C. Hakenkamp, 2001. The functional role of burrowing bivalves in freshwater ecosystems. Freshwater Biology 46: 1431–1446.
Venetis, C., I. Theologidis, E. Zouros & G. C. Rodakis, 2006. No evidence for presence of maternal mitochondrial DNA in the sperm of Mytilus galloprovincialis males. Proceedings of the Royal Society B: Biological Sciences 273: 2483–2489.
Wacker, S., F. Fossøy, B. M. Larsen, H. Brandsegg, R. Sivertsgård & S. Karlsson, 2019. Downstream transport and seasonal variation in freshwater pearl mussel (Margaritifera margaritifera) eDNA concentration. Environmental DNA 1: 64–73.
Wiepkema, P. R., 1961. An ethological analysis of the reproductive behaviour of the bitterling (Rhodeus amarus Bloch). Archives Néerlandaises De Zoologie 14: 103–199.
Ye, J., G. Coulouris, I. Zaretskaya, I. Cutcutache, S. Rozen & T. L. Madden, 2012. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics 13: 134.
Yoshimura, C., T. Omura, H. Furumai & K. Tockner, 2005. Present state of rivers and streams in Japan. River Research and Applications 21: 93–112.
Acknowledgements
We are grateful to Jun-ichi Kitamura and Rintaro Taniguchi for supplying unionid specimens, Shoichiro Yamamoto and Atsushi Maruyama for eDNA assay instructions, Shin-ichi Kitamura and Kei Nakayama for letting us use StepOnePlus, Mikio Inoue and Yume Imada for their support in field and laboratory work, and Takaki Kondo and Manuel Lopes-Lima for taxonomic information on unionids. This research was supported by JSPS KAKENHI (18KK0208, 20K06814).
Funding
This study was funded by Japan Society for the Promotion of Science (Grant Nos.: 18KK0208, 20K06814)
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Handling editor: Xavier Pochon
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Hata, H., Ogasawara, K. & Yamashita, N. Population decline of an endangered unionid, Pronodularia japanensis, in streams is revealed by eDNA and conventional monitoring approaches. Hydrobiologia 849, 2635–2646 (2022). https://doi.org/10.1007/s10750-022-04852-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10750-022-04852-6