Abstract
Here, we combined archived mitochondrial sequences for Ponto-Caspian gobiids with new sequences from the south Caspian basin to assess and evaluate its gobioid diversity and taxonomy, and to provide a first mitochondrial-based phylogenetic and phylogeographic framework. We demonstrate that: (i) Proterorhinus nasalis is the tubenose goby taxon in the saline waters of the southern Caspian Sea, whereas the name Pr. semipellucidus for the Azov/northern Caspian Sea/Volga River populations is likely be resurrected depending on the outcome of an integrative taxonomical approach; (ii) the deep-water goby Ponticola bathybius should be re-assigned to the genus Neogobius, as it is the sistergroup of N. melanostomus; (iii) specimens previously identified as Po. cyrius and Po. iljini from the south Caspian basin appear conspecific with Po. iranicus and Po. gorlap, respectively, and should be omitted from the checklist of Iranian and south Caspian freshwater fishes; (iv) the low stand of the Caspian Sea during the Tyurkyanian regression is inferred to have led to the isolation and evolution of Po. iranicus; and (v) similarities in genetic background, and invasion history of Rhinogobius sp. and Pseudorasbora parva in Iran and Turkmenistan indicate that the initial introduction of both species into the region possibly originated from Japan in the 1980s.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The Ponto-Caspian region is the stage of a highly diverse endemic evolution of two main lineages of Gobiidae sensu Gill & Mooi 2012: (i) the endemic Ponto-Caspian benthophiline gobies (Agorreta et al., 2013; referred to by Neilson and Stepien 2009a as a own subfamily, Benthophilinae Beling & Iljin, 1927), and (ii) a branch of the Pomatoschistus-lineage or sand gobies and related genera (Agorreta et al., 2013; Schwarzhans et al., 2017).
Benthophiline gobies represent a monophyletic freshwater and brackish water radiation of Paratethyan gobies currently classified in nine genera and two major lineages subdivided in three tribes (Neilson & Stepien, 2009a): (1) the neogobiine-lineage with the two tribes Neogobiini (with the genus Neogobius Iljin, 1927), and Ponticolini (with the genera Ponticola Iljin, 1927, Mesogobius Bleeker, 1874, Proterorhinus Smitt, 1899, and Babka Iljin, 1927); and (2) the tadpole gobies lineage or Benthophilini (with the genera Anatirostrum Iljin, 1930, Benthophilus Eichwald, 1831, Benthophiloides Beling & Iljin, 1927, and Caspiosoma Iljin, 1927). Their adaptation to brackish and freshwaters most likely evolved during the process of isolation of the Paratethys from the Tethys in the Middle Miocene and the subsequent freshening of the Paratethys in the latest Miocene (Popov et al., 2004). In this diverse group, there are several real freshwater endemics, several species restricted to the brackish waters of the seas, and several euryhaline species, but no species has ever been recorded as inhabiting strictly marine conditions (Miller, 2003, 2004b; Kottelat & Freyhof, 2007; Freyhof, 2012). Its evolutionary history has been driven by the dynamic geologic and hydrologic evolution of the region (Rögl, 1999; Reid & Orlova, 2002; Neilson & Stepien, 2009a). Neilson and Stepien (2009a) provided the first comprehensive phylogenetic and biogeographic analysis of the group based on mitochondrial and nuclear markers, and addressed classification issues and phylogenetic relationships within the group. They presented a revised taxonomy and nomenclature that does not, however, aptly fit with the current understanding of gobioid family and subfamily phylogenetics, as, e.g., presented in Agorreta et al. (2013), and recently confirmed by McCraney et al. (2020); nevertheless, their basic phylogenetic conclusions and tribe designations remain useful as they are not in conflict with any familial and subfamilial taxonomy.
The Pomatoschistus-sand goby lineage is primarily represented by several species of Knipowitschia Iljin, 1927 and the Caspian-endemic Hyrcanogobius Iljin, 1928 in the Ponto-Caspian basin. Hyrcanogobius Iljin, 1928 (type H. bergi Iljin, 1928) was originally distinguished as a separate genus because of the reduced condition of the head lateral-line canal system (Iljin, 1928), but later suggested as congeneric with Knipowitschia by Economidis and Miller (1990). However, following the last examination of actual material, Miller (2004c) would now agree that this genus warrants separation from Knipowitschia. Sand gobies have their highest diversity in the Mediterranean, but there are several endemics in the Black and Caspian Seas (Freyhof, 2012; Thacker et al., 2019).
Furthermore, there is an introduced Rhinogobius species in the Ponto-Caspian basin with established populations in the inland waters of Iran (e.g., in the Anzali Wetland and Zarivar Lake; Coad, 2016; Sadeghi et al., 2019), Turkmenistan (Kara-Kum Canal; Aliev et al., 1988), and the Caucasus (Epitashvili et al., 2020; Japoshvili et al., 2020), which has appeared under several scientific names over the last 2 decades [i.e., Rhinogobius similis Gill, 1859, R. cheni (Nichols, 1931), and currently R. lindbergi Berg, 1933]. There have been controversial debates about the taxonomic status of this introduced species to the Ponto-Caspian basin (Coad & Abdoli, 2000; Vasil’eva & Kuga, 2008; Sadeghi et al., 2019), and its invasion history, spread pattern, and genetic background remain unknown.
The Caspian Sea basin is ecologically split into three sub-basins (Naseka & Bogutskaya, 2009): (i) a northern shallow sub-basin (less than 10 m); (ii) a middle sub-basin with an average and maximum depth of 200 m and 790 m, respectively; and (iii) the southern and deepest sub-basin with a maximum depth of 980–1,025 m and an average of 325 m. It is a basin with high degree of species-level endemism up to 80% (Dumont 1998, 2000) as 99 out of 159 Caspian Sea fish species (62%) are endemic and restricted to specific areas (Naseka & Bogutskaya, 2009). Gobiidae sensu Gill & Mooi 2012 is the second most species-rich fish family in the basin with 36 species in 11 genera, 31 (97.2%) of which are Caspian endemic (Miller, 2003, 2004b; Kottelat & Freyhof, 2007; Naseka & Bogutskaya, 2009). The high endemism of gobies in the Caspian Sea basin is highlighted by the fact that a large number of gobiid species have not been included yet in any large- or small-scale phylogenetic, biogeographic, or phylogeographic analysis (Neilson & Stepien, 2009a; Thacker & Roje, 2011; Agorreta et al., 2013; Medvedev et al., 2013; Thacker et al., 2019). Most importantly, gobiids of the southern and eastern parts of the Caspian Sea have not been included in any molecular study of gobiid systematics (Fig. S1, Online Resources), despite the fact that their documentation and analysis could provide in depth insights into yet unaddressed evolutionary biology questions. Field ichthyology remains to accomplish basic research in the Caspian Sea basin, as, e.g., the presence of a large number of gobiid species listed for the southern Caspian Sea basin has to be confirmed by any specimen (Miller, 2003, 2004b; Boldyrev & Bogutskaya, 2007; Kottelat & Freyhof, 2007; Esmaeili et al., 2018).
Nevertheless, several recently published sources of mitochondrial cytochrome c oxidase subunit 1 (COI) sequence information for several Ponto-Caspian gobiids (e.g., Neilson & Stepien, 2009a, b; Keskİn & Atar, 2013; Geiger et al., 2014; Knebelsberger et al., 2015; Thalinger et al., 2016; Thacker et al., 2019) provide results of various recent efforts to obtain DNA barcodes for these taxa, which are useful for evolutionary analyses, species identification and application for conservation. The partial COI barcode fragment generally does not provide adequate phylogenetic resolution for large-scale analyses (for a review, see Rubinoff & Holland, 2005), but the taxonomic scope in this study is narrow enough that COI is useful as a first step for integrative taxonomy (see Thacker et al., 2019). Here, we combine archived COI data for Ponto-Caspian gobiids with new sequences from the southern Caspian Sea basin to assess its gobioid diversity, to provide a first mitochondrial-based phylogenetic and phylogeographic study, and to evaluate, from a mitochondrial viewpoint, the taxonomy of gobiid fishes in the deepest and oldest southern part of the Caspian Sea. The outcome will be a very first deep insight into the southern Caspian Sea’s gobiid diversity and evolution, and it will be important for the conservation and management programs, too.
Materials and methods
Study area, specimen collection, DNA extraction, amplification, and sequencing
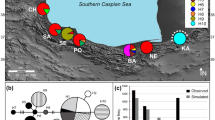
During numerous field works between 2014 and 2020, we collected gobiid specimens from shallow and deep waters of the southern Caspian Sea and its associated waterbodies and river drainages (30 localities; Table 1) using beach seining, scuba diving, deep-water bottom beam trawls, and electrofishing devices. Information on sample codes, species, and sampling localities are given in Table 1, and a map of localities is shown in Fig. 1. We took photographs of live specimens, and after anesthesia with 1% clove oil solution or Quinaldine Sulfate (Ross & Ross, 2009), muscle tissue or the right pectoral fin of each individual was separated and fixed in 96% ethanol and subsequently kept at − 20 °C until DNA extraction. Species-level identification of the specimens was carried out using major taxonomic keys and primary taxonomic literature (Miller, 2003, 2004b; Boldyrev & Bogutskaya, 2007; Kottelat & Freyhof, 2007; Vasil’eva et al., 2015). The specimens were fixed in 10% formalin, transferred to 70% ethanol, and deposited in the Zoological Museum of Shiraz University, Collection of Biology Department (ZM-CBSU) and in the SNSB-Bavarian State Collection of Zoology, Munich (SNSB-ZSM) as voucher specimens.
A map of the collection sites in southern Caspian Sea. The location codes correspond to those in Table 1
Genomic DNA samples were extracted from muscle tissues or fin clips according to the salt method protocol described by Bruford et al. (1992). Partial COI gene sequences were amplified using primer pairs FishF1 (5′-TCAACCAACCACAAAGACATTGGCAC-3′) and FishR1 (5′-TAGACTTCTGGGTGGCCAAAGAATCA-3′) (Ward et al., 2005). Amplification was performed on a Bioer XP Thermal Cycler (Bioer Technology Co. Ltd., Hangzhou, China), programmed as following: 94 °C for 1 min for initial denaturing, 35 cycles of 94 °C for 30 s, 52–56 °C for 45 s, and 72 °C for 45 s, followed by 72 °C for 5 min as the final extension. An alternative primer pair, FISH-BCL (5′-TCAACYAATCAYAAAGATATYGGCAC-3′) and FISH-BCH (5′-TAAACTTCAGGGTGACCAAAAAATCA-3′) (Baldwin et al., 2009), was used for Knipowitschia caucasica. The thermal cycler program for PCR was: initial denaturation step of 94 °C for 5 min, 35 cycles of 94 °C for 1 min, 58.4 °C for 45 s, 72 °C for 1 min, and one cycle of 5 min at 72 °C. After purification of the PCR products with the ExoASP-IT® (usb) kit, they were sent out for Sanger sequencing with BigDye® Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA) on an ABI PRISM 3730xl DNA Analyzer (Applied Biosystems, Foster City, CA) to the Faghihi Lab., Shiraz, Iran.
Phylogenetic, biogeographic, and species delimitation analyses
Sequence chromatograms were viewed and edited in BioEdit 7.0.4 (Hall, 1999). We generated COI sequence data (652 bp) from 68 representatives of the species Ponticola gorlap (Iljin, 1949), Po. iranicus Vasil’eva, Mousavi-Sabet & Vasil’ev, 2015, Po. syrman (Nordmann, 1840), Po. bathybius (Kessler, 1877), Neogobius caspius (Eichwald, 1831), N. pallasi (Berg, 1916), N. melanostomus (Pallas, 1814), Benthophilus leobergius Berg, 1949, Protherorhinus nasalis (De Filippi, 1963), K. caucasica (Berg, 1916), and R. lindbergi Berg, 1933 (Table 2). Sequences were tagged with species names following traditional taxonomic identification and submitted to GenBank (Table 2). In addition to the newly determined sequences from the southern Caspian Sea basin, we used 346 archived DNA barcode sequences (members of both benthophiline sublineages and Knipowitschia; Table S1, Online Resources) obtained from GenBank and largely derived from the works of Neilson and Stepien (2009a, b), Keskİn and Atar (2013), Geiger et al. (2014), Knebelsberger et al. (2015), Thalinger et al. (2016), and Thacker et al. (2019). Furthermore, we integrated 18 COI barcodes of K. caucasica obtained from digital catalogue of the Zoologisches Forschungsmuseum Alexander Koenig (ZFMK, www.collections.zfmk.de). GenBank numbers, ZFMK museum numbers, and geographical origins of these archived sequences are given in Table S1. Regarding the benthophilines, we assembled a total of 294 individual sequences (61 newly developed plus 233 archived) from 27 nominal Ponto-Caspian gobiid species, with sampling comprising the entire Ponto-Caspian region (except for the eastern parts of the Caspian Sea basin), and the European and North American invasive ranges. To assess the taxonomic status of the introduced freshwater goby of the genus Rhinogobius to Iran, 9 BOLD (derived from Japoshvili et al., 2020) and ZFMK-deposited COI barcodes of R. lindbergi sampled from Georgia (Ozaani Stream) and Azerbaijan (Kura River basin), and 74 archived COI sequences of related species (Table S2) largely derived from the works of Yamasaki et al. (2015), Chang et al. (2017), Shen et al. (2016), Xia et al. (2018), and Chen et al. (2015) were included in analysis along with the four new sequences collected from the southern Caspian Sea basin (Anzali Wetland) and the Tigris River system (Zarivar Lake).
DNA sequences were aligned using ClustalW multiple alignment accessory application implemented in Mega 7 (Kumar et al., 2016). The best-fit nucleotide substitution models for the COI datasets were selected using the jModelTest 2.1.3 (Darriba et al., 2012) based on the Bayesian Information Criterion (BIC; Schwarz, 1978). PopART 1.7 (Leigh & Bryant, 2015) was used to depict the evolutionary relationships among haplotypes and evaluate the phylogeographic depth (Avise, 2000) based on the TCS method. Substitution saturation in the COI barcode region was examined with DAMBE 7 (Xia, 2018) using the Xia’s et al. (2003) nucleotide substitution saturation test. For phylogenetic reconstruction, the Bayesian method (BI) was run based on four simultaneous runs of four Markov chains for 100,000,000 generations and a burn‐in of 15% of the initial trees in MrBayes 3.2.6 (Ronquist et al., 2012). Phylogeny of benthophiline gobies was rooted with Gobius (G. niger, FJ526837; and G. ophiocephalus, FJ526797) and Chromogobius zebratus (FJ526797). The Rhinogobius tree was rooted with Awaous guamensis (HQ639035). Resulting COI alignments were further analyzed phylogenetically using the Maximum Likelihood (ML) method in RAxML 7.2.5 (Stamatakis, 2006) with 10,000 bootstrap replicates. Phylogenetic hypothesis testing [Shimodaira–Hasegawa (SH) test; Shimodaira & Hasegawa, 1999; Goldman et al., 2000] was performed in CONSEL v.1.20 (Shimodaira & Hasegawa, 2001) to test for statistical significance of topological differences between the BI and ML trees.
Species delimitation analyses estimate the number of lineages (i.e., putative species) supported by molecular sequences or gene trees without a priori assignment of individuals to species, and to generate a preliminary hypothesis of species limits (Carstens et al., 2013). Five single-locus species delimitation analyses were performed on the benthophiline COI dataset: Automatic Barcode Gap Discovery (ABGD; Puillandre et al., 2012), reversed Statistical Parsimony (SP; Hart & Sunday, 2007), Bayesian Poisson Tree Process (bPTP; Zhang et al., 2013), multiple rate PTP (mPTP; Kapli et al., 2017), and Bayesian General Mixed Yule-Coalescent (bGMYC; Reid & Carstens, 2012). COI data set was tested on the ABGD webserver (https://bioinfo.mnhn.fr/abi/public/abgd/) with combinations of ABGD settings within the parameter range of Pmin = 0.001, Pmax = 0.1, all for a total of 10 steps and applying a K2P-corrected genetic distance matrix calculated in Mega. TCS 1.21 (Clement et al., 2000) was used to calculate a Statistical Parsimony network, using a 95% connection probability threshold to delineate putative species. The bPTP server (http://species.h-its.org/) was used with a Bayesian tree produced in MrBayes as input tree and analyses were run under default settings. Convergence was visualized on the MCMC interactions plots vs. log-likelihood (Fig. S2). The mPTP analysis was run using the online server (http://mptp.h-its.org), under the same parameters as for bPTP. The bGMYC method is conceptually similar to bPTP and mPTP, and uses a tree topology to infer species hypotheses, but unlike these methods, it applies an ultrametric tree as an input topology. This analysis was performed using the bGMYC package (Reid & Carstens, 2012) for R ver. 3.6.1 (R Core Team, 2013) on an time-calibrated tree produced in BEAST ver. 1.8.2 (Drummond et al., 2012) as an input file. To achieve a first and preliminary time calibration of the benthophiline tree, we reduced the data set to limited haplotypes of each species to avoid loss of computational efficiency due to the intraspecific polytomies. We reconstructed a Bayesian phylogeny as outlined above and performed the calibration analysis on a matrix of 56 haplotypes (including the 3 outgroup exemplars) with BEAST, run with an uncorrelated lognormal relaxed clock model and a birth–death speciation prior. Although not yet fully scrutinized, we followed Neilson and Stepien (2009a) and set the age for the most recent common “neogobiin” ancestor to 10 Mya, at date derived from fossil otoliths of Neogobius inferred to be of Late Miocene–Early Pliocene origin (Rückert-Ülkümen, 2006). We assigned a secondary calibration of 6.25 Mya to the node subtending Proterorhinus + Mesogobius (derived from the analysis of Neilson & Stepien, 2009a) and ran the BEAST analysis in four independent runs of 100,000,000 generations, with trees sampled every 1,000 generations; the first 10% were discarded as burn-in. At the end, convergence and sufficient effective sampling sizes (ESS; ESS > 200) were confirmed using the Tracer ver. 1.6 (Rambaut et al., 2014), and a maximum clade credibility consensus tree was constructed in Tree Annotator 1.8.2 (Drummond et al., 2012). Phylogenetic trees were edited in FigTree 1.4.4 (Rambaut & Drummond, 2012), and the species delimitation results were depicted as grey bars on the tree.
Results
JModelTest determined GTR + I + G as the best-fitting substitution model for the benthophiline data set (294 individuals, 652 bp length). The nucleotide substitution pattern showed that the benthophiline sequences have not reached substitution saturation and are, therefore, well suitable for phylogenetic analyses (Iss < Iss.c S/Iss.c A; Table S3). Sequence analysis of this dataset detected 253 variable nucleotide sites (13 singleton variable and 240 parsimony informative sites), which allowed the definition of 106 haplotypes (H1-H106; see Tables 2, S1). Phylogenies inferred from the BI and ML (Fig. S3) analyses of benthophilines were not significantly different (SH test, P = 0.43), and the BI tree is presented in Fig. 2 for clarity. In total, the resulting benthophiline phylogenetic hypothesis includes novel sequences for 61 individuals from southern Caspian Sea basin combined with 233 archived sequences, defining 106 haplotypes, and three outgroup individuals (Fig. 2). Most genera and nominal species are highly supported, and three clades correlating with previously identified sublineages within the benthophilines are highly resolved.
Bayesian phylogenetic hypothesis of the endemic Ponto-Caspian gobies (tribes Benthophilini, Neogobiini and Ponticolini), including 294 individuals (106 haplotypes) plus 3 outgroups. Each species names is followed by the designated haplotype and its frequency. The new haplotypes sampled from the southern Caspian Sea basin (61 individuals) are shown in blue. Support values are indicated beside the nodes (BI posterior probability/ML bootstrap)
(i) The first basally diverging clade includes the tadpole gobies (Benthophilus and Caspiosoma) (97% bootstrap support, 1.00 PP), comprising the tribe Benthophilini. The deepest split within Benthophilini is between C. caspium and the remainder of the species. The two tadpole goby species, Be. granulosus and Be. abdurahmanovi comprise a group (100% bootstrap support, 1.00 PP) constituting the sister clade to the clade containing Be. stellatus, Be. mahmudbejovi, and Be. leobergius sampled from southern Caspian Sea (100% bootstrap support, 1.00 PP).
(ii) A clade corresponding to the tribe Neogobiini (100% bootstrap support, 1.00 PP) includes a now-restricted Neogobius (N. fluviatilis, N. pallasi, N. caspius, and N. melanostomus) and Po. bathybius. Neogobius fluviatilis and N. pallasi are sister species (91% bootstrap support, 0.98 PP), forming a clade sister to the remainder of the species (100% bootstrap support, 1.00 PP). The mitochondrial haplotype of Po. bathybius, sampled from southern Caspian Sea, is closely related to N. melanostomus with robust statistical support (100% bootstrap support, 1.00 PP).
(iii) A larger clade comprising the tribe Ponticolini (89% bootstrap support, 1.00 PP) contains the genera Proterorhinus, Mesogobius, Babka, and Ponticola. Mesogobius and Proterorhinus are strongly supported as sister groups (87% bootstrap, 1.00 PP), and the Mesogobius + Proterorhinus clade is then the sister to the clade containing Babka and Ponticola (except for Po. bathybius) (89% bootstrap support, 1.00 PP). Our trees show that Ponticola (comprising Po. cephalargoides, Po. constructor, Po. cyrius, Po. eurycephalus, Po. gorlap, Po. kessleri, Po. platyrostris, Po. ratan, Po. rhodioni, Po. syrman, and Po. iranicus) is strongly supported as a separate clade (100% bootstrap support, 1.00 PP), which includes, however, Babka gymnotrachelus (with low support; 48% bootstrap, 0.44 PP). Interestingly, the combined analysis of a concatenated alignment of two mitochondrial and two nuclear gene fragments analysis of Neilson and Stepien (2009a) strongly supported a Babka–Ponticola sister group relationship, each with strongly supported clades clearly divergent from the other genera, thus supporting the recognition of Babka and Ponticola as distinct genera. Four clades are highly resolved within Ponticola. The deepest split is between Po. ratan and the remainder of the species (100% bootstrap support, 1.00 PP). The second clade contains Po. syrman and Po. iranicus with shallow divergence, in which the placement of one Po. iranicus haplotype from Siah Darvishan with the Po. syrman haplotypes renders Po. iranicus paraphyletic. The other two clades comprise the “platyrostris” (Po. cephalargoides, Po. cyrius, Po. constructor, Po. platyrostris, and Po. rhodioni), and the “kessleri” (Po. eurycephalus, Po. gorlap, and Po. kessleri) species groups, respectively. Furthermore, two reciprocally monophyletic clades with deep divergence are strongly supported within Proterorhinus: a freshwater Proterorhinus clade including Pr. semilunaris, Pr. semipellucidus and Proterorhinus sp., and a marine/brackish water Proterorhinus clade including Pr. marmoratus and Pr. nasalis. Neogobiini are strongly supported (83% bootstrap, 0.97 PP) as the sister clade of Ponticolini.
Our hypothesis based on a single mitochondrial locus is very similar to the results derived from combined four gene analysis of Neilson and Stepien (2009a), except for the slightly different placement of Babka, and with regard to the sister group relationship of Ponticolini with Neogobiini rather than with Benthophilini; this grouping is, although, not robustly supported in the study of Neilson and Stepien (2009a).
The species delimitation analysis was performed using one distance-based (ABGD), one network-based (SP) and three tree/topology-based species delimitation methods (mPTP, bPTP, and bGMYC). Results are depicted as grey bars in Fig. 2. ABGD was the most conservative method, since it found support for 12 out of 27 nominal species included here, and lumped Po. eurycephalus and Po. kessleri, the “platyrostris” group elements (Po. cephalargoides, Po. cyrius, Po. constructor, Po. platyrostris, and Po. rhodioni), Po. syrman and Po. iranicus, the freshwater Proterorhinus species (Pr. semilunaris, Pr. semipellucidus, and Proterorhinus sp.), and Benthophilus taxa (Be. granulosus, Be. abdurahmanovi, Be. stellatus, Be. Mahmudbejovi, and Be. leobergius) into single entities. The other four methods retrieved similar results, providing robust supports for 18 out of 27 nominal species, and lumped Po. eurycephalus and Po. kessleri, Po. constructor, Po. platyrostris and Po. rhodioni, Po. syrman and Po. iranicus, and Be. stellatus, Be. mahmudbejovi, and Be. leobergius into single entities.
Using samples from the southern Caspian Sea basin for the first time enables us to reveal refined phylogeographic patterns of several Ponto-Caspian gobiid species. Sequence analysis of 49 marine and freshwater Proterorhinus specimens, spanning its native and invasive ranges, detected 126 variable nucleotide sites (4 singleton, 122 parsimony informative), leading to the definition of 16 haplotypes in 2 independent evolutionary clades with marked phylogenetic divergence (92% bootstrap, 1.00 PP; 5.4 Mya; Figs. 2, 3, 4), the marine and freshwater Proterorhinus clade. The marine clade comprises 2 deeply divergent lineages (100% bootstrap, 1.00 PP; 4.6 Mya) separated by 79 fixed substitutions: one from the Caspian Sea basin corresponding to Pr. nasalis (collected from the southern Caspian Sea basin); and the second from Black Sea basin corresponding to Pr. marmoratus. The freshwater clade comprises three primary lineages: one from the freshwater Caspian Sea basin corresponding to Pr. semipellucidus (Fig. 3a); the second from freshwater Black Sea basin (native range), Central Europe and the North American Great Lakes locations (invasive range) corresponding to Pr. semilunaris; and the third lineage with a single specimen corresponding to Proterorhinus sp. (Neilson & Stepien, 2009a) from the Kuma-Manych Depression. Among basin differences (Black Sea, Caspian Sea, Kuma-Manych Depression) within the freshwater clade showed very high supports in phylogenetic analyses (> 80% bootstrap, 1.00 PP; Fig. 2). ABGD lumped these freshwater lineages into a single putative species; however, the four other delimitation methods considered Pr. semilunaris + Proterorhinus sp., and Pr. semipellucidus as separate entities (Fig. 2). In addition, if the K2P distance species threshold value is accepted (> 2%: Ward, 2009; Kartavtsev, 2011), Pr. semilunaris and Pr. semipellucidus (3.1% sequence divergence) would represent separate species (Table S4).
TCS haplotype networks and maternal phylogeographies of 16 haplotypes (49 individuals) in Proterorhinus spp. (a), and 12 haplotypes (85 individuals) in Neogobius melanostomus (b). Each line without hatch marks/number of mutations between two neighboring haplotypes represents one mutational step. Circle sizes depict proportions of haplotypes; the smallest corresponds to one. Small black circles correspond to missing/hypothetical haplotypes. Distribution of Pr. semilunaris and the Black Sea lineage of N. melanostomus in their North American invasive ranges are not depicted
a Phylogeny of benthophiline gobies, based on a reduced data set of 53 haplotypes plus 3 outgroups. The phylogeny is calibrated with a legacy date of 6.25 Mya at the base of Proterorhinus + Mesogobius (from Neilson & Stepien, 2009a), and fossil calibration for the origin of “neogobiin” (10 Mya) derived from Rückert-Ülkümen (2006). Error bars indicate 95% highest posterior density. b Schematic reconstruction of the Caspian Sea water-level curve during the Pleistocene to Holocene
We analyzed 85 individuals of N. melanostomus and recovered 12 haplotypes in 3 lineages with shallow genealogical separations (Fig. 3b): 1 from the Caspian Sea basin corresponding to described subspecies N. m. affinis; one from the Black Sea basin (native range), European and North American locations (invasive ranges) corresponding to N. m. melanostomus; and one undescribed lineage confined to the southern Black Sea basin at Sinop, northern Turkey. All delimitation methods lumped these lineages into a single entity (Fig. 2).
Phylogeographic analysis of 25 individuals of Po. gorlap from 17 localities (15 in the southern Caspian Sea basin) detected 30 variable nucleotide sites (10 singleton, 23 parsimony informative), defining 19 haplotypes in 4 lineages with shallow separations (5–9 mutational steps; Fig. 5a): 1 from Lenkoran (brackish water); 2 sympatric lineages in the Babolrud (freshwater); and the most common and diverse lineage (17 individuals, 12 haplotypes) distributed across the southern Caspian Sea basin (fresh and brackish waters). All delimitation methods lumped these lineages into a single entity (Fig. 2).
TCS haplotype networks and maternal phylogeographies of 19 haplotypes (25 individuals) in Ponticola gorlap (a), and eight mitochondrial haplotypes (12 individuals) in Po. iranicus (b). Each line without hatch marks/number of mutations between two neighboring haplotypes represents one mutational step. Circle sizes depict proportions of haplotypes; the smallest corresponds to one. Small black circles correspond to missing/hypothetical haplotypes. The geographic locations of Narban (middle Caspian Sea) and Karpovska Reservoir (northern Caspian Sea) are not depicted on the map
A preliminary phylogeographic analysis of Po. iranicus is shown in Fig. 5b [ZM-CBSU P2782 (haplotype H3; see Fig. 2) was excluded from this analysis due to the possibility of mitochondrial introgression from Po. syrman]. Within its entire range in the southern Caspian Sea basin, 22 variable nucleotide sites (13 singletons, 9 parsimony informative) were detected among 11 Po. iranicus individuals, leading to the definition of eight mitochondrial haplotypes in 2 main lineages (1 with additional substructure) with shallow separation (8 mutational steps; Fig. 5b): one lineage confined to the Nowshahr River, the most distant locality; and a larger lineage (7 haplotypes) with further phylogeographic substructure in the study area. All delimitation methods lumped these two lineages into a single entity (Fig. 2).
Sequence analysis of K. caucasica detected 45 variable nucleotide sites (15 singleton and 30 parsimony informative) among 52 individuals, leading to the definition of 30 haplotypes in four main lineages (Hg1-Hg4) with shallow genealogical separations (Fig. 6): a diverse lineage (Hg1) in the Aegean Sea basin (Greece) with little substructure; a second lineage (Hg2) confined to Axios (Greece); a third lineage (Hg3) found in Rihios and Volvi (Greece), and other localities in the Black Sea basin [i.e., Simav (Turkey), Pomorijsko (Bulgaria), Sinoe (Romania), Kuban (Russia), Kodori (Georgia), and Don River (Russia)]; and a fourth lineage (Hg4) in the southern Caspian Sea basin (Anzali and Qarah Su). The mean K2P genetic distance between these four lineages varied between 0.01 (Hg3/Hg4) and 0.022 (Hg2/Hg4) (Table S5).
TCS haplotype network and maternal phylogeography of 30 mitochondrial haplotypes (52 individuals) in Knipowitschia caucasica. Hg1–Hg4 represent major lineages. Each line without hatch marks/number of mutations between two neighboring haplotypes represents one mutational step. Circle sizes depict proportions of haplotypes; the smallest corresponds to one. Small black circles correspond to missing/hypothetical haplotypes
For the Rhinogobius data set (87 sequences, 669 bp length), GTR + I + G was detected as the best-fitting substitution model. The saturation test of Xia’s et al. (2003) showed that the Rhinogobius sequences have not reached substitution saturation (Iss < Iss.c S/Iss.c A; Table S3), and their BI and ML phylogenies were not significantly different (SH test, P = 0.14). The BI phylogeny is presented in Fig. 7, and it includes novel sequences for 4 Rhinogobius individuals from Iran combined with 83 archived sequences (GenBank, ZFMK and BOLD-deposited) including 9 Rhinogobius individuals from Azerbaijan and Georgia. Several major clades are highly supported as being distinct within this phylogeny. Rhinogobius similis is the most distant species within the genus (100% bootstrap, 1.00 PP). Within the largest and most diverse clade, the four COI sequences developed for the Rhinogobius individuals collected from the Anzali Wetland (southern Caspian Sea basin) and the Zarivar Lake (Tigris River system) along with all Rhinogobius samples from Azerbaijan and Georgia defined one haplotype, closely related to the archived R. nagoyae haplotypes from Japan main islands, Honshuu (Nomura River at Akita, Seto River at Shizuoka, Migiaidu River at Wakayama, and Satsu River at Hyogo) and Kyuushuu (Saigou River at Fukuoka) (100% bootstrap, 1.00 PP). The R. nagoyae haplotypes from Okinawa (Okinawa and Iriomote islands) are placed in a separate clade with R. brunneus from Okinawa Island and the three other undescribed species (see Akihito et al., 2013; Yamasaki et al., 2015; Suzuki et al., 2020): Rhinogobius sp. YB (Kibara-Yoshinobori or yellow belly medium-egg type, distributed in Ryukyu Archipelago), Rhinogobius sp. DL (Hira-Yoshinobori, distributed in Yakushima-Iriomote-jima Island), and Rhinogobius sp. MO (Aya-Yoshinobori, distributed in Amamioshima-Kume-jima Island).
Discussion
Diversity, systematics, and ichthyogeography of tubenose gobies in the Ponto-Caspian basin
The expanded geographical area of molecular genetic investigation by adding tubenose goby samples from estuarine habitats of the southern Caspian Sea for the first time, as well as including modern statistical, phylogenetic, and species delimitation analyses allowed us to reassess hypotheses on the taxonomy, zoogeography, and evolution of tubenose gobies.
All populations of Proterorhinus were once classified as Pr. marmoratus (Pallas, 1814), originally described from Sevastopol, Ukraine (Kottelat, 1997; Pinchuk et al., 2004). Stepien and Tumeo (2006) used cytochrome b (Cyt b) data and considered two species in the genus, Pr. marmoratus as the marine species in the Black Sea and Pr. semilunaris (Heckel, 1837), as the freshwater species in other Ponto-Caspian habitats; however, Caspian Sea samples were not included in their study. Freyhof and Naseka (2007) restricted Pr. marmoratus to brackish waters in Sevastopol, and, based on samples primarily from marine regions of northern and middle Caspian Sea basin, concluded that the Caspian Sea basin specimens likely constitute another separate species, suggesting resurrection of Pr. nasalis (De Filippi, 1863), the oldest available name for the tubenose gobies in the Caspian Sea basin, originally described from near Baku, Azerbaijan. Neilson and Stepien (2009b) examined a concatenated nuclear and mitochondrial dataset and inferred the presence of at least three separate species, Pr. marmoratus from marine and estuarine habitats of the Black Sea; Pr. semilunaris from the freshwater Black Sea basin (also introduced to the North American Great Lakes); and they tentatively advocated for the name Pr. cf. semipellucidus as available taxon for another freshwater species inhabiting the Caspian Sea/Volga River basins. They further predicted Pr. nasalis to be the taxon distributed in the more saline waters of southern Caspian Sea. Sorokin et al. (2011) used Cyt b data and consider Pr. nasalis as the taxon widely distributed in the Caspian/Azov Sea basins, with Pr. semipellucidus as its synonym.
We found two primary Proterorhinus clades with marked genetic divergence, i.e., the marine and the freshwater clade. The marine clade comprises two independent evolutionary lineages with deep divergence: Pr. marmoratus in marine and estuarine habitats of the Black Sea basin, and Pr. nasalis in marine and estuarine habitats of the southern Caspian Sea. The freshwater clade comprises three lineages: Pr. semilunaris in the freshwater Black Sea basin (also introduced to the European rivers and North American Great Lakes); another freshwater lineage inhabiting the Sea of Azov and northern Caspian Sea/Volga River basins, which Neilson and Stepien (2009b) tentatively identified as Pr. cf semipellucidus; and the Proterorhinus sp. lineage from the Kuma-Manych Depression. The species delimitation analyses including SP, mPTP, bPTP, bGMYC, and the K2P distance species threshold value of > 2% congruently inferred Pr. semilunaris + Proterorhinus sp., and Pr. semipellucidus as two separate species. The pronounced phylogenetic distinction between Pr. semilunaris and Pr. semipellucidus is also registered in the sequence of mitochondrial Cyt b, and nuclear recombination activating gene 1 (RAG1) and S7 ribosomal protein gene (Neilson & Stepien, 2009b; Sorokin et al., 2011). Accordingly, we hypothesize Pr. nasalis to be the taxon in the more saline waters of the southern Caspian Sea, and, we hypothesize that the name Pr. semipellucidus for the Azov/northern Caspian Sea/Volga River basin populations will be resurrected depending on the outcome of additional morphological and molecular data in an integrative taxonomical approach. Morphological divergence among tubenose goby lineages appears not as marked as their genetic divergence (Freyhof & Naseka, 2007; Neilson & Stepien, 2009b), but pending additional molecular and/or morphological data, we would support taxonomic distinction of morphologically similar Proterorhinus species separated by deep molecular differences, since significant genetic divergence among morphologically cryptic species is not rare in Gobiidae (e.g., Lima et al., 2005; Victor, 2010, 2014; Hashimoto et al., 2014).
Based on mixed nature of samples at several localities, Sorokin et al. (2011) rejected the freshwater/marine Black Sea Proterorhinus species hypothesis erected by Neilson and Stepien (2009b). They suggested that Pr. semilunaris and Pr. marmoratus are two euryhaline species evolved in northwestern vs. northeastern part of the Black Sea basin, followed by recent expansion and secondary contact. Our results not only support the Neilson and Stepien (2009b) scenario, but also extend it to the Caspian Sea basin, because: (i) Sorokin’s et al. (2011) hypothesis entails a sister relationship between Pr. marmoratus and Pr. semilunaris, which our phylogeny does not support; (ii) a freshwater/marine Caspian Sea Proterorhinus species pattern is also present; (iii) the observed northwestern vs. northeastern distribution of Pr. semilunaris and Pr. marmoratus in the Black Sea basin might have resulted from unbalanced sampling, as samples from many parts of the Black Sea basin have not yet investigated in any genetic study; and (iv) only a freshwater/marine species hypothesis could better explain the successful invasion of freshwater preadapted Pr. semilunaris and the wider distributional range of Pr. semipellucidus, whereas Pr. marmoratus and Pr. nasalis, adapted to higher salinities would explain their predominant restriction to the marine and estuarine habitats of the Black Sea and southern Caspian Sea.
Based on our extended taxon sampling and preliminary time tree analysis, a revised hypothesis for the evolutionary and diversification history for tubenose gobies is possible. According to our preliminary node age estimates, Proterorhinus and Mesogobius separated approximately 6.3 Mya in the Pontian Lake-Sea (Neilson & Stepien, 2009b). In Late Miocene (Pontian) times, the Caspian and Black Sea basins were still connected (Popov et al., 2004; Krijgsman et al., 2010). About 5.4 Mya (Fig. 4), the first major division occurred, and due to decrease in salinity of the Late-Pontic and Kimmerian Lake-Seas (proto-Black Sea basin), the marine and freshwater tubenose goby clades were separated (Zaitsev & Mamaev, 1997; Reid & Orlova, 2002; Neilson & Stepien, 2009b). In the earliest Pliocene, the two basins became isolated after a major drop in water level in the far southward retreat of lacustrine environments (Van Baak et al., 2016), leading to a division between the Black and Caspian Sea marine tubenose goby lineages approximately 4.6 Mya (Fig. 4). The Black Sea and Caspian Sea freshwater tubenose goby lineages were separated about 1.9 Mya (early Pleistocene; Fig. 4) when freshwater tubenose gobies migrated during the Aspheronian transgression from the Gurian Lake-Sea into the Apsheron Lake-Sea across the Kuma-Manych Depression (Reid & Orlova, 2002; Cristescu et al., 2003; Neilson & Stepien, 2009b).
Generic assignment and systematics of the Caspian deep-water goby
The Caspian deep-water goby (Fig. 8) was originally described as Gobius bathybius by Kessler (1877) from Svinoi Island, south of Baku, Caspian Sea, Azerbaijan. The genus name Chasar appears in print for the first time in Berg (1949) as a subgenus of Neogobius to accommodate G. bathybius Kessler, 1877. Berg (1949) attributed this name to Iljin but without reference to any publication by the latter author. Berg (1949) provided a brief description of the species as Neogobius (Chasar) bathybius, but did not define the genus-group category. Although Vasil’eva (1996) stated that Iljin (1927, 1930) had used this subgeneric name for the classification of bathybius, a search of the latter publication by Miller (2004a) found this species mentioned only as being “incertae sedis” but without reference to any previous use of the name Chasar or to a definition by Iljin. Both Iljin (1956) and Ragimov (1967) used the name at a subgeneric level, but again provided no diagnosis. Pinchuk and Ragimov (1985), in their redescription of bathybius, placed this species in Neogobius without comment about a possible subgeneric location. The first generic diagnosis of Chasar, with indication of the type and only species, thus appears to be that by Vasil’eva (1996). The monotypic genus Chasar was recognized as a valid taxon by Miller (2004a) on the basis of the head sensory papillae patterns noted by Pinchuk and Ragimov (1985) and Vasil’eva (1996). The resulting paraphyly of Neogobius sensu lato (Berg, 1949) was changed in Neilson and Stepien’s (2009a) revised classification, by elevating two of Iljin’s (1927) subgenera to genus rank, i.e., Babka and Ponticola for the remainder of the ‘neogobiin’ species. The new classification thus recognized five genera, Proterorhinus, Mesogobius, Neogobius, Babka, and Ponticola. Neilson and Stepien (2009a) included bathybius in Ponticola in their molecular study without further justification, since they had not included any specimen of bathybius nor any discussion of the nominal genus. Here, for the first time, we presented sequence data from five specimens of bathybius from southern Caspian Sea deep waters, allowing us to suggest a revised generic assignment hypothesis, which should be tested with additional morphological and/or nuclear DNA data. The five bathybius sequences defined one haplotype, and in our extended phylogeny, the clade corresponding to the tribe Neogobiini (100% bootstrap support, 1.00 PP) comprises a now-restricted Neogobius (N. fluviatilis, N. pallasi, N. caspius, and N. melanostomus) and bathybius, as a distant sister species of N. melanostomus (100% bootstrap support, 1.00 PP). Therefore, bathybius is likely to be assigned to the genus Neogobius sensu stricto (Neilsen & Stepien, 2009a) as the fifth species, and the species would achieve a new combination as N. bathybius (Kessler, 1877). Neogobius caspius, N. pallasi, and N. bathybius are Caspian endemics, N. fluviatilis in the Black Sea is a sister species of N. pallasi, and N. melanostomus is native to both basins. Neogobius bathybius differs from other Neogobius species in their cheek sensory papillae pattern by featuring one additional transverse row before row b (Pinchuk & Ragimov, 1985). The presence of five transverse infraorbital rows before row b might be interpreted as a synapomorphy with Mesogobius, but N. bathybius does not have more than two such rows below row b, a plesiomorphic feature shared with the Neogobius and Ponticola (except for Po. syrman) but not with Mesogobius, which has three. As an alternative to our mitochondrial DNA (mtDNA)-based hypothesis, an ancient hybridization scenario with N. melanostomus as one partner and either a Ponticola or Mesogobius as another lineage might have led to the same mitochondrial clade phylogenetic pattern as a sister group relationship of bathybius with N. melanostomus, hereby highlighting the necessity of an integrative taxonomic approach to revise the generic classification of Neogobini and Ponticolini tribes. In the meantime, assignment of bathybius to Neogobius is nevertheless the most parsimonious solution, given the first phylogenetic evidence provided here.
South Caspian riverine species of Ponticola and the evolutionary history of Po. iranicus
The taxonomic composition in the genus Ponticola has been variable, unstable, and frequently uncertain especially in the group Po. cephalarges (Pallas, 1811), Po. platyrostris (Pallas, 1811), and Po. kessleri (Giinther, 1861); this is not only because of slight morphological and genetic differences and a mosaic pattern of morphological and karyological features, but also because of distinctive migratory and resident populations in some species. Thus, the presence of two Ponticola species in the southern Caspian Sea basin had become dubious. Ahnelt and Holcik (1996) collected gobies from four rivers, Massuleh, Siah Darvishan, Pasikhan, and Baham Bar, forming a part of watershed of the Anzali Wetland. They reported Po. cyrius from Massuleh and Pasikhan, and Po. iljini Vasil’eva and Vasil’ev, 1996 from Siah Darvishan and Baham Bar as new for Iran and the southern Caspian Sea, as well. Coad (1998) placed both species in context with the Iranian ichthyofauna. Neilson and Stepien (2009a) placed Po. iljini as a synonym of Po. gorlap in their revised classification, but they had not studied any material of this species. Vasil’eva et al. (2016) reestablished the validity of Po. iljini based on karyological data, but also limited its distribution to the coast of the Mangyshlak Peninsula, western Kazakhstan. Morphological as well as karyological analyses of specimens collected by Vasil’eva et al. (2015) from localities in Sefidrud and Gisum River revealed noticeable differences between those fishes and other known Ponticola species including Po. cyrius, leading to the description of Po. iranicus as the only endemic riverine gobiid species in the southern Caspian Sea basin. Esmaeili et al. (2010, 2014, 2017, 2018) followed Kottelat (1997) and Neilson and Stepien (2009a) in placing Po. iljini as a synonym of Po. gorlap, but listed Po. cyrius as a member of the Iranian and southern Caspian Sea freshwater fish fauna. Our extensive fieldwork between 2014 and 2020 and the phylogenetic results presented here can only support the presence of two species Po. gorlap and Po. iranicus in the riverine system and reservoirs of the southern Caspian Sea. In the rivers of the Anzali Wetland watershed that were investigated by Ahnelt and Holcik (1996), we only collected Po. gorlap and Po. iranicus from Siah Darvishan, Po. gorlap and Po. iranicus from Massuleh, and Po. iranicus from Baham Bar and Pasikhan (Figs. 9, 10). Here, we infer accordingly that fishes previously identified by Ahnelt and Holcik (1996) as Po. cyrius and Po. iljini from the Anzali Wetland watershed are conspecific with Po. iranicus and Po. gorlap, respectively; thus, P. iljini and P. cyrius should thus be omitted from the checklist of Iranian freshwater fish and the southern Caspian Sea.
The placement of one Po. iranicus haplotype from Siah Darvishan within the Po. syrman clade suggests possible introgression. Ponticola syrman differs noticeably from Po. iranicus in distribution, ecological requirements, coloration. Its morphological identification is unambiguously possible, especially with regard to differentiation from Po. syrman due to the presence of ctenoid scales on the nape (vs. nape scaled completely with cycloid scales), narrow upper lip of uniform width, never swollen (vs. upper lip expanding in middle and slightly swollen, with sharpened end), head depth usually greater than head width (vs. head width slightly larger than depth), anterior membrane of pelvic disc with very shallow, rounded lateral lobes (vs. anterior pelvic membrane with clearly pointed, but short lateral lobes), and three transverse infraorbital papillae rows below longitudinal hyomandibular row b (vs. two rows) (Miller, 2004b; Vasil’eva et al., 2015). In the southern Caspian Sea, Po. syrman has never been recorded from freshwaters, but it is found in inshore habitats, and less saline estuaries of Sefidrud, Shalmanrud, and Anzali Wetland, where Po. iranicus is also present (Abbasi, 2017). According to our preliminary node age estimate, Ponticola iranicus and Po. syrman separated at about 0.9 Mya (Fig. 4), a date almost coinciding with the Tyurkyanian regression at the end of the Pleistocene Apsheronian (Svitoch, 2012; Krijgsman et al., 2019). This low stand, around 150 m below sea level and with a temporal extent of ca. 100 ka, was one of the most severe regressions in the history of the Caspian Sea. We hypothesize that during this low stand, ancestors of contemporary Po. iranicus were isolated in freshwaters of the Anzali Wetland watershed, and, because of evolution under isolation, a high degree of morphological and ecological divergence was achieved rather quickly. Following the early Bakunian transgression, Po. iranicus has remained confined to the rivers of Anzali Wetland watershed. Therefore, it is possible that original reproductive barriers formed during speciation secondarily broke down during secondary contact in the estuaries after the early Bakunian transgression, allowing mitochondrial introgression. However, our result could simply be a result of incomplete lineage sorting (Pollard et al., 2006), although multilocus data will be required to test this hypothesis (for a review, see Rubinoff & Holland, 2005).
Taxonomic status, source, and dispersal pattern of the introduced Rhinogobius species into Iran, Turkmenistan and the Caucasus
The genus Rhinogobius with more than 86 species is the largest genus of freshwater gobies (Yamasaki et al., 2015; Fricke et al., 2020) widely distributed from East to Southeast Asia. The large variations in their life history and egg size resulted in radiation via colonization of novel habitats associated with the ecology of migration. An introduced Rhinogobius species has been reported from Iranian inland waters from the Kashaf and Hari Rivers in the Hari River basin (Feb 1996: Abdoli et al., 2000; Coad & Abdoli, 2000), the Anzali Wetland in the Caspian Sea basin [July 2007: K. Abbasi’s unpub. data in Coad (2016)], the Zarrineh River in Urmia Lake basin (July 2013: Eagderi & Moradi, 2017), the Jajrud River in the Namak Lake basin (2016: Eagderi et al., 2017), the Zarivar Lake (May 2017: Sadeghi et al., 2019), the Gaveh River (Sep 2016: Eagderi et al., 2018), the Eivashan River (Oct 2018: Eagderi et al., 2018), and the Tange-Hamam River (May 2018; Mousavi-Sabet et al., 2019), all in the Tigris River basin; and, most recently, from the Aras River in the Caspian Sea basin (Jouladeh-Roudbar et al., 2020). Aliev et al. (1988), Shakirova and Sukhanova (1994), and Sal’nikov (1995) refer to a Rhinogobius species from the Kara-Kum Canal in Turkmenistan without confirming the species identity. There have been controversial debates on the correct species identification of introduced Rhinogobius populations into Iran and Central Asia. Abdoli et al. (2000), and Coad and Abdoli (2000) provisionally identified the Hari River basin samples as R. similis Gill, 1859 and considered them conspecific with the species found in the Kara-Kum Canal. Based on this identification, these authors considered it an accidental introduction from the Amur River basin in eastern Asia penetrated into Iran via the Tedzhen/Hari River, although R. similis is more widely distributed (Suzuki et al., 2016). Vasil’eva and Kuga (2008) consider the Rhinogobius introduced to Central Asia as R. cheni (Nichols, 1931), a Chinese species of the Yangtze River drainage. The taxonomic work of Sadeghi et al. (2019) following a redescription of R. similis by Suzuki et al. (2016) show that the Iranian populations differ from both R. cheni and R. similis, and rather identified it R. lindbergi, the northernmost species of the genus described from Russia (Amur and Ussuri Rivers).
Rhinogobius taxonomy is plagued with considerable confusion; and molecular and morphological phylogenetic studies have targeted only a small set of species and have yet failed to obtain a robust phylogenetic hypothesis. Despite its limitations, the phylogeny presented here contains significant information about the Iranian and Caucasian Rhinogobius populations; (i) they belong to the same species and mitochondrial haplotype; (ii) it supports the taxonomic recognition by Sadeghi et al. (2019), indicating that the Iranian samples do not belong to R. similis; and (iii) Iranian and Caucasian samples are placed within in a particular clade (the R. brunneus species complex) with most, but not all, species being endemic to Japan. Within this clade, the Iranian and Caucasian samples are closely related to the R. nagoyae haplotypes collected by Yamasaki et al. (2015) from the mainland Japan. A total of 18 species of Rhinogobius are known from Japanese waters (all included here), 15 of which are endemic to Japan (Suzuki & Chen, 2011; Akihito et al., 2013; Yamasaki et al., 2015). In spite of the fact that their species status has been strongly supported by morphological, ecological, and genetic studies, scientific names of more than half of the species have not yet been published (Mizuno, 2001; Suzuki et al., 2011); rather, species codes consisting of two alphabet characters have been commonly used for such species (Fig. 6) (e.g., BB, YB, DL, MO, BW, TO, CO, OM, BF, KZ, and OR; Mizuno, 2001; Akihito et al., 2013). We were not able to include autochthonous R. lindbergi in our analysis; however, allozyme comparisons with seven Japanese congeners by Sakai et al. (2000) indicate that R. lindbergi is genetically only distantly related to these Japanese congeners. Therefore, the species identity of Iranian and Central Asian populations as R. lindbergi is ambiguous and must remain provisional.
Based on our results, an alternative hypothesis for the origin and colonisation of this Rhinogobius species in the Iranian, Central Asian, and Caucasian waters is possible, and strong support may come from another non-native fish in Iranian waters, the topmouth gudgeon Pseudorasbora parva (Temminck & Schlegel, 1846). Pseudorasbora parva (Teleostei: Gobionidae), native to East Asia, is one of the most successful invasive fish species in the world. The invasion history, historical records, and genetic background of the Iranian Rhinogobius species are similar to those of P. parva. In Iran, P. parva was first recorded on May 1991 from “Ab Bandans” of Avaness (vs. Feb 1996 for Rhinogobius in Hari River), and soon later from “Hesen Tebib” and “Shaheed Ziaee”, “Tir Tash”, all in the Caspian Sea basin with an approximate distance of 45–70 km to the Atrak River on the Iran–Turkmenistan border (Coad & Abdoli, 1993). Sal’nikov (1995, 1998) considers that fishes in the Tedzhen/Hari, Murgab, and Amu Darya Rivers of Turkmenistan may reach the Caspian Sea basin and conversely via the Atrek River basin. Following the initial discovery of P. parva in the Caspian Sea basin, specimens were found at fish ponds in Arak (central) and Mashhad (northeastern Iran; 140 km distance to Hari River at Sarakhs). Coad and Abdoli (1993), Coad and Abdoli (2000), Coad (1996), and Abdoli et al. (2000) recorded P. parva along with Rhinogobius in the Hari River in Iran. Welcomme (1981, 1988 in: Courtenay & Stauffer, 1984) also reported P. parva from the Kara-Kum Canal in Turkmenistan. Nowadays, P. parva is widely distributed and established in Iranian inland waters, and it has been found in all localities where Rhinogobius has been recorded (Coad, 2016; Eagderi et al., 2017, 2018; Eagderi & Moradi, 2017; Mousavi-Sabet et al., 2019; Sadeghi et al., 2019; Jouladeh-Roudbar et al., 2020). Recently, based on mtDNA control region and COI sequence variation of 161 samples collected from 15 Iranian localities and additional sites in Europe and Asia, Ganjali et al. (2020) characterized the pattern of genetic diversity and colonisation history of P. parva in Iran. Ganjali et al. (2020) show the presence of three P. parva haplotypes in Iran belonging to two distinct lineages: (ii) a Chinese lineage represented by a single haplotype found in the Shafarood River of the Caspian Sea basin; it penetrated recently to northern Iran through natural dispersal from Azerbaijan as an extension of the European wave of invasion; and (i) a widespread lineage with a common haplotype found throughout the country and one private haplotype confined to the Mashkid basin, originated from Japan through a single introduction of a small number of propagules and corresponded to the Iranian introduction of P. parva in the 1980s. After the Iran–Iraq war in 1988, the Iranian government began paying more attention to its fisheries and aquaculture industry (Karimpour et al., 2013). During this period, Japan was a good trading partner for Iran and the volume of Japanese exports to Iran was the second largest in the world, behind Germany (Nobuaki, 2012). Therefore, similarities in the genetic background, invasion history, and historical records in Iranian waters might indicate that the initial and simultaneous introduction of P. parva and Rhinogobius sp. into the Iranian inland waters originated from Japan as a by-product of Asian carp imports for aquaculture, possibly associated with the strong commercial link between Japan and Iran in the 1980s (Fig. 11). Following this initial introduction to the northeastern part of the southern Caspian Sea basin, both species penetrated eastward through the Atrak River basin into the Tedzhen/Hari River and Kara-Kum Canal in Turkmenistan. In addition, these species spread toward west and southwest in Iranian inland waters and historical records for both species support this conclusion. The Iranian Rhinogobius species has penetrated into Azerbaijan and recently, into Georgia (Epitashvili et al., 2020; Japoshvili et al., 2020), and is expected to be found in Armenian, and to enter the Black Sea basin very soon (Kuljanishvili et al., 2020). Similar to P. parva, the presence of only one Rhinogobius sp. mitochondrial haplotype in the inland waters of Iran and the Caucasus as well most likely refers to a single introduction of a small number of propagules.
Proposed invasion scenario for Rhinogobius sp. in the inland waters of Iran, Turkmenistan and the Caucasus based on genetic background and historical records of Rhinogobius sp. and Pseudorasbora parva. The yellow zone shows the area of initial introduction from Japan in the 1980s. Circled letters (a–g) represent the first recording sites of P. parva in the region in chronological order: a: Kara-Kum Canal (Welcomme, 1981, 1988); b: on 6 May 1991 from “Ab Bandans” of Avaness (37°03′ N, 54°47′ E); c: “Hesen Tebib” and “Shaheed Ziaee” (37°1′6″ N, 54°47′11″ E); d: “Tir Tash” and “Lemrask” (36°42′43.1382″ N, 53°44′25.4508″ E); e: Arak (34°05′ N, 49°41′ E); and f: Hari River (36°30′00.0″ N, 61°10′00.0″ E) (Coad & Abdoli, 1993). Circled numbers (1–10) represent the first recording sites of Rhinogobius sp. in the region in chronological order: 1: Kara-Kum Canal of Turkmenistan (Aliev et al., 1988); 2: Kashaf and Hari Rivers in the Hari River basin (Feb 1996: Abdoli et al., 2000; Coad & Abdoli, 2000); 3: Anzali Wetland in the Caspian Sea basin [July 2007: K. Abbasi unpub. data in Coad (2016)]; 4, Azerbaijan (Oct 2012; ZFMK, www.collections.zfmk.de); 5: Zarrineh River in Urmia Lake basin (July 2013: Eagderi & Moradi, 2017); 6: Jajrud river in the Namak Lake basin (2016: Eagderi et al., 2017); 7: Gaveh River (Sep 2016: Eagderi et al., 2018); 8: Georgia (Apr 2017; Japoshvili et al., 2020); 9: Zarivar Lake (May 2017: Sadeghi et al., 2019); 10: Eivashan River (Oct 2018: Eagderi et al., 2018); 11: Tange-Hamam River (May 2018; Mousavi-Sabet et al., 2019); and 12: Aras River (Jouladeh-Roudbar et al., 2020)
Phylogeography of the Caucasian dwarf goby
The Caucasian dwarf goby, K. caucasica, is a widespread Ponto-Caspian species (Berg, 1949; Svetovidov, 1964), in the Caspian Sea, Sea of Azov, and Black Sea, and also introduced to the Aral Sea but in addition found outside this region, in the Aegean and eastern Ionian catchments (Miller, 2004b). The specific name of caucasicus, proposed by Kawrajsky for museum use, was originally published as a nomen nudum by Radde (1899) and not until 1916 was the taxon described by Berg (with the type locality being a swamp near Batum and Lake Inkit near Pitzunda, Georgia, Black Sea), the name being attributed to Kawrajsky. The phylogeographic outcome retrieved here for K. caucasica is a shallow genealogy with major lineages mainly allopatric. Four main clades were identified among the Caucasian dwarf goby COI haplotypes, corresponding to three major hydrogeographic basins, the Aegean (Hg1–Hg2), Black (Hg3), and Caspian Sea (Hg4) basins. The implication is that contemporary gene flow has been low enough in relation to population size to have permitted lineage sorting and random drift (or, perhaps, diversifying selection) to promote genetic divergence among basins/populations that nonetheless were in historical contact recently. The phylogeographic break between the Caspian and Black Sea lineages (either population, subspecies, or species-level separations) was also documented for some mysid crustaceans (Audzijonyte et al., 2006), Pontogammarus amphipods and onychopod cladocerans (Cristescu et al., 2003), the round goby N. melanostomus (this study; Brown & Stepien, 2008), the cyprinid fish Rutilus frisii (Kotlik et al., 2008), the tubenose gobies of genus Proterorhinus (this study; Neilson & Stepien, 2009b), the chub Leuciscus cephalus (Durand et al., 1999), and the Eurasian monkey gobies N. fluviatilis and N. pallasi (Neilson & Stepien, 2011). These cases characterize what is implied by the principles of genealogical concordance; concordance in the geography of gene-tree partitions across multiple co-distributed taxa implicates shared historical biogeographic factors in shaping genealogies (Avise & Ball, 1990). This fact corresponds to the geological history of the Ponto-Caspian region, as the historic Black and Caspian Sea basins have been intermittently separated and connected over the past 5 Ma associated with Pliocene and Pleistocene glaciations (Reid & Orlova, 2002), promoting isolation, adaptation, and divergence into localized, distinct lineages in many taxonomic groups (Dumont, 1998, 2000; Naseka & Bogutskaya, 2009).
Management units (MUs) can be distinguished by considerable divergence in allele frequencies, regardless of depth in a genealogy (Moritz, 1994). Mitochondrial haplotypes are particularly important for distinguishing MUs because of their typical fourfold smaller effective population size (compared to haplotypes at autosomal loci), and because of their special relevance to demographic and reproductive connections among populations (Avise, 2000). Even shallow matrilineal subdivisions can be relevant to conservation efforts. Accordingly, we may consider populations of K. caucasica in the Caspian, Black, and Aegean Sea basins and Axios as separate MUs. However, since this phylogeographic outcome is established based on a limited number of K. caucasica specimens, one cautionary point should be made. As larger numbers of individuals are assayed, the power to detect a different pylogeographic pattern increases (Avise, 2000). For this reason, the shallow but allopatric phylogeographic pattern retrieved here for the Caucasian dwarf goby should be considered as preliminary.
Data availability
The COI sequence dataset generated and analyzed during this study is available in the GenBank repository. Specimens used in the present study are deposited as voucher specimens in the Zoological Museum of Shiraz University, Collection of Biology Department (ZM-CBSU), and the SNSB-Bavarian State Collection of Zoology, Munich (SNSB-ZSM).
References
Abbasi, K., 2017. Fishes of Guilan. Iliya Culture Publication, Rasht.
Abdoli, A., B. W. Coad & M. Naderi, 2000. First record of Rhinogobius similis, Gill 1895 in Iran. Iranian Journal of Fisheries Sciences 9: 73–76.
Agorreta, A., D. San Mauro, U. K. Schliewen, J. L. Van Tassell, M. Kovačić, R. Zardoya & L. Rüber, 2013. Molecular phylogenetics of Gobioidei and phylogenetic placement of European gobies. Molecular Phylogenetics and Evolution 69: 619–633.
Ahnelt, H. & J. Holcik, 1996. Distribution of two species of the genus Neogobius (Pisces: Gobiidae) in the catchment area of the southern Caspian Sea. Acta Universitatis Carolinae Biologica 40: 99–114.
Akihito, S. K., Y. Ikeda & M. Aizawa, 2013. Gobioidei. In: Nakabo, T. (ed), Fishes of Japan With Pictorial Keys to the Species. Tokai University Press, Tokyo, pp 1347–1608.
Aliev, D. S., A. I. Sukhanova & F. M. Shakirova, 1988. Fishes of the Inland Waters of Turkmenistan. Ylym, Ashkhabad.
Audzijonyte, A., M. E. Daneliya & R. Väinölä, (2006). Comparative phylogeography of Ponto‐Caspian mysid crustaceans: Isolation and exchange among dynamic inland sea basins. Molecular Ecology 15: 2969–2984.
Avise, J. C., 2000. Phylogeography: The History and Formation of Species. Harvard University Press, Cambridge.
Avise, J. C. & R. M. Ball, (1990). Principles of genealogical concordance in species concepts and biological taxonomy. Oxford Surveys in Evolutionary Biology 7: 45–67.
Baldwin, C. C., J. H. Mounts, D. G. Smith & L. A. Weigt, 2009. Genetic identification and color descriptions of early life-history stages of Belizean Phaeoptyx and Astrapogon (Teleostei: Apogonidae) with comments on identification of adult Phaeoptyx. Zootaxa 26: 1–22.
Berg, L. S., 1949. Freshwater Fishes of the USSR and Adjacent Countries. Program for Scientific Translations, Jerusalem.
Boldyrev, V. S. & N. G. Bogutskaya, 2007. Revision of the tadpole-gobies of the genus Benthophilus (Teleostei: Gobiidae). Ichthyological Exploration of Freshwaters 18: 31–96.
Brown, J. E. & C. A. Stepien, (2008). Ancient divisions, recent expansions: Phylogeography and population genetics of the round goby Apollonia melanostoma. Molecular Ecology 17: 2598–2615.
Bruford, M. W., O. Hanotte, J. F. Y. Brookfield & T. A. Burke, 1992. Single-locus and multilocus DNA fingerprinting. In: Hoezel, C. (ed), Molecular Genetics Analysis of Populations: A Practical Approach. Oxford University Press, New York, pp 225–269.
Carstens, B. C., T. A. Pelletier, N. M. Reid & J. D. Satler, 2013. How to fail at species delimitation. Molecular Ecology 22: 4369–4383.
Chang, C. H., K. T. Shao, H. Y. Lin, Y. C. Chiu, M. Y. Lee, S. H. Liu & P. L. Lin, 2017. DNA barcodes of the native ray‐finned fishes in Taiwan. Molecular Ecology Resources 17: 796–805.
Chen, W., X. Ma, Y. Shen, Y. Mao & S. He, 2015. The fish diversity in the upper reaches of the Salween River, Nujiang River, revealed by DNA barcoding. Scientific Reports 5: 1–12.
Clement, M., D. Posada & K. A. Crandall, 2000. TCS: A computer program to estimate gene genealogies. Molecular Ecology 9: 1657–1659.
Coad, B. W., 1996. Exotic and transplanted fishes in Southwest Asia. Publicaciones Especiales Instituto Español de Oceanografía 21: 81–106.
Coad, B. W., 1998. Systematic biodiversity in the freshwater fishes of Iran. Italian Journal of Zoology 65: 101–108.
Coad, B. W., 2016. Freshwater Fishes of Iran [available on internet at www.briancoad.com]. Accessed 12 Oct 2016.
Coad, B. W. & A. Abdoli, 1993. Exotic fish species in the fresh waters of Iran. Zoology in the Middle East 9: 65–80.
Coad, B. W. & A. Abdoli, 2000. Rhinogobius cf. similis Gill, 1859, a goby new to the fish fauna of Iran and the problem of alien invasions. Zoology in the Middle East 20: 55–59.
Courtenay, W. R. & J. R. Stauffer, 1984. Distribution, Biology, and Management of Exotic Fishes. Johns Hopkins University Press, Baltimore.
Cristescu, M. E. A., P. D. N. Hebert & T. M. Onciu, 2003. Phylogeography of Ponto‐Caspian crustaceans: A benthic–planktonic comparison. Molecular Ecology 12: 985–996.
Darriba, D., G. L. Taboada, R. Doallo & D. Posada, 2012. jModelTest 2: More models, new heuristics and parallel computing. Nature Methods 9: 772–772.
Drummond, A. J., M. A. Suchard, D. Xie & A. Rambaut, 2012. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Molecular Biology and Evolution 29: 1969–1973.
Dumont, H. J., 1998. The Caspian Lake: History, biota, structure, and function. Limnology and Oceanography 43: 44–52.
Dumont, H. J., 2000. Endemism in the Ponto-Caspian fauna, with special emphasis on the Onychopoda (Crustacea). Advances in Ecological Research 31: 181–196.
Durand, J. -D., H. Persat & Y. Bouvet, (1999). Phylogeography and postglacial dispersion of the chub (Leuciscus cephalus) in Europe. Molecular Ecology 8: 989–997.
Eagderi, S., A. Jouladeh-Roudbar, A. Soleymani & T. Hosseinpour, 2017. The first record of Rhinogobius similis Gill, 1859 from the Namak Basin, Iran. Shil 5: 39–46.
Eagderi, S. & M. Moradi, 2017. Range extension of the lake goby Rhinogobius similis Gill, 1859 (Teleost: Gobiidae) to Urmia Lake basin in northwestern Iran. Biharean Biologist 11: 123–125.
Eagderi, S., M. Nasri & E. Çiçek, 2018. First record of the Amur goby Rhinogobius lindbergi Berg 1933 (Gobiidae) from the Tigris River drainage, Iran. International Journal of Aquatic Biology 6: 202–207.
Economidis, P. S. & P. J. Miller, 1990. Systematics of freshwater gobies from Greece (Teleostei: Gobiidae). Journal of Zoology 221: 125–170.
Epitashvili, G., M. Geiger, J. J. Astrin, F. Herder, B. Japoshvili & L. Mumladze, 2020. Towards retrieving the Promethean treasure: A first molecular assessment of the freshwater fish diversity of Georgia. Biodiversity Data Journal 8: e57862.
Esmaeili, H. R., B. W. Coad, A. Gholamifard, N. Nazari & A. Teimory, 2010. Annotated checklist of the freshwater fishes of Iran. Zoosystematica Rossica 19: 361–386.
Esmaeili, H. R., W. C. Brian, H. R. Mehraban, M. Masoudi, R. Khaefi, K. Abbasi, H. Mostafavi & S. Vatandoust, 2014. An updated checklist of fishes of the Caspian Sea basin of Iran with a note on their zoogeography. Iranian Journal of Ichthyology 1: 152–184.
Esmaeili, H. R., H. Mehraban, K. Abbasi, Y. Keivany & W. C. Brian, 2017. Review and updated checklist of freshwater fishes of Iran: Taxonomy, distribution and conservation status. Iranian Journal of Ichthyology 4: 1–114.
Esmaeili, H. R., G. Sayyadzadeh, S. Eagderi & K. Abbasi, 2018. Checklist of freshwater fishes of Iran. FishTaxa 3: 1–95.
Freyhof, J., 2012. Diversity and distribution of freshwater gobies from the Mediterranean, the Black and Caspian Seas. In: Patzner, R. (ed), The Biology of Gobies. CRC Press, Boca Raton, pp 279–288.
Freyhof, J. & A. M. Naseka, 2007. Proterorhinus tataricus, a new tubenose goby from Crimea, Ukraine (Teleostei: Gobiidae). Ichthyological Exploration of Freshwaters 18: 325–334.
Fricke, R., W. N. Eschmeyer & R. Van der Laan, 2020. Eschmeyer’s Catalog of Fishes: Genera, Species, References [available on internet at http://researcharchive.calacademy.org/research/ichthyology/catalog/fishcatmain.asp]. Accessed 2 Feb 2020.
Ganjali, Z., H. R. Esmaeili, F. Zarei, G. Sayyadzadeh, S. Eagderi & R. E. Gozlan, 2020. West Asian colonisation of topmouth gudgeon, Pseudorasbora parva (Teleostei: Gobionidae): Genetic admixture at the crossroad of Europe and East Asia. Freshwater Biology 66: 1–17.
Geiger, M. F., F. Herder, M. T. Monaghan, V. Almada, R. Barbieri, M. Bariche, P. Berrebi, J. Bohlen, M. Casal‐Lopez, G. B. Delmastro, G. P. J. Denys, A. Dettai, I. Doadrio, E. Kalogianni, H. Kärst, M. Kottelat, M. Kovačić, M. Laporte, M. Lorenzoni, Z. Marčić, M. Özuluğ, A. Perdices, S. Perea, H. Persat, S. Porcelotti, C. Puzzi, J. Robalo, R. Šanda, M. Schneider, V. Šlechtová, M. Stoumboudi, S. Walter & J. Freyhof, 2014. Spatial heterogeneity in the Mediterranean Biodiversity Hotspot affects barcoding accuracy of its freshwater fishes. Molecular Ecology Resources 14: 1210–1221.
Goldman, N., J. P. Anderson & A. G. Rodrigo, 2000. Likelihood-based tests of topologies in phylogenetics. Systematic Biology 49: 652–670.
Hall, T. A., 1999. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series 41: 95–98.
Hart, M. W. & J. Sunday, 2007. Things fall apart: Biological species form unconnected parsimony networks. Biology Letters 3: 509–512.
Hashimoto, S., I. Koizumi, K. Takai & S. Higashi, 2014. Different habitat salinity between genetically divergent groups of a worm-like goby Luciogobius guttatus: An indication of cryptic species. Environmental Biology of Fishes 97: 1169–1177.
Iljin, B. S., 1927. A guide to the gobies (family Gobiidae) of the Azov and Black Seas. Trudy Azovo-Chernomorsky Nauchno Ekspeditsii 2: 128–143.
Iljin, B. S., 1928. Two new genera and a new species of Gobiidae from the Caspian Sea. Reports of the Astrakhan Scientific Fishery Station 6: 1–14.
Iljin, B. S., 1930. Le Système des Gobiidés. Ministerio de Fomento. Instituto Español de Oceanografía.
Iljin, B. S., 1956. Remarks and corrections to suborder Gobioidei in the book of L.S. Berg “Freshwater fishes of the USSR and neighbouring countries”, edit. 4, 1948–1949. Voprosy Ikhtyologii 7: 85–192.
Japoshvili, B., T. Lipinskaya, H. Gajduchenko, A. Sinchuk, A. Bikashvili & L. Mumladze, 2020. First DNA-based records of new alien freshwater species in the Republic of Georgia. Acta Zoologica Bulgarica 72: 545–551.
Jouladeh-Roudbar, A., H. R. Ghanavi & I. Doadrio, 2020. Ichthyofauna from Iranian freshwater: Annotated checklist, diagnosis, taxonomy, distribution and conservation assessment. Zoological Studies 59: 21.
Kapli, P., S. Lutteropp, J. Zhang, K. Kobert, P. Pavlidis, A. Stamatakis & T. Flouri, (2017). Multi-rate Poisson tree processes for single-locus species delimitation under maximum likelihood and Markov Chain Monte Carlo. Bioinformatics 33: 1630–1638.
Karimpour, M., M. M. Harlioglu, A. A. Khanipour, S. Abdolmalaki & Ö. Aksu, 2013. Present status of fisheries in Iran. Journal of FisheriesSciences.com 7: 161–177.
Kartavtsev, Y. P., 2011. Divergence at Cyt-b and Co-1 mtDNA genes on different taxonomic levels and genetics of speciation in animals. Mitochondrial DNA 22: 55–65.
Keskİn, E. & H. H. Atar, 2013. DNA barcoding commercially important fish species of Turkey. Molecular Ecology Resources 13: 788–797.
Kessler, K. F., 1877. Fishes distributed and found in the Aral-Caspian-Pontic ichthyological region. Trudy Aralo-Kaspiiskoi Ekspeditsii 4: 1–360.
Knebelsberger, T., A. R. Dunz, D. Neumann & M. F. Geiger, 2015. Molecular diversity of Germany’s freshwater fishes and lampreys assessed by DNA barcoding. Molecular Ecology Resources 15: 562–572.
Kotlik, P., S. Markova, L. Choleva, N. G. Bogutskaya, F. G. Ekmekci & P. P. Ivanova, (2008). Divergence with gene flow between Ponto‐Caspian refugia in an anadromous cyprinid Rutilus frisii revealed by multiple gene phylogeography. Molecular Ecology 17: 1076–1088.
Kottelat, M., 1997. European freshwater fishes. An heuristic checklist of the freshwater fishes of Europe (exclusive of former USSR), with an introduction for non-systematists and comments on nomenclature and conservation. Biologia (Bratislava) 52: 1–271.
Kottelat, M. & J. Freyhof, 2007. Handbook of European Freshwater Fishes. Kottelat, Cornol and Freyhof, Berlin.
Krijgsman, W., M. Stoica, I. Vasiliev & V. V. Popov, 2010. Rise and fall of the Paratethys Sea during the Messinian Salinity Crisis. Earth and Planetary Science Letters 290: 183–191.
Krijgsman, W., A. Tesakov, T. Yanina, S. Lazarev, G. Danukalova, C. G. C. Van Baak, J. Agustí, M. C. Alçiçek, E. Aliyeva & D. Bista, 2019. Quaternary time scales for the Pontocaspian domain: Interbasinal connectivity and faunal evolution. Earth-Science Reviews 188: 1–40.
Kuljanishvili, T., G. Epitashvili, J. Freyhof, B. Japoshvili, L. Kalous, B. Levin, N. Mustafayev, S. Ibrahimov, S. Pipoyan & L. Mumladze, 2020. Checklist of the freshwater fishes of Armenia, Azerbaijan and Georgia. Journal of Applied Ichthyology 36: 501–514.
Kumar, S., G. Stecher & K. Tamura, 2016. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution 33: 1870–1874.
Leigh, J. W. & D. Bryant, 2015. Popart: Full‐feature software for haplotype network construction. Methods in Ecology and Evolution 6: 1110–1116.
Lima, D., J. E. P. Freitas, M. E. Araujo & A. M. Solé-Cava, 2005. Genetic detection of cryptic species in the frillfin goby Bathygobius soporator. Journal of Experimental Marine Biology and Ecology 320: 211–223.
McCraney, W. T., C. E. Thacker & M. E. Alfaro, 2020. Supermatrix phylogeny resolves goby lineages and reveals unstable root of Gobiaria. Molecular Phylogenetics and Evolution 151: 106862.
Medvedev, D. A., P. A. Sorokin, V. P. Vasil’ev, N. V. Chernova & E. D. Vasil’eva, 2013. Reconstruction of phylogenetic relations of Ponto-Caspian gobies (Gobiidae, Perciformes) based on mitochondrial genome variation and some problems of their taxonomy. Journal of Ichthyology 53: 702–712.
Miller, P. J., 2003. The Freshwater Fishes of Europe, Vol. 8/I Mugilidae, Atherinidae, Atherinopsidae, Blenniidae, Odontobutidae, Gobiidae 1. AULA-Verlag GmbH Wiebelsheim, Verlag fur Wissenschaft und Forschung.
Miller, P. J., 2004a. Chasar Vasil’eva, 1996. In Miller, P. J. (ed), The Freshwater Fishes of Europe Vol. 8/II Gobiidae 2. AULA-Verlag GmbH Wiebelsheim, Verlag fur Wissenschaft und Forschung: 94–96.
Miller, P. J., 2004b. The Freshwater Fishes of Europe Vol. 8/II Gobiidae 2. AULA-Verlag GmbH Wiebelsheim, Verlag fur Wissenschaft und Forschung.
Miller, P. J., 2004c. Knipowitschia Iljin, 1927. In Miller, P. J. (ed), The Freshwater Fishes of Europe Vol. 8/II Gobiidae 2. AULA-Verlag GmbH Wiebelsheim, Verlag fur Wissenschaft und Forschung: 331–337.
Mizuno, N., 2001. Rhinogobius. In: Kawanabe, H., Mizuno N. & K. Hosoya (eds), Fresh Water Fishes of Japan. Yama-Kei Publishers, Tokyo, p 584.
Moritz, C., 1994. Applications of mitochondrial DNA analysis in conservation: A critical review. Molecular Ecology 3: 401–411.
Mousavi-Sabet, H., M. Amouei, M. Salehi, A. Salehi-Farsani & A. Heidari, 2019. Range extension and a new locality for the lake goby Rhinogobius lindbergi Berg, 1933 in the Upper Tigris River drainage, Iran. FishTaxa 4: 9–12.
Naseka, A. M. & N. G. Bogutskaya, 2009. Fishes of the Caspian Sea: Zoogeography and updated check-list. Zoosystematica Rossica 18: 295–317.
Neilson, M. E. & C. A. Stepien, 2009a. Escape from the Ponto-Caspian: Evolution and biogeography of an endemic goby species flock (Benthophilinae: Gobiidae: Teleostei). Molecular Phylogenetics and Evolution 52: 84–102.
Neilson, M. E. & C. A. Stepien, 2009b. Evolution and phylogeography of the tubenose goby genus Proterorhinus (Gobiidae: Teleostei): Evidence for new cryptic species. Biological Journal of the Linnean Society 96: 664–684.
Neilson, M. E. & C. A. Stepien, (2011). Historic speciation and recent colonization of Eurasian monkey gobies (Neogobius fluviatilis and N. pallasi) revealed by DNA sequences, microsatellites, and morphology. Diversity and Distributions 17: 688–702.
Nobuaki, K., 2012. Japan II. Diplomatic and commercial relations with Iran. Encyclopædia Iranica 14: 547–556.
Pinchuk, V. I. & D. B. Ragimov, 1985. The lateral line system in two endemic species of gobies of the Caspian Sea. Zoologichesky Zhurnal 64: 562–567.
Pinchuk, V. I., E. D. Vasil’eva, V. P. Vasil’ev & P. J. Miller, 2004. Proterorhinus marmoratus (Pallas, 1814). In Miller, P. J. (ed), The Freshwater Fishes of Europe Vol. 8/II Gobiidae 2. AULA-Verlag GmbH Wiebelsheim, Verlag fur Wissenschaft und Forschung: 72–93.
Pollard, D. A., V. N. Iyer, A. M. Moses & M. B. Eisen, 2006. Widespread discordance of gene trees with species tree in Drosophila: Evidence for incomplete lineage sorting. PLoS Genetics 2: e173.
Popov, S. V., F. Rögl, A. Y. Rozanov, F. F. Steininger, I. G. Shcherba & M. Kovac, 2004. Lithological-paleogeographic maps of Paratethys-10 maps Late Eocene to Pliocene. Courier Forschungsinstitut Senckenberg 250: 1–46.
Puillandre, N., A. Lambert, S. Brouillet & G. Achaz, 2012. ABGD, Automatic Barcode Gap Discovery for primary species delimitation. Molecular Ecology 21: 1864–1877.
R Core Team, 2013. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna [available on internet at http://www.R-project.org].
Radde, G., 1899. Die Sammlungen des Kaukasischen Museums. Band I. Zoologie, von Dr. Gustav Radde, Tiflis.
Ragimov, D. B., 1967. On the systematics of fishes belonging to the genus Gobius in the Caspian Sea. In Khalilov, R. A. (ed), Biological Productivity of the Kurinsk-Caspian Fishing Region. Trudy Zoologicheskogo Instituta Akademii Nauk SSSR: 252–277.
Rambaut, A. & A. J. Drummond, 2012. FigTree version 1.4.0. Institute of Evolutionary Biology, University of Edinburgh [available on internet at http://tree.bio.ed.ac.uk/]. Accessed 2 Feb 2020.
Rambaut, A., M. A. Suchard, D. Xie & A. J. Drummond, 2014. Tracer v1.6 [available on internet at http://beast.bio.ed.ac.uk/Tracer]. Accessed 2 Feb 2020.
Reid, D. F. & M. I. Orlova, 2002. Geological and evolutionary underpinnings for the success of Ponto-Caspian species invasions in the Baltic Sea and North American Great Lakes. Canadian Journal of Fisheries and Aquatic Sciences 59: 1144–1158.
Reid, N. M. & B. C. Carstens, 2012. Phylogenetic estimation error can decrease the accuracy of species delimitation: A Bayesian implementation of the general mixed Yule-coalescent model. BMC Evolutionary Biology 12: 196.
Rögl, F., 1999. Mediterranean and Paratethys. Facts and hypotheses of an Oligocene to Miocene paleogeography (short overview). Geologica Carpathica 50: 339–349.
Ronquist, F., M. Teslenko, P. Van Der Mark, D. L. Ayres, A. Darling, S. Höhna, B. Larget, L. Liu, M. A. Suchard & J. P. Huelsenbeck, 2012. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61: 539–542.
Ross, L. G. & B. Ross, 2009. Anaesthetic and Sedative Techniques for Aquatic Animals. Blackwell Publishing Ltd., Oxford.
Rubinoff, D. & B. S. Holland, 2005. Between two extremes: Mitochondrial DNA is neither the panacea nor the nemesis of phylogenetic and taxonomic inference. Systematic Biology 54: 952–961.
Rückert-Ülkümen, N., 2006. Otolithen aus dem Mio-Pliozän von Yalova bei Istanbul, Türkei. Neues Jahrbuch für Geologie und Paläontologie-Monatshefte 2006: 577–594.
Sadeghi, R., H. R. Esmaeili, F. Zarei, A. Esmaeili & K. Abbasi, 2019. The taxonomic status of an introduced freshwater goby of the genus Rhinogobius to Iran (Teleostei: Gobiidae). Zoology in the Middle East 65: 51–58.
Sakai, H., K. Ikoma, S. V. Frolov, Y. Yamazaki, H. Takahashi & H. Ida, 2000. Morphological features of a Russian freshwater goby, Rhinogobius lindbergi (Pisces: Gobiidae), and its genetic relationships to Japanese species. Biogeography 2: 51–61.
Sal'nikov, V. B., 1995. Possible changes in the composition of the ichthyofauna after completion of the Karakum Canal in Turkmenistan. Journal of Ichthyology 35: 108–121.
Sal’nikov, V. B., 1998. Anthropogenic migration of fish in Turkmenistan. Journal of Ichthyology 38: 591–602.
Schwarz, G., 1978. Estimating the dimension of a model. The Annals of Statistics 6: 461–464.
Schwarzhans, W., H. Ahnelt, G. Carnevale, S. Japundžić, K. Bradić & A. Bratishko, 2017. Otoliths in situ from Sarmatian (Middle Miocene) fishes of the Paratethys. Part III: Tales from the cradle of the Ponto-Caspian gobies. Swiss Journal of Palaeontology 136: 45–92.
Shakirova, F. M. & A. I. Sukhanova, 1994. Iktiofauna Turkmenistana (sostav i rasprostranenie) [Ichthyofauna of Turkmenistan (composition and distribution)]. Izvestiya Akademii Nauk Turkmenistana, Seriya Biologicheskikh Nauk 3: 35–45.
Shen, Y., L. Guan, D. Wang & X. Gan, 2016. DNA barcoding and evaluation of genetic diversity in Cyprinidae fish in the midstream of the Yangtze River. Ecology and Evolution 6: 2702–2713.
Shimodaira, H. & M. Hasegawa, 1999. Multiple comparisons of log-likelihoods with applications to phylogenetic inference. Molecular Biology and Evolution 16: 1114–1116.
Shimodaira, H. & M. Hasegawa, 2001. CONSEL: For assessing the confidence of phylogenetic tree selection. Bioinformatics 17: 1246–1247.
Sorokin, P. A., D. A. Medvedev, V. P. Vasil’ev & E. D. Vasil’eva, 2011. Further studies of mitochondrial genome variability in Ponto-Caspian Proterorhinus species (Actinopterygii: Perciformes: Gobiidae) and their taxonomic implications. Acta Ichthyologica et Piscatoria 41: 95–104.
Stamatakis, A., 2006. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22: 2688–2690.
Stepien, C. A. & M. A. Tumeo, 2006. Invasion genetics of Ponto-Caspian gobies in the Great Lakes: A ‘cryptic’ species, absence of founder effects, and comparative risk analysis. Biological Invasions 8: 61–78.
Suzuki, T. & I. S. Chen, 2011. Redescriptions of three species of genus Rhinogobius (Perciformes, Gobiidae) described by Dr. Shigeho Tanaka. Bulletin of the Osaka Museum of Natural History 65: 9–24.
Suzuki, T., I.-S. Chen & H. Senou, 2011. A new species of Rhinogobius Gill, 1859 (Teleostei: Gobiidae) from the Bonin Islands, Japan. Journal of Marine Science and Technology 19: 693–701.
Suzuki, T., N. Oseko, S. Kimura & K. Shibukawa, 2020. Two new species of torrential gobies of the genus Rhinogobius from the Ryukyu Islands, Japan. Bulletin of the Kanagawa Prefectural Museum Natural Science 2020: 7–28.
Suzuki, T., K. Shibukawa, H. Senou & I.-S. Chen, 2016. Redescription of Rhinogobius similis Gill, 1859 (Gobiidae: Gobionellinae), the type species of the genus Rhinogobius Gill, 1859, with designation of the neotype. Ichthyological Research 63: 227–238.
Svetovidov, A. N., 1964. Fishes of the Black Sea. Nauka, Moscow.
Svitoch, A. A., 2012. The Caspian Sea shelf during the Pleistocene regressive epochs. Oceanology 52: 526–539.
Thacker, C. E. & D. M. Roje, 2011. Phylogeny of Gobiidae and identification of gobiid lineages. Systematics and Biodiversity 9: 329–347.
Thacker, C. E., C. Gkenas, A. Triantafyllidis, S. Malavasi & I. Leonardos, 2019. Phylogeny, systematics and biogeography of the European sand gobies (Gobiiformes: Gobionellidae). Zoological Journal of the Linnean Society 185: 212–225.
Thalinger, B., J. Oehm, H. Mayr, A. Obwexer, C. Zeisler & M. Traugott, 2016. Molecular prey identification in Central European piscivores. Molecular Ecology Resources 16: 123–137.
Van Baak, C. G. C., M. Stoica, A. Grothe, E. Aliyeva & W. Krijgsman, 2016. Mediterranean-Paratethys connectivity during the Messinian salinity crisis: The Pontian of Azerbaijan. Global and Planetary Change 141: 63–81.
Vasil’eva, E. D., 1996. Skull morphology of the deep-sea goby Gobius bathybius Kessler in connection with its systematic position within the genus Gobius sensu lato (Gobiidae). Voprosy Ikhtyologii 36: 448–453.
Vasil’eva, E. D. & T. I. Kuga, 2008. Gobies of the genus Rhinogobius (Gobiidae) of Primorye and water bodies of Central Asia and Kazakhstan: II. Comparative craniological analysis of gobies introduced to Central Asia. Journal of Ichthyology 48: 29–36.
Vasil’eva, E. D., H. Mousavi-Sabet & V. P. Vasil’ev, 2015. Ponticola iranicus sp. nov. (Actinopterygii: Perciformes: Gobiidae) from the Caspian Sea basin. Acta Ichthyologica et Piscatoria 45: 189–197.
Vasil’eva, E. D., W. W. Schwarzhans, D. A. Medvedev & V. P. Vasil’ev, 2016. Cryptic species of Ponto-Caspian bighead goby of the genus Ponticola (Gobiidae). Journal of Ichthyology 56: 1–18.
Victor, B. C., 2010. The Redcheek Paradox: The mismatch between genetic and phenotypic divergence among deeply-divided mtDNA lineages in a coral-reef goby, with the description of two new cryptic species from the Caribbean Sea. Journal of the Ocean Science Foundation 3: 1–16.
Victor, B. C., 2014. Three new endemic cryptic species revealed by DNA barcoding of the gobies of the Cayman Islands (Teleostei: Gobiidae). Journal of the Ocean Science Foundation 12: 25–60.
Ward, R. D., 2009. DNA barcode divergence among species and genera of birds and fishes. Molecular Ecology Resources 9: 1077–1085.
Ward, R. D., T. S. Zemlak, B. H. Innes, P. R. Last & P. D. N. Hebert, 2005. DNA barcoding Australia’s fish species. Philosophical Transactions of the Royal Society B: Biological Sciences 360: 1847–1857.
Welcomme, R. L., 1981. Register of International Transfers of Inland Fish Species. FAO, Rome.
Welcomme, R. L., 1988. International Introductions of Inland Aquatic Species. FAO, Rome.
Xia, J.-H., H.-L. Wu, C.-H. Li, Y.-Q. Wu & S.-H. Liu, 2018. A new species of Rhinogobius (Pisces: Gobiidae), with analyses of its DNA barcode. Zootaxa 4407: 553–562.
Xia, X., 2018. DAMBE7: New and improved tools for data analysis in molecular biology and evolution. Molecular Biology and Evolution 35: 1550–1552.
Xia, X., Z. Xie, M. Salemi, L. Chen & Y. Wang, 2003. An index of substitution saturation and its application. Molecular Phylogenetics and Evolution 26: 1–7.
Yamasaki, Y. Y., M. Nishida, T. Suzuki, T. Mukai & K. Watanabe, 2015. Phylogeny, hybridization, and life history evolution of Rhinogobius gobies in Japan, inferred from multiple nuclear gene sequences. Molecular Phylogenetics and Evolution 90: 20–33.
Zaitsev, Y. & V. Mamaev, 1997. Marine Biological Diversity in the Black Sea. United Nations Publications, New York
Zhang, J., P. Kapli, P. Pavlidis & A. Stamatakis, 2013. A general species delimitation method with applications to phylogenetic placements. Bioinformatics 29: 2869–2876.
Acknowledgements
The authors thank Marcelo Kovačić (Natural History Museum Rijeka), Dirk Erpenbeck (Ludwig-Maximilians-Universität München), R. Sadeghi, Y. Bakhshi, H. Mehraban, M. Masoudi, and staff of the Anzali’s Inland Waters Aquaculture Research Center (IFSRI) for helping with fish collection in southern Caspian Sea basin. Our thanks are also due to Daisuke Kageyama (NARO, Japan) and Mahmood Soofi (University of Aberdeen) for sending us some old publications, and to M. Neilson (U.S. Geological Survey) and C. E. Thacker (Natural History Museum of Los Angeles County) for providing additional information on some archived materials in the GenBank repository. We thank two anonymous reviewers for their constructive remarks, which greatly enhanced the quality of manuscript. The research work was funded by Shiraz University (SU-9630190).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Ethical approval
All applicable international, national, and/or institutional guidelines for the care and use of animals were followed by the authors. All necessary permits for sampling have been obtained by the authors from the Iranian authorities. The research work was approved by Ethics Committee of Biology Department, Shiraz University (SU-9630190).
Additional information
Handling editor: Christian Sturmbauer.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zarei, F., Esmaeili, H.R., Schliewen, U.K. et al. Mitochondrial phylogeny, diversity, and ichthyogeography of gobies (Teleostei: Gobiidae) from the oldest and deepest Caspian sub-basin and tracing source and spread pattern of an introduced Rhinogobius species at the tricontinental crossroad. Hydrobiologia 848, 1267–1293 (2021). https://doi.org/10.1007/s10750-021-04521-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10750-021-04521-0