Abstract
Fully or semi-automated methods are becoming viable, cost-effective alternatives to manual approaches for characterizing zooplankton. The goal of this study was to compare a semi-automated approach (FlowCAM®) and a traditional microscopy method for characterizing zooplankton body size, density, and community structure. We demonstrate that estimating mass from FlowCAM® profile area had similar accuracy to a commonly used length to mass regression model. FlowCAM® and microscopy produced related length measurements for Daphnia, Calanoida, and Cyclopoida. Length measurements of rotifers, nauplii, and Sididae were not significantly related between the two methods, likely because of high morphological variation within taxa. Density comparisons between methods indicated high correlation between the semi-automated approach and microscopy-derived densities with a subtle bias of lower densities with the semi-automated method. After applying a correction factor, independent samples showed similar density estimates between methods, with community composition also not differing between methods. Comparison of processing time between the two methods showed that the semi-automated approach was 11 min (33%) faster per sample. With corrections, semi-automated methods represent a viable and cost-effective alternative to traditional microscopy methods for the processing of zooplankton samples.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Accurately characterizing plankton communities often requires large sample sizes and extensive processing time to account for spatial and temporal heterogeneity in freshwater (e.g., Lewis, 1979; Pinel-Alloul et al., 1999; Detmer et al., 2017a) and marine environments (e.g., Calbet et al., 2001; Siokou-Frangou et al., 2008). The characterization of plankton communities has traditionally been accomplished using microscopy techniques, which can be laborious and subject to a degree of variation among individuals processing samples (Culverhouse et al., 2003). To reduce sample processing time and improve accuracy, automated (i.e., no post-processing) and semi-automated (i.e., post-processing required) methods using novel hardware and software technologies have been developed (e.g., Davis et al., 1992; Sieracki et al., 1998; Grosjean et al., 2004; Culverhouse et al., 2006; Benfield et al., 2007; Gorsky et al., 2010). Automated and semi-automated plankton imaging methods are now widely available with the capacity to quickly extract large amounts of information from imaged data sets. Although technological advances have the potential to expedite plankton sample processing, the utility of the method depends on reliability—here defined as a machine’s ability to accurately quantify plankton body dimensions, densities, and community composition—in a timely manner.
Accurate measures of plankton body size and abundance are necessary to estimate and characterize many important metrics including size frequency distributions, individual and standing crop biomass, secondary production, filtration capacity, and resource availability to higher trophic levels (e.g., Makarewicz & Likens, 1979; Hamilton et al., 1990; Detmer et al., 2017b; Collins et al., 2018). Flow-through imaging systems coupled with novel machine learning algorithms can simultaneously identify and calculate multiple metrics of imaged particles, potentially yielding rapid and accurate estimates of individual body characteristics across the plankton size spectrum. Yet, like any tool or method, there is the potential for methodological biases that, if unaccounted for, can influence the interpretability of data. For phytoplankton, several studies have provided evidence for, mechanisms to explain, and methods to resolve biases in particle size and abundance estimates between automated and microscopy methods (e.g., Jakobsen & Carstensen, 2011; Moberg & Sosik, 2012; Álvarez et al., 2014). In contrast, methodological comparisons for assessing larger planktonic particles such as mesozooplankton taxa (i.e., > 200 μm) are less prevalent (but see Kydd et al., 2017; Wong et al., 2017). Complex body morphology, variation in organism orientation during imaging (e.g., portrait vs. side profile), errors during the instrument imaging process, and the capture of partial body images have been cited as potentially introducing biases through flow cytometer imaging errors (Kydd et al., 2017; Wong et al., 2017). If unaccounted for, such biases can skew particle size estimates, frequency distributions, and other important ecological metrics.
Taxon-specific and community-level analyses are important for understanding population demographics, food web dynamics, and ecosystem function (e.g., Makarewicz & Likens, 1979; Saunders & Lewis, 1988). Thus, biases concerning the numbers and types of particles imaged may affect population- and community-level indices. Studies comparing density estimates of phytoplankton and zooplankton between microscopy and image analysis methods are numerous and have identified congruence between traditional and novel approaches in some instances (e.g., Sieracki et al., 1998; Busky & Hyatt, 2006; Ide et al., 2008; Zarauz & Irigoien, 2008) and a lack of congruence in others (e.g., Jakobsen & Carstensen, 2011; Kydd et al., 2017; Álvarez et al., 2014; Garmendia et al., 2013). Inconsistencies between methods for mesozooplankton density estimates warrant exploration and rectification (Ide et al., 2008; LeBourg et al., 2015; Kydd et al., 2017; Stanislawczyk et al., 2018). To the knowledge of the authors, no community-level comparison between microscopy and image analysis methods has been published for zooplankton. Because community-level analyses depend on accurate density estimates of populations, corrections for any density biases should also improve confidence in zooplankton community metrics, as has been addressed in macroinvertebrate gear comparisons (Turner & Trexler, 1997; García-Criado & Trigal, 2005).
We evaluated the reliability of a semi-automated method by comparing it to traditional microscopy. Our goals were to (1) characterize detection rates and body sizes, (2) assess the accuracy of population and community analyses, and (3) determine processing efficiency in terms of processing time per sample. In the first section, we evaluated the influence of zooplankton body size on instrument detection rates, the number of individuals imaged relative to the number of individuals that passed through the flow cell. We further quantified and contrasted body mass estimates derived from two-dimensional profile area (i.e., the area of the zooplankton delineated from the image generated by FlowCAM®) and microscopy-derived body length. We also compared lengths of several zooplankton taxa from Illinois reservoirs between FlowCAM® and microscopy. In the second section, to assess population and community metrics, we first evaluated the relationship between taxa-specific density estimates from FlowCAM® to density estimates from a standard microscopy method. We next determined if a systematic correction was required to adjust for any methodological biases. We used taxa-specific densities to evaluate congruence between FlowCAM® and microscopy characterization of zooplankton community structure and to determine if taxonomic biases exist between methods. Finally in the third section, processing efficiency (i.e., time per sample) of FlowCAM® was compared with microscopy to estimate the time–cost savings of the semi-automated method.
Methods
Organisms
Zooplankton in the present study were obtained from mesocosms and a routine reservoir sampling program in the state of Illinois. From aquatic mesocosms housed at the Kaskaskia Biological Station (Sullivan, Illinois, USA), Daphnia pulex Leydig, 1860 were collected, narcotized with sodium bicarbonate, and processed. D. pulex in the mesocosms originated from several nearby reservoirs and have been self-sustaining for approximately 9 years. D. pulex was chosen as a model organism because of its widespread use in laboratory studies (Pennak, 1973; Krueger & Dodson, 1981; Thorp & Covich, 1991). In addition, zooplankton were collected from 12 central Illinois reservoirs, sampled from May to September of 2016 and 2017 and preserved in 4% Lugols solution. Eight samples were collected per reservoir (four nearshore and four offshore) using a 63-µm zooplankton net with a diameter of 0.5 m and width to depth ratio of 1:3. Collections occurred at inshore sites at a depth of 1 m and at offshore sites to a depth where dissolved oxygen dropped below 3 ppm (see Detmer et al., 2019).
Instrument configuration and procedure for FlowCAM® and microscopy
All analyses were completed using the FlowCAM® VS-1 benchtop model and its paired particle analysis software (VisualSpreadsheet© version 4.1.95) to enumerate and measure body characteristics of individuals. Samples were run using one of two configurations: one for mesozooplankton taxa (mean taxon size > 200 µm; e.g., Daphnia, Copepoda, Bosminidae) and one for microzooplankton taxa (mean taxon size < 200 µm; e.g., nauplii and Rotifera). For objectives characterizing a range of zooplankton sizes, samples were first separated as mesozooplankton and microzooplankton using a layered sieve consisting of 200 µm and 63 µm, respectively. Mesozooplankton runs used a configuration of a 2× objective lens, D12 thick wall collimator, FC1000FV Field of View flow cell (1000 μm depth, 3000 μm width), 0.5-cm inner diameter tubing, an imaging rate of 20 frames/second, and a flow rate of 7.5 ml/min. The D12 thick collimator was used for even illumination and the FC1000FV Field of View flow cell was used to permit organisms up to 2.5 mm in size to be reliably processed, according to the manufacturer’s specifications. Microzooplankton runs used a 4× objective lens, FC300 flow cell (300 μm depth, 3000 μm width), 0.16 cm inner diameter tubing, an imaging rate of 20 frames/s, and a flow rate of 2 ml/min. AutoImage mode was used to process both groups as this setting is recommended for both high density (Fluid Imaging Technologies Inc., 2011) and preserved samples (Poulton, 2016). Both processing methods used a 12.5-ml internal syringe pump (model C-70) and a magnetic stir plate was used to ensure uniform sample distribution. Samples were diluted (20–400 ml depending on sample density) to reduce zooplankton density and decrease the likelihood of clogging the device during processing. Samples were diluted to > 50 ml when the density was estimated at > 1000 individuals per liter (~10% of samples), between 20 and 50 ml where density was estimated at 400–1000 individuals per liter (~40% of samples), and 20 ml where density was estimated at < 400 individuals per liter (~ 50% of samples). Processing included imaging 10% of the volume for mesozooplankton runs and 5% for microzooplankton runs, except in evaluations of detection rate when known numbers of individuals were passed through.
Once the sample was photographed, images were auto-identified by the VisualSpreadsheet© software and categorized by taxonomic group. Image libraries of organisms categorized by taxa were compiled during a pilot period and were used as a reference for the auto-categorization of organisms processed (see Supplemental Table 1). An additional step of manual post-processing validation was included because sorting accuracy can vary between runs. Once images were correctly sorted, the total number of images per taxa was recorded and densities derived. Zooplankton identification was based on keys from Thorp & Covich (1991). Particles lengths were then estimated for each identified organism. Length estimates were the maximum Feret diameter, or the maximum distance between two tangents made by the outline of the particle, and area-based diameter (ABD), or the diameter of the area of the particle when shaped as a circle (Fluid Imaging Technologies Inc., 2011). Length estimates for both particle metrics in a sample represented the sample mean for each taxon.
Sample processing by microscopy for field-based samples began with subsampling by 1 ml aliquots from concentrated samples described above using a Hensen-Stemple pipette (see Detmer et al., 2019). Zooplankton were then identified (Thorp & Covich, 1991) and whole subsamples counted until at least 200 organisms of all taxa (except water mites and mussel veligers) were enumerated or until 10% of the total sample was counted (Welker et al., 1994). For each taxon, body lengths (i.e., total body length excluding spines; mm) were measured for all individuals when fewer than 20 were encountered or 20 random individuals for more numerous taxa using a digitizing pad and the Java image processing software ImageJ (version 1.45). All statistical analyses were performed in R version 3.0 statistical program (R Core team, 2013) and effects were considered significant at α = 0.05.
Characterization of detection rate and body size
Evaluation of the instrument’s capacity to detect and measure zooplankton was evaluated in three components. First, we evaluated detection rate, defined as the proportion of organisms imaged to the number of organisms passed through the flow cell, of the FlowCAM® for different size classes of D. pulex. Second, we explored whether two-dimensional profile area, which can easily be measured with semi-automated methods such as FlowCAM®, could be used as an alternate and more rapid approach to estimating dry-weight body mass. Specifically, we compared FlowCAM® and traditional approaches for estimating dry-weight body mass of D. pulex by relating measured dry-weight body mass to two-dimensional profile area derived from FlowCAM as well as to estimates of body mass using an established, widely used length to dry-weight body mass relationship (Dumont et al., 1975). Third, we compared lengths derived from FlowCAM® and microscopy for several taxa from samples collected in Illinois reservoirs.
For the first component, to assess detection rates of FlowCAM® across a range of D. pulex body sizes, we measured, sorted, and enumerated similarly sized individuals into aggregated size classes, described here as bins because there were multiple replicates in each size class. Detection rate processing was unique from general processing of zooplankton because a known number of individuals were processed independent of volume. Size class binning of zooplankton for analyses is described by Dumont et al. (1975). Each size class bin replicate included 10–16 individual zooplankton. There were 4–6 replicates per each size class bin and designated size class bins were 0.25 mm and ranged from 0.50 to 1.75 mm. Mean microscopy body lengths were calculated per bin. Each bin was independently processed through the FlowCAM® and the proportion of individuals imaged relative to the number of individuals in the aggregate bin was recorded as the detection rate. We related detection rate (dependent variable) to mean FlowCAM® D. pulex body length (independent variable).
In the second component, we examined if individual profile area metrics of D. pulex derived from FlowCAM® provide accurate estimates of individual dry-weight body mass. We compared relationships of measured dry-weight body mass (i.e., balance-derived weights of D. pulex) to profile area and estimated mass from a traditional microscopy method that uses length measurements to estimate mass. Because dry-weight masses of individual zooplankton at the lower end of the size spectrum evaluated were below the limit of the scale, we followed a method similar to the previous section where all D. pulex were measured under a compound microscope and individuals of similar sizes sorted into 0.25-mm-size bins ranging from 0.5 to 1.75 mm. For this component, there were four replicates per each size class bin and in each size class bin replicate, count and mean length of each size-aggregated sample were recorded with the number of individuals, which ranged from 2 to 19 individual zooplankton. Because of risk of physical damage to individuals during FlowCAM® processing and subsequent alteration of individual D. pulex body metrics, we split the 4 replicate size class bins into two paired, equivalent groups where D. pulex were either (1) dried and weighed for group mass (dry-weight), or (2) processed for profile area (ABD; µm2) with FlowCAM® (profile group). Thus, each size class bin had two replicates for each method and there were 12 replicates in total for each method. Pairs of size groups that were divided to be dried and weighed or processed by FlowCAM® did not differ in microscopy body length (Student’s t test, P > 0.05) and method of processing was randomly assigned. For the dry-weight group, we followed the methods of Dumont et al., (1975) for drying and weighing, using a Cahn C-35 microbalance, and report mean dry-weight per individual (µg), which was calculated by dividing the group mass by the number of individuals. Mean individual dry-weight mass for each individual bin was also predicted from microscopy lengths using a length to dry-weight regression equation (Dumont et al., 1975), and linear regression was used to evaluate the relationship between measured and predicted mass (Dumont et al., 1975). Similarly, mean D. pulex dry-weight body mass from the group that was weighed was related to FlowCAM® mean individual profile area. Data were log10 transformed for lengths from both methods to meet the assumptions of simple linear regression. The two linear models were then compared using Akaike’s information criterion corrected for small sample size (AICc; Burnham and Anderson, 2002) to determine whether FlowCAM® body profile area was superior, inferior, or similar in predicting body mass as a commonly used length to mass regression model from Dumont et al. (1975). Models with the lowest AICc values have the most relative support from the data (Burnham & Anderson, 2002), and we considered models within 2 AICc units as having similar levels of support.
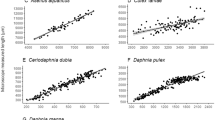
In the third and final component of this section, we compared body size measurements between the two methods using zooplankton samples, 35 total, from a routine reservoir sampling program in Illinois. Body sizes of common and abundant zooplankton taxa were estimated via the FlowCAM® and microscopy. Common and abundant was defined as being present in two or more reservoirs and in samples where ten or more individuals were measured for both methods for each taxon (Table 1). Mean body length estimates between methods were tested with model II regression where FlowCAM® length was related to microscopy length. Model II regression was used because size was not experimentally controlled.
Characterization of populations and communities
Samples collected from Illinois reservoirs were used for population and community analyses. Meso- and microzooplankton methodologies for density were compared with an analysis of covariance (ANCOVA), where the two techniques were used as the covariate, to test whether the semi-automated approach processed zooplankton-size categories differently. Thirty of the original 35 samples were initially included in the analysis, with the remaining five samples withheld as an independent data set for assessing correction factors derived from linear regression models relating density estimates from FlowCAM® and microscopy approaches. If an interaction or difference in intercept was observed, each method would be characterized separately. Model II regression models were used to algebraically derive coefficients (i.e., slope, intercept) to develop model corrections for each method separately. Coefficients were used to adjust for bias so that the relationship between the FlowCAM® and microscopy methods was 1:1 and the intercept to zero (e.g., if the slope of the relationship between the two methods indicated that FlowCAM®-derived density was only 50% of organisms evaluated with the microscope, the correction would be 2×). To quantify the validity of the correction, accuracy was estimated for the five samples that were withheld as test data. Accuracy here is defined as the percentage difference from 100% (a 1:1 ratio) for the corrected FlowCAM® density value divided by the density derived from microscopy. Accuracy was evaluated with a one sample t test to evaluate whether densities were consistent with an ideal ratio of 1:1 for FlowCAM® density to microscopy density. Density data for these analyses were log10 (x + 10) transformed.
Densities from microscopy and corrected densities from FlowCAM® were used to compare community composition estimates between the two methods. To visualize community composition differences between methods, non-metric multidimensional scaling was used (NMDS; Bray & Curtis, 1957; Minchin, 1987). If microscopy and FlowCAM® characterize zooplankton communities similarly, we predict no significant differences between methods. This was evaluated with a permutational analysis of variance (PERMANOVA) via the “adonis” function within the vegan package in R. Data for this analysis were log10 (x + 10) transformed.
Processing efficiency
We recorded the total processing time with a stopwatch of both microscopy and FlowCAM® methods. Total processing times were recorded beginning from taking the sample off the shelf to the completion of the data sheet (the same data sheet was used for both methods); this included instrument setup, pre-processing preparation of samples, post-processing validation of automated classifications, enumeration, and measurements. Simple linear regression was used to relate processing times of FlowCAM® and microscopy.
Results
Characterization of detection rate and body size
Detection rates of FlowCAM® ranged from a low of 19% to a high of 80% (Fig. 1), and declined with increasing body length at a rate of 30% per unit length (P < 0.01, adjusted R2 = 0.40). The flow cell used in our configuration has a maximum suggested size threshold of 2.5-mm particles that can pass through the system. Yet, the relationship between body length and detection rate suggests that detection efficiencies are low (18%) for larger particles, even when below this maximum threshold. Specific values reported here are valid for the frames per second and flow rate configuration of our device and may vary slightly given alternative configurations.
Relationship between FlowCAM® detection rate, the number of individuals imaged relative to the number of individuals that passed through the flow cell, and mean lengths of Daphnia pulex (detection rate = 0.81 − 0.30 × microscopy length). Mean lengths were calculated from each 0.25-mm-size bin between 0.5 and 1.75 mm, with 10 – 16 D. pulex per bin. Detection rates were determined with a device configuration of 20 frames/s and a flow rate of 7.5 ml/min
A strong positive relationship was detected between the measured dry-weight mass per individual and mean profile area derived from FlowCAM® (adjusted R2 = 0.91, P < 0.01; Fig. 2A). Similarly, a strong positive relationship was detected between mean measured and predicted dry-weight masses of D. pulex obtained from published length–mass relationships (Dumont et al., 1975; adjusted R2 = 0.89, P < 0.01; Fig. 2B). Model comparison with AICc showed that the Dumont et al. (1975; AICc = − 17.99) and the semi-automated (AICc = − 20.32) models were nearly indistinguishable, indicating that both methods are equally qualified at predicting dry-weight body mass.
Taxa-specific relationships between microscopy and FlowCAM® were variable for length estimation. Calanoida, Cyclopoida, and Daphnia each exhibited strong positive relationships between each of the FlowCAM® particle properties (maximum feret diameter and ABD) and microscopy measurements, whereas no relationships were detected for nauplii, Rotifera, and Sididae (Table 1). For all taxonomic groups, FlowCAM® maximum feret lengths were similar to or exceeded those from microscopy, indicating that maximum feret length measures from FlowCAM® was likely including measurements of appendages and/or spines not included in standard microscopy-derived length estimations. Conversely, FlowCAM® ABD lengths were consistently lower than microscopy measurements for all taxonomic groups (Table 1) indicating that body shape diverged strongly from circular. Despite variation in the relationships between the two methods for the various taxa, all differences from microscopy to FlowCAM® maximum feret lengths were ≤ 10%, except for nauplii (13%), Rotifera (31%), and Sididae (15%). Collectively, zooplankton body lengths for all taxa from all reservoir samples were strongly similar between the feret length from FlowCAM® and microscopy (Fig. 3; Table 1; P < 0.01, R2 = 0.93). Agreement between methods across taxa and relatively small differences within the taxonomic groups evaluated indicate that although precision is not always high for a particular taxon, community body size structure estimates are likely reliable and corrections possible to improve estimates.
Characterization of populations and communities
Because of interaction between macro- and microzooplankton methods (F1,253 = 1.50, P = 0.03), densities of each size group were analyzed independently. Regression of density estimates for mesozooplankton (adjusted R2 = 0.69, P < 0.01; Fig. 4) and microzooplankton (adjusted R2 = 0.51, P < 0.01) methods were positively related between the semi-automated and microscopy techniques.
Comparison between microscopy and semi-automated density estimates for microzooplankton (empty triangles; log10 (FlowCAM® density + 10) = − 0.51 + 1.06 × log10 (microscopy density + 10)) and mesozooplankton (filled circles; log10 (FlowCAM® density + 10) = − 0.05 + 0.80 × log10 (microscopy density + 10)), with a dotted line for the 1:1 relationship
Accuracy after the correction was high for both methods. The percentage difference between FlowCAM®-corrected and microscopy values for mesozooplankton was 4.5% ± 4.9% (mean ± SE) and did not differ significantly from an ideal ratio of 1:1 (one sample t test, t32 = 1.10, P = 0.28). A similar pattern was observed for microzooplankton, where the percentage difference between FlowCAM®-corrected and microscopy-derived density estimates was low, with a mean difference of 2.1% (± 3.5% SE) that did not differ from an ideal 1:1 ratio (t9 = − 0.61, P = 0.56).
After applying the density correction, zooplankton community structure did not differ between microscopy and FlowCAM® (PERMANOVA; F1,59 = 0.87, P = 0.51). The NMDS supported the PERMANOVA and showed that reservoir samples processed by FlowCAM® had a very similar community structure to those derived by microscopy and no systematic biases were observed, as evidenced by the close proximity of data points between methods for a given sample (Fig. 5). Strong congruence between methods suggests the absence of taxonomic bias.
NMDS plot of zooplankton community structure derived from microscopy and FlowCAM® methods applied to samples collected from Illinois reservoirs. Open circles are microscopy-derived values and closed circles represent community structure quantified from corrected FlowCAM® densities. Each sample-method couplet is connected by a line to show the distance between methods in their sample-specific community measurement. NMDS 2D stress was 0.20
Processing efficiency
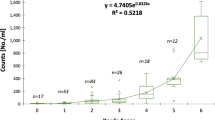
Total sample processing time for microscopy was greater (34 ± 3 min; mean ± 95% CI) than the FlowCAM® method (23 ± 3 min; mean ± 95% CI). Total processing time via the FlowCAM® approach, including the post-processing validation step, was 11 min (± 2 min, 95% CI) or 33% (± 5%, 95% CI) faster, on average, than the standard microscopy method. There was a moderate relationship in the time to process samples using both methods (Fig. 6; adjusted R2 = 0.55, P < 0.01) indicating that samples that are more time intensive with the microscopy method are also more time intensive with the FlowCAM® method.
Discussion
We found congruence between microscopy and FlowCAM® methods across several tests and levels of biological organization. Comparison of body mass estimates from profile area were shown to be equivalent to the length-to-mass equation developed by Dumont et al. (1975). Notably, we also identified a methodological bias involving individuals near the upper size threshold of the instrument that affected density estimates (Kydd et al., 2017; Wong et al., 2017). We also demonstrated that the application of a correction factor to biased density estimates improved comparability of data between the semi-automated and microscopy methods, resulting in strong congruence for several zooplankton taxa and overall community structure. Our findings highlight that the reliability of semi-automated methods requires validation before usage. After a thorough vetting, semi-automated methods can increase processing efficiency of zooplankton samples by reducing processing times without sacrificing accuracy.
Detection rates of individual zooplankton were inversely related to body size. In general, detection rates were lower than 100% for all organisms, despite optimization of the device, with a notable decline for organisms larger than 1 mm. Large organisms that approach the processing size threshold of the device have a disproportionally greater probability of being excluded or partially imaged from datasets depending on the system configuration. The acceptable imaging region of the camera field of view is typically specified as ‘constrained,’ the recommended configuration by the manufacturer. In this setting, the maximum range of the camera field of view (1024 × 768 resolution or 1280 × 960 pixels) is reduced slightly by the user to create a buffer (i.e., unacceptable imaging region) between the acceptable imaging region and the edge of the flow cell. Particles that overlap with the buffer area during the imaging process are excluded because they could not be fully imaged (Fluid Imaging Technologies Inc., 2011). For field of view flow cell models, this occurs at the upper and lower edges of the camera field of view because the left and right edges of the flow cell are constrained by the physical walls of the flow cell. Therefore, this specific device configuration ensures datasets include only individuals imaged in their entirety, but the exclusion of individuals is related to body size. As body size increases, organisms occupy a larger proportion of the acceptable imaging region, increasing the probability of overlap between a particle and the buffer area leading to exclusion from imaged datasets. Unequal detection probabilities of individuals by size may potentially skew size frequency distributions by over-representing small individuals or particles and reducing overall density estimates (e.g., Kydd et al., 2017; Wong et al., 2017). Alternatively, if the imaging region of the camera field of view is specified as ‘unconstrained,’ the acceptable imaging region spans the maximum range of the camera field of view, and particles that lie across the edge of the field of view are imaged as partial particles (Fluid Imaging Technologies Inc., 2011). The inclusion of partially imaged particles in the development of reliable datasets is sometimes undesirable, particularly in studies considering metrics of particle size. Although inclusion of partial particle images may be warranted in some studies interested in zooplankton density or rare taxa, we recommend greater consideration of the configuration (i.e., constrained or unconstrained) of the imaging region of the camera field of view and how each can influence imaged datasets.
For D. pulex, two-dimensional body profile area was as closely related to individual mass as body length. Our findings indicate that profile area derived from imaging systems can be used as a reliable alternative and more rapid approach to accurately estimate individual biomass (Billones et al., 1999; Alcaraz et al., 2003; Hernández-León & Montero, 2006). Consequently, estimates of body mass derived from profile area can be used to estimate other ecological metrics, including standing crop biomass and secondary production.
For multiple taxa, we detected positive relationships between mean body size estimates from the two methods. Similar to previous studies, however, FlowCAM® length estimates for several zooplankton taxa were slightly greater than microscopy-based estimates (Kydd et al., 2017; Wong et al., 2017). Length-estimation algorithms of FlowCAM® are likely affected by complexities in body morphology, such that appendages and spines that extend beyond the body are included in length measurements by FlowCAM®, while these components are excluded in microscopy length measurements (See Wong et al., 2017). For some taxa (i.e., Calanoida, Cyclopoida, Daphnia), these differences in length measurements were systematic between methods (e.g., exclusion of a single tail spine), suggesting that length estimates could be corrected for comparability, while other taxa (i.e., nauplii, Rotifera, Sididae) exhibited no such relationship despite similar measurement methods. The lack of a relationship between methods for these taxa is likely from the morphological diversity present within these coarse taxonomic categories. High variation in aspect ratio among species of nauplii and rotifers could also have influenced the lack of relationship between methods as some taxa, such as Kellicottia longispina (Kellicot, 1879), have a very low aspect ratio driven by long spines and an elongate form while others have a higher aspect ratio, such as Keratella testudo (Ehrenberg, 1832). Variation in aspect ratio may cause different taxa to move through the cytometer differently with a greater or lower likelihood of rolling, which could influence the likelihood of dorsal profile images versus side profile images (see supplemental Fig. 1 for example). Data in the present study suggest that FlowCAM® users should use caution when generating taxon-specific metrics because systematic error can be propagated when calculating other biometrics.
Our study identified a potential mechanism responsible for the previously documented underestimation of zooplankton densities by semi-automated methods (Jakobsen & Cartensen, 2011; Kydd et al., 2017). The present study suggests that although the micro- and mesozooplankton techniques yielded similar results and biases, the slopes were significantly different with mesozooplankton being shallower. Different relationships for the two methods indicate that size frequency distribution of the zooplankton community under consideration and device setup need to be taken into account in order to reduce bias. As hypothesized by Kydd et al. (2017), our findings suggest that body size can contribute to enumeration biases due to patterns associated with detection rate and the configuration of the imaging system. After applying a density correction, however, we observed that semi-automated methods provided reliable estimates of densities and community structure congruent with those provided by traditional manual microscopy, with no taxonomic-specific biases. Given these results, we recommend that before using FlowCAM® or other similar technologies to produce estimates of zooplankton biomass, density, or community structure, potential size- and taxonomic-related biases need to be quantified in order to develop appropriate correction factors and an understanding of potential uncertainties where correction factors are not possible.
One major consideration when deciding whether to invest in a new technology is the time and resulting cost savings. The present study provides evidence that there can be a reduction in processing effort associated with the use of the semi-automated approach. We found that the identification software was fairly accurate at identifying zooplankton correctly and it rarely misclassified zooplankton as non-zooplankton particles. As a result, a short period of post-processing time was needed to correct misidentifications (i.e., detrital material identified as zooplankton, or zooplankton that had been misclassified as other zooplankton or detrital material). In addition, when the number of imaged debris particles was large (e.g., turbid samples), a long post-processing time was often required because there were simply more imaged particles to sort through. Our experience suggests that samples with low quantities of suspended particulate material (e.g., offshore locations with oligotrophic conditions) would require less post-processing time and potentially be much more efficient than manual microscopy methods.
References
Alcaraz, M., E. Saiz, A. Calbet, I. Trepat & E. Broglio, 2003. Estimating zooplankton biomass through image analysis. Marine Biology 143: 307–315.
Álvarez, E., M. Moyano, A. Lopez-Urrutia, E. Nogueira & R. Scharek, 2014. Routine determination of plankton community composition and size structure: a comparison between FlowCAM and light microscopy. Journal of Plankton Research 36: 170–184.
Benfield, M. C., P. Grosjean, P. F. Culverhouse, X. Irigoien, M. E. Sieracki, A. Lopez-Urrutia, H. G. Dam, Q. Hu, C. S. Davis, A. Hansen, C. H. Pilskaln, E. M. Riseman, H. Schultz, P. E. Utgoff & G. Gorsky, 2007. RAPID: research on automated plankton identification. Oceanography 20: 172–187.
Billones, R. G., M. L. M. Tackx, A. T. Flachier, L. Zhu & M. H. Daro, 1999. Image analysis as a tool for measuring particulate matter concentrations and gut content, body size, and clearance rates of estuarine copepods: validation and application. Journal of Marine Systems 22: 179–194.
Bray, J. R. & J. T. Curtis, 1957. An ordination of upland forest communities of southern Wisconsin. Ecological Monographs 27: 325–349.
Burnham, K. P. & D. R. Anderson, 2002. Model selection and multimodel inference: a practical information theoretic approach, 2nd ed. Springer-Verlag, New York.
Buskey, E. J. & C. J. Hyatt, 2006. Use of the FlowCAM for semi-automated recognition and enumeration of red tide cells (Karenia brevis) in natural plankton samples. Harmful Algae 5: 685–692.
Calbet, A., S. Garrido, E. Saiz, M. Alcaraz & C. M. Duarte, 2001. Annual zooplankton succession in coastal NW Mediterranean waters: the importance of the smaller size fractions. Journal of Plankton Research 23: 319–331.
Collins, S. F., T. M. Detmer, K. A. Kelson, M. A. Nannini, G. G. Sass & D. H. Wahl, 2018. The release and regulation of rotifers: examining the predatory effects of invasive juvenile common and bighead carp. Hydrobiologia 813: 199–211.
Culverhouse, P. F., R. Williams, B. Reguera, V. Henry & S. González-Gil, 2003. Do experts make mistakes? A comparison of human and machine identification of dinoflagellates. Marine Ecological Progress Series 247: 17–25.
Culverhouse, P. F., R. Williams, M. Benfield, P. R. Flood, A. F. Sell, M. G. Mazzochi, I. Buttino & M. Sieracki, 2006. Automatic image analysis of plankton: future perspectives. Marine Ecological Progress Series 312: 297–309.
Davis, C. C., S. M. Gallager & A. R. Solow, 1992. Microaggregations of oceanic plankton observed by towed video microscopy. Science 257: 230–232.
Detmer, T. M., J. H. McCutchan Jr. & W. M. Lewis Jr., 2017a. Trophic interactions across lake-stream boundaries in mountain lakes. Inland Waters 7: 440–448.
Detmer, T. M., J. H. McCutchan Jr. & W. M. Lewis Jr., 2017b. Predator driven changes in prey size distribution stabilize secondary production in lacustrine food webs. Limnology and Oceanography 62: 592–605.
Detmer, T. M., M. J. Diana & D. H. Wahl, 2019. Season and presence of Gizzard Shad influence horizontal spatial distribution of zooplankton in reservoirs of the Midwestern United States. Freshwater Sciences 38: 183–192.
Dumont, H. K., I. Van de Velde & S. Dumont, 1975. The dry weight estimate of biomass in a selection of Cladocera, Copepoda, and Rotifera from the plankton, periphyton and benthos of continental waters. Oecologia 19: 75–97.
Fluid Imaging Technologies Inc., 2011. FlowCAM manual version 3.0 [online]. [available on internet at http://www.ihb.cas.cn/fxcszx/fxcs_xgxz/201203/P020120329576952031804.pdf].
García-Criado, F. & C. Trigal, 2005. Comparison of several techniques for sampling macroinvertebrates in different habitats of a North Iberian pond. Hydrobiologia 545: 103–115.
Garmendia, M., M. Revilla & L. Zarauz, 2013. Testing the usefulness of a simple automatic mathod for particles abundance and size determination to derive cost-effective biological indicators in large monitoring networks. Hydrobiologia 704: 231–252.
Gorsky, G., M. D. Ohman, M. Picheral, S. Gasparini, L. Stemmann, J. Romagnan, A. Cawood, S. Pesant, C. García-Comas & F. Prejger, 2010. Digital zooplankton image analysis using the ZooScan integrated system. Journal of Plankton Research 32: 285–303.
Grosjean, P., M. Picheral, C. Warembourg & G. Gorsky, 2004. Enumeration, measurement, and identification of net zooplankton samples using the ZOOSCAN digital imaging system. ICES Journal of Marine Sciences 61: 518–525.
Hamilton, S. K., S. J. Sippel, W. M. Lewis Jr. & J. F. Saunders III, 1990. Zooplankton abundance and evidence for its reduction by macrophyte mats in two Orinoco floodplain lakes. Journal of Plankton Research 12: 345–363.
Hernández-León, S. & I. Montero, 2006. Zooplankton biomass estimated from digitized images in Antarctic waters: a calibration exercise. Journal of Geophysical Research 111: 3.
Ide, K., K. Takahashi, A. Kuwata, M. Nakamachi & H. Saito, 2008. A rapid analysis of copepod feeding using FlowCAM. Journal of Plankton Research 30: 275–281.
Jakobsen, H. H. & J. Carstensen, 2011. FlowCAM: sizing cells and understanding the impact of size distributions on biovolume of planktonic community structure. Aquatic Microbial Ecology 65: 75–87.
Krueger, D. A. & S. I. Dodson, 1981. Embryological induction and predation ecology in Daphnia pulex. Limnology and Oceanography 26: 219–223.
Kydd, J., H. Rajakaruna, E. Briski & S. Bailey, 2017. Examination of a high resolution laser optical plankton counter and FlowCAM for measuring plankton concentration and size. Journal of Sea Research 133: 2–10.
Le Bourg, B., V. Cornet-Barthaux, M. Pagano & J. Blanchot, 2015. FlowCAM as a tool for studying small (80–1000μm) metazooplankton communities. Journal of Plankton Research 37: 666–670.
Lewis, W. M., 1979. Zooplankton community analysis: studies on a tropical system. Springer-Verlag, New York New York.
Makarewicz, J. C. & G. E. Likens, 1979. Structure and function of the zooplankton community of Mirror Lake, New Hampshire. Ecological Monographs 49: 109–127.
Minchin, P. R., 1987. An evaluation of the relative robustness of techniques for ecological ordination. Theory and Models in Vegetation Science 69: 89–107.
Moberg, E. A. & H. M. Sosik, 2012. Distance maps to estimate cell volume from two-dimensional plankton images. Limnology and Oceanography: Methods 10: 278–288.
Pennak, R. W., 1973. Some evidence for aquatic macrophytes as repellents for a limnetic species of Daphnia. Internationale Revue der gesamten Hydrobiologie 58: 569–576.
Pinel-Alloul, B., C. Guay, N. Angeli, P. Legendre, P. Dutilleul, G. Balvay, D. Gerdeaux & J. Guillard, 1999. Large-scale spatial heterogeneity of macrozooplankton in Lake of Geneva. Canadian Journal of Fisheries and Aquatic Sciences 56: 1437–1451.
Poulton, N. J., 2016. FlowCAM: quantification and classification of phytoplankton by imaging flow cytometry. Imaging Flow Cytometry: Methods and Protocols 1389: 237–247.
R Core Team, 2013. T: a language and environment for statistical computing. R Foundation for statistical computing, Vienna, Austria. [available on internet at http://www.R-project.org/.
Saunders III, J. F. & W. M. Lewis Jr., 1988. Composition and seasonality of the zooplankton community of Lake Valencia, Venezuela. Journal of Plankton Research 10: 957–985.
Sieracki, C. K., M. E. Sieracki & C. S. Yentsch, 1998. An imaging-in-flow system for automated analysis of marine microplankton. Marine Ecological Progress Series 168: 285–296.
Siokou-Frangou, I., U. Christaki, M. G. Mazzocchi, M. Montresor, M. Riberad’Alcala, D. Vaque & A. Zingone, 2008. Plankton in the open Mediterranean Sea: a review. Biogeosciences 7: 1543–1586.
Stanislawczyk, K., M. L. Johansson & H. L. MacIsaac, 2018. Microscopy versus automated imaging flow cytometry for detecting and identifying rare zooplankton. Hydrobiologia 807: 53–65.
Thorp, J. H. & A. P. Covich (eds), 1991. Ecology and classification of North American freshwater invertebrates. Academic Press, San Diego.
Turner, A. M. & J. C. Trexler, 1997. Sampling aquatic invertebrates from marshes: evaluating the options. Journal of the North American Benthological Society 16: 694–709.
Welker, M. T., C. L. Pierce & D. H. Wahl, 1994. Growth and survival of larval fishes: roles of competition and zooplankton abundance. Transactions of the American Fisheries Society 123: 703–717.
Wong, E., A. R. Sastri, F. S. Lin & C. H. Hsieh, 2017. Modified FlowCAM procedure for quantifying size distribution of zooplankton with sample recycling capacity. PLoS One 12: e0175235.
Zarauz, L. & X. Irigoien, 2008. Effects of Lugol’s fixation on the size structure of natural nano-microplankton samples, analyzed by means of an automatic counting method. Journal of Plankton Research 30: 1297–1303.
Acknowledgements
We thank M. Diana and B. Diffen for initial setup of the FlowCAM® device and laboratory assistance. We also thank the D. Soucek lab for the use of a microbalance. This study was supported by the Great Lakes Restoration Initiative (CAFWS-93) and the Federal Aid in Sporfish Restoration Act (project F-185-R-6) with funding administered through the Illinois Department of Natural Resources (IDNR). We would like to thank K. Irons and M. McClelland for coordination of this project with the IDNR Division of Fisheries. We would also like to thank three anonymous reviewers for providing constructive comments and helping us to improve our methodology.
Disclaimer
Any use of trade, firm, or product names is for descriptive purposes only and does not imply endorsement by the University of Illinois, Prairie Research Institute, or the Illinois Natural History Survey.
Author information
Authors and Affiliations
Corresponding author
Additional information
Handling editor: Piet Spaak
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Detmer, T.M., Broadway, K.J., Potter, C.G. et al. Comparison of microscopy to a semi-automated method (FlowCAM®) for characterization of individual-, population-, and community-level measurements of zooplankton. Hydrobiologia 838, 99–110 (2019). https://doi.org/10.1007/s10750-019-03980-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10750-019-03980-w