Abstract
Brown trout (Salmo trutta L.) is an important resource both for conservation and recreational fisheries in Iberian Peninsula, but the species is threatened in Llobregat River, an important river drainage in Catalonia (NE Spain), due to the high anthropological influence. In order to ascertain the native genetic diversity, phylogenetic relationships, population structure and stocking impact in this drainage, 295 individuals collected from eight different locations distributed throughout the main river course and the main tributary (i.e. Llobregat River and Cardener River, respectively) were analysed for the period 1990–2017. The complete mitochondrial DNA (mtDNA) control region and the LDH-C* diagnostic locus for monitoring the stocking were analysed. Moderate-high levels of genetic introgression were detected (frequency ~ 0.300), affecting especially the main course. Despite the stocking introgression and the expected genetic depletion by genetic drift due to population isolation, a new mtDNA haplotype group pertaining to the Mediterranean lineage was found restricted to the Cardener River. This haplogroup is responsible of the high population differentiation observed between rivers in the Llobregat drainage (ΦCT = 0.456, P < 0.001) and highlights the conservation interest of that region. Management strategies to achieve fisheries sustainability and conserve endemic resources in the region are proposed.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Genetic diversity makes feasible the adaptation of wild populations to environmental changes and thus, facilitates the long-term conservation of the species. Knowledge on the amount of genetic diversity existing within and among populations, its distribution through the species range and its temporal stability over generations are key issues to design appropriate management and conservation strategies (Frankham et al., 2002). Connectivity ensures gene flow among populations to mitigate diversity losses by genetic drift, which is high in small and isolated populations. Loss of connectivity, sometimes due to natural barriers (e.g. waterfalls), leads to population fragmentation, but since the last century the fragmentation of wild populations has been intensified worldwide by artificial barriers built by humans (e.g. roads, hydroelectric dams, railway lines). For species inhabiting rivers, individuals are confined within hydrological networks (Arthington et al., 2016). In these habitats, fragmentation by dams, together with pollution, invasive aquatic species, unsustainable water extraction and fisheries overexploitation have contributed to the decline of fish populations worldwide (Saura & Faria, 2011; Kohout et al., 2012; Bernas et al., 2014; Vera et al., 2018), particularly for migratory species (around 40% since 1970; WWF, 2016). In addition, the progressive climatic change can also compromise the sustainability of freshwater plant and animal species (Ormerod, 2009; Almodovar et al., 2012).
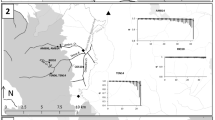
The Llobregat River is one of the main rivers (4,948 km2 basin surface) of Catalonia (NE Spain). This river is born in the Castellar de N’Hug village at an altitude lower than 1300 m in the Sierra del Cadí pre-Pyrenean Mountains and, after 175 km, flows into the Mediterranean Sea in Prat de Llobregat city (see Fig. 1). The Llobregat River is under Mediterranean climate conditions, commonly characterized by irregular hydrological regimes, including periodical severe droughts and floods (Trigo et al., 2004). This river basin has been highly disturbed by human activities, including around 300 artificial barriers, being 228 of them impassable obstacles for fish (ACA, 2017). Water extraction includes domestic consumption for the city of Barcelona and its metropolitan area (3.2 million of inhabitants), and for an important concentration of industries (tannery, textile, food products and paper mills) and agricultural activities along its main course and tributaries (Marce et al., 2012). These human activities have also released into the river a wide variety of pollutants such as pesticides, surfactants, pharmaceuticals and personal care products (Gonzalez et al., 2012). Of the 33 fish species identified in the Llobregat River basin, 21 are introduced, the brown trout (Salmo trutta Linnaeus, 1758) being the only autochthonous species inhabiting the headwaters of the basin (Sostoa et al., 2010). Nevertheless, in some sections, brown trout populations are additionally threatened by hybridization and genetic introgression due to releases of trout from foreign domesticated stocks (Aparicio et al., 2005; Araguas et al., 2009).
Map of locations analysed in the Llobregat drainage. Codes are shown in Table 1. The name of the main rivers (Llobregat, Cardener, Anoia) are also included. The dotted area represents the genetic refuge in the zone, while grey polygons show the Special Areas of Conservation (SACs) defined by NATURA 2000 European network. Pie charts show the haplotype composition for each location
Because of its high value for recreational fisheries, stocking of trout populations has been carried out for many decades in Iberian Peninsula and other European countries to counterbalance the decline of wild trout populations (Vera et al., 2013; Fabiani et al., 2018). Seven distinct brown trout lineages (major branches of the phylogenetic tree) have been described throughout the native brown trout range based on mitochondrial DNA (mtDNA) analyses (Bernatchez, 2001; Susnik et al., 2005; Vera et al., 2010). Athough only two of these lineages, the Adriatic (AD) and the Mediterranean (ME), are native to the Iberian Mediterranean basins (Cortey et al., 2004), they hold an important portion of the trout genetic diversity of the Iberian Peninsula, including groups of haplotypes (haplotype groups or haplogroups) inside those lineages, because of the long-time persistence of trout populations in this region (Suarez et al., 2001; Cortey et al., 2004, 2009; Vera et al., 2010). The genetic differentiation among lineages has contributed to the detection of molecular diagnostic markers to monitor and evaluate stocking impact in Mediterranean trout populations. Thus, the nuclear lactate dehydrogenase C (LDH-C*) locus and the mtDNA control region (CR) sequencing have been extensively used for this purpose (e.g. Araguas et al., 2009; Vera et al., 2013; Splendiani et al. 2016; Araguas et al., 2017). The LDH-C*100 allele is fixed in native Mediterranean populations, while the Central-European hatchery stocks, often used to release fish in the region, are fixed for the LDH-C*90 allele (García-Marín et al., 1991). These stocks also show mtDNA CR haplotypes characteristic of the non-native Atlantic (AT) lineage (Cortey & García-Marín, 2002). Stocking practices in Southern European peninsulas have shown different outcomes, but a higher impact of stocking in Mediterranean rivers has been described (Almodovar et al., 2006; Splendiani et al., 2006; Vera et al., 2013), threatening native genetic resources and local adaptations.
The brown trout inhabits in the headwaters of Llobregat River, commonly placed below 1300 m above sea level (a.s.l.), being therefore very threatened by the climatic change in progress. Almodovar et al. (2012) predicted the near extinction of brown trout populations at quite similar altitudes by 2100 in the Western Pyrenees. Since 2013, some headwaters of the Llobregat River have been included as Special Areas for Conservation (SACs) in the NATURA 2000 network (EU organization responsible to ensure the survival of the most valuable and threatened European species and habitats). Nevertheless, the knowledge on the genetic status and evolutionary relevance of these brown trout populations is very scarce (see Aparicio et al., 2005; Araguas et al., 2009). The overall aim of this study was to evaluate the spatial and temporal monitoring of brown trout lineages in the Llobregat River using diagnostic mtDNA and LDH-C* markers to: (i) describe their phylogenetic relationships, genetic diversity and population structure, (ii) assess the genetic impact of hatchery releases of foreign origin and (iii) discuss fisheries management actions for the conservation of brown trout in this river drainage.
Materials and methods
Sampling sites
Eight locations placed in the headwaters of the Llobregat River and its main tributary, the Cardener River, were studied (Fig. 1, Table 1), two of them being temporally sampled. Thus, temporal collections from 1990 to 2017 of the Riutort stream (Llobregat River, N = 26.5 ± 5.4) were available (years 1990, 1999, 2002 and 2017). In the Cardener River, the Aiguadora stream at Castellar village was also twice sampled in years 2002 and 2017 (N = 20.0 ± 8.5). Comparison of collections at these two streams was used to check for temporal genetic stability in the studied area. Adipose fin clips were collected from each wild brown trout after sedation of fish with tricaine methanesulphonate (MS-222), excluding RT90 specimens previously collected by Cortey et al. (2004). Finally, a sample of the Bagà hatchery collected in 2002 (N = 40) was included as reference for the hatchery used for stocking. Legal authorizations limited sample size to a maximum of 35 specimens per collection in the wild, but numbers of some collections were further restricted by the availability of fish at the time of capture.
DNA extraction and molecular analyses
Genomic DNA was obtained from all individuals using either the standard phenol–chloroform protocol (Sambrook et al., 1989) or the Chelex® protocol (Walsh et al., 1991). Molecular analyses included the genotyping of the diagnostic LDH-C* locus to assess introgression due to stocking in the studied area, and the sequencing of the complete mtDNA control region (CR) to assess the evolutionary relationships of the Llobregat River brown trout with other studied populations in Mediterranean rivers, and as a complementary evaluation of maternal genetic introgression. The LDH-C* genotypes for all sampled fish were obtained following the PCR Restriction Fragment Length Polymorphism (PCR–RFLP) protocol described by McMeel et al. (2001) using the BseLI restriction enzyme. The amplification and sequencing of the complete CR (1,013 bp) was carried out following Cortey & García-Marín (2002) and Sanz et al. (2006). Sequences were obtained using the ABI PRISM BigDye™ Terminator v3.1 Cycle Sequencing Kit protocol (Applied Biosystems, Foster City, CA) on an ABI PRISM® 3730xl Genetic Analyzer (Applied Biosystems, Foster City, CA). Variable sites were checked by hand with the programme SEQSCAPE 2.5 (Applied Biosystems, Foster City, CA) using the MEcs1 haplotype as reference (GenBank Accession number: AY836350). The different haplotypes were identified using the programme MEGA 7.0 (Kumar et al., 2016).
Genetic diversity, phylogenetic relationships and population structure
For LDH-C* locus, allele frequencies and deviation from Hardy–Weinberg (HW) expectations in each location were estimated using GENEPOP 4.0 (Rousset, 2008). Genetic differentiation between location pairs and temporal replicates was estimated using the exact G test implemented in GENEPOP 4.0 with default parameters (10,000 dememorisation steps, 20 batches and 500 iterations per batch).
Diversity among haplotypes and among haplogroups (i.e. genetic distances, pairwise nucleotide differences) was estimated using the programme MEGA 7.0. The nucleotide substitution was adjusted to the Tamura-Nei model (Tamura & Nei, 1993) with a gamma parameter of 0.9253, as suggested for the brown trout control region (Cortey et al., 2009). Phylogenetic relationships among haplotypes were analysed by a Median-Joining network (Bandelt et al., 1999) using NETWORK 4.5.1.2 (http://www.fluxus-engineering.com/sharenet.htm). Detected haplotypes along with all other complete mtDNA control region haplotypes from native ME (15) and AD (20) brown trout lineages present in the Eastern Pyrenees and Iberian Mediterranean drainages available in the GenBank database (Cortey et al., 2004; GenBank Accession Numbers for MEcs1-MEcs15: AY836350-AY836364; GenBank Accession Numbers for ADcs1-ADcs20: AY836330-AY836349) were included in the network. Network loops (which represent ambiguities) involving three or more haplotypes were solved using the frequency, the topological and the geographical criteria as proposed by Pfenninger & Posada (2002). Divergence times among clusters of haplotypes were estimated using a mtDNA divergence rate of 1–2% per million years (My) applied for S. trutta (Cortey et al., 2004; Schenekar et al., 2014).
The haplotypes ATcs1, ATcs2, ATcs3 and ATcs4 of the Atlantic brown trout lineage are specific of the foreign stocks used in Spanish hatcheries (Cortey & García-Marín, 2002). Therefore, their presence in the wild is considered the result of stocking in the region (Almodovar et al., 2006), and the sum of the observed frequencies for all these AT haplotypes is a further estimate of the impact of hatchery releases. In each location, the mtDNA variability was estimated using haplotype (h) and nucleotide (π) diversity estimators (Nei & Tajima, 1981). Pairwise population differentiation coefficient (\(\Phi\)ST) between locations and temporal replicates was estimated using haplotype frequencies and their divergence involving a gamma Tamura-Nei parameter of 0.9253. Significance was estimated by 10,000 permutations using ARLEQUIN 3.5 (Excoffier et al., 2005). Hierarchical analysis of the molecular variance (AMOVA; Excoffier et al., 1992) was used to estimate the percentage of genetic variation distributed within (\(\Phi\)SC) and among (\(\Phi\)CT) groups (i.e. Llobregat and Cardener). This test was carried out using ARLEQUIN 3.5 and significance was estimated by 10,000 permutations.
Results
Genetic diversity, stocking impact and temporal stability
As expected, the sampled fish at the Bagà hatchery (BA) were all homozygous for the LDH-C*90 allele. The LDH-C* locus was in HW equilibrium at almost all studied wild collections and only for the Riera Monells location (MON02; P = 0.0447) there was a significant deviation from HW proportions due to heterozygote deficit (FIS = 0.433), suggestive of a Wahlund effect related to released hatchery fish (Table 2). Overall, the frequency of the LCH-C*90 allele ranged from 0.000 in CT02 and AVI02 to 1.000 in AJ02 (Arija stream), where a pure hatchery trout population seems to be established, as confirmed by mtDNA results (see below). Excluding the Arija stream collection, samples in 2002 were less affected by stocking (mean = 0.196 ± 0.242) than more recent 2017 collections (mean = 0.435 ± 0.219), but this difference was not significant due to the wide standard deviation (SD) and the reduced number of collections in each group (t test = − 1.527, P = 0.156). At a local scale, a significant increase of the LDH-C*90 allele frequency was detected on the Castellar location (CT02 vs CT17, P = 0.009) and on the Riutort location since 1990 versus all latter collections (all pairwise P values < 0.010). A significant higher mean introgression value was detected when comparing the Llobregat River (mean = 0.586 ± 0.221) and the Cardener River (0.052 ± 0.068) (t test = 4.612, P = 0.002).
Ten different mtDNA CR haplotypes were detected in the studied zone. Seven of them belonged to the native ME lineage, while the other three were hatchery haplotypes of the AT lineage (Table 2). No AD haplotypes were detected. Four ME haplotypes are described for the first time in this study (MEcs23, MEcs25, MEcs26 and MEcs27) and they have been submitted to GenBank (Accession Numbers: MG970273-MG970276). The most frequent haplotype in the whole region was MEcs1 (116 out of 295 individuals) followed by the hatchery ATcs4 haplotype (65 individuals), but the haplotype MEcs23 was the most frequent in the Cardener River (Table 2). Haplotype diversity (h) ranged from 0.0000 in locations where a single haplotype was fixed (CT17, AVI02) to 0.7500 in AVII02 (mean h = 0.3963 ± 0.2627), while nucleotide diversity (π) ranged from 0.0000 to 0.0056 in RT99 where a high number of haplotypes from different lineages were detected (mean π = 0.0024 ± 0.0022; Table 2). Introgression impact from mtDNA data (measured as the total frequency of AT haplotypes in the sample) ranged from 0.000 in Cardener locations to 1.000 in AJ02 and GRE17 (mean = 0.298 ± 0.365; Table 2), confirming a higher stocking impact in the Llobregat River than in the Cardener River. No significant differences in AT haplotype frequency for the whole region were found between 2002 and 2017 (Mann–Whitney U test = 17.00, P = 0.352). Despite the LDH-C*90 frequency was often higher at each location than the total frequency of hatchery AT haplotypes, a positive correlation was found between these two estimates of stocking impact (Spearman’s ρ coefficient = 0.881, P = 0.001). When AT haplotypes were removed from the analysis, genetic diversity decreased in all locations (mean h = 0.1408 ± 0.2675, mean π = 0.0004 ± 0.0008, Table 2). No statistically significant differences in native haplotype distribution were detected between temporal replicates at the Castellar location (χ = 1.136, P = 0.567), but the MEcs26 and MEcs27 haplotypes were not found in 2017, which can be explained by their low occurrence in the 2002 larger collections (each of them only detected in one individual, see Table 2). Significant differences were detected in all pairwise comparisons involving RT90 collection on the Riutort location, where increased abundance of hatchery AT haplotypes occurred since 1990 (ΦST_RT90-RT99 = 0.283, P = 0.001; ΦST_RT90-RT02 = 0.115, P = 0.040; ΦST_RT90-RT17 = 0.263, P = 0.003).
Phylogenetic relationships and population structure
The total number of variable sites among detected haplotypes was 21 and increased to 24 when ATcs1 and ADcs1 were included in the multiple alignment (Fig. 2). The number of differences among haplotypes ranged from 1 to 16 (between ATcs4 and MEcs27). Network analysis clearly distinguished among haplotypes of AD, AT and ME lineages (Fig. 3). In addition, four new ME haplotypes (MEcs23, MEcs25, MEcs26 and MEcs27) were found in the Cardener River, all these haplotypes differing from other S. trutta lineages by a double nucleotide deletion in the positions 931 and 932 (Fig. 2). These two deletions resulted in the appearance of a new group in the haplotype network within the ME brown trout lineage (MEcs23 haplogroup, Fig. 3). Comparing with sequences previously described in the extensive study for Iberian Mediterranean trout by Cortey et al. (2004), the MEcs25 (found in two individuals) differed by two mutational steps from the MEcs1 haplotype and the MEcs23 (the more abundant new haplotype detected in 49 individuals) differed by five mutational steps from MEcs25, indicating the extinction of intermediate haplotypes in the region. The Tamura-Nei genetic distance between the two detected ME haplogroups (i.e. MEcs1 haplogroup and MEcs23 haplogroup, see Fig. 3) was 0.0024 ± 0.0010 (0.0029 ± 0.0012 when only haplotypes detected in this study were used). Therefore, the divergence time between these haplogroups applying a divergence rate of 1–2% per My would be ~ 120,000–290,000 years.
Variable sites in the complete mtDNA CR haplotypes detected in the present study. Dots denote identical sites and dashes indels. The two most representative haplotypes for AT and AD lineages (ATcs1 and ADcs1, respectively) have been included as a reference. GenBank accession numbers are shown in parentheses
Median-Joining network of detected haplotypes in Llobregat River drainage. Black squares represent median vectors needed to connect all the haplotypes. Numbers besides lines indicate the number of mutational steps when more than one mutational step is involved. Dashed lines show alternative connections among haplotypes. Size of the circles is related with the haplotype abundance. Polygons indicate the different haplogroups and lineages. Black, white and grey colours in the circles represent the presence of haplotypes in Llobregat River, Cardener River and other haplotypes present in the Iberian Mediterranean drainages, respectively
The distribution of mitochondrial haplotypes indicated a highly significant population differentiation in the studied area (ΦST = 0.706, P < 0.001). Pairwise \(\Phi\)ST values were higher in Llobregat versus Cardener pairwise comparisons (Table 3), mainly due to the presence of different haplogroups in both rivers (i.e. MEcs1 haplogroup mainly present in Llobregat and MEcs23 haplogroup exclusively found in Cardener). Such pattern of differentiation between the Llobregat and Cardener trout was even more evident when hatchery AT haplotypes were removed from the analyses. This high differentiation between streams of both rivers was also depicted by AMOVA analyses where a 45.61% of genetic differentiation was due to differences between rivers (\(\Phi\)CT = 0.456, P < 0.001) and a 25.03% to differences among locations within river (\(\Phi\)SC = 0.460, P < 0.001). When AT hatchery haplotypes were removed, the percentage of genetic differentiation associated to differences between rivers increased up to 90.20% (\(\Phi\)CT = 0.902, P < 0.001) while differences among locations within rivers only explained 4.70% of genetic differentiation (\(\Phi\)SC = 0.479, P < 0.001) by the presence of different ME haplogroups within both rivers.
Discussion
Brown trout is a valuable ecological and fisheries resource in the Iberian Peninsula (Vera et al., 2018), being the only native salmonid species in the Iberian Mediterranean drainages. To counterbalance population declines, in many European countries administrations in charge of inland fisheries have released hatchery reared trout during the past century until the beginning of 2000’s. The foreign origin of the hatchery stocks have compromised the conservation of the native gene pools due to genetic introgression, especially throught all the Mediterranean basins (e.g. Almodovar et al., 2006; Vera et al., 2013; Berrebi et al., 2017; Fabiani et al., 2018). The average level of introgression due to hatchery releases estimated in our regional study in 2017 was moderate—high (frequency of hatchery variants ~ 0.3 for both studied diagnostic markers), much higher than in the Atlantic Spanish populations (Almodovar et al., 2006), but with a broad variation among locations. Thus, among introgressed populations, the CT17 collection showed a pure hatchery origin according to diagnostic markers and GRE17 showed fixed mtDNA haplotypes of hatchery origin, but without fixation of the LDH-C*90 allele (see Table 2). Despite evidences indicating male overrepresentation among reared trout (Rasmussen, 1986), the result at GRE17 suggests either assortative mating between native males and hatchery females or female bias for survival or reproduction success among released fish (Weiss & Schmutz, 1999; Hansen et al., 2006). Conversely, previous studies in Mediterranean drainages suggested that often mtDNA results underestimate introgression (Nonnis Marzano et al., 2003). These observations highlight the need of use of both mtDNA and nuclear markers for introgression studies (Vera et al., 2013). Furthermore, mtDNA AT haplotypes were not detected in our collections from the Cardener River, but the hatchery impact in some sections was disclosed according to the LDH-C*90 frequency since 2002 (Table 2). The introgression estimates were similar to those observed in other Western Mediterranean populations (0.200–0.500; Aparicio et al., 2005; Splendiani et al., 2013; Vera et al., 2013; Araguas et al., 2017) and in Atlantic Iberian drainages where climatic Mediterranean conditions characterize river flows (Vera et al., 2018). Currently, the Autonomous Government of Catalonia is still restocking with Atlantic hatchery fish (including the Llobregat River drainage), but with sterile triploids, and releases are performed into controlled river sections under high recreational fishery pressure (see in: http://agricultura.gencat.cat/es/ambits/medi-natural/pesca-continental/repoblacions/index.html). Our temporal comparisons at Riutort and Aiguadora streams indicated that introgression increased over time, although statistically significant differences were only observed in the Riutort stream.
Despite some authors consider that increased diversity resulting from hybridization with foreign stocks could improve the long-term viability of the populations (Sonstebo et al., 2008), introgression can lead to outbreeding depression and loss of local adaptations and native population structure (Hansen et al., 2009; Fernandez-Cebrian et al., 2014). Some strategies to conserve native resources include the eradication of all exotic and introgressed fish (Allendorf et al., 2004). Except for locations such as the Arija (AJ02) stream inhabited only by naturalized hatchery fish, eradication seems to be pointless for Iberian Mediterranean drainages due to the current scarcity of the species and the admixture between native and hatchery gene pools over several generations (Araguas et al., 2008; Vera et al., 2013). Although only the LDH-C* nuclear marker was tested, our studied locations were in HW equilibrium, fishes harbouring different genomic proportions of native and hatchery constitution. In spite of the stocking impact, the maintenance of native genetic diversity is one of the main goals for conservation genetics because local gene pools and genetic diversity ensures the current and future adaptation of the species (Ryman et al., 1995; Frankham et al., 2002). Based on phenotype differences between native, hybrid and hatchery fish reported by Aparicio et al. (2005), selective removal of hybrid and hatchery fish by anglers might contribute to restore native gene pools in the studied locations. Such actuation should require continuous monitoring of populations to prevent census declines resulting in increased genetic drift on remnant gene pools.
Two important evolutionary features should be highlighted from this study. The first one is the presence of a haplogroup of the ME lineage endemic to the Cardener River streams (i.e. MEcs23 haplogroup). The abundance of this haplogroup in the Cardener River locations contrasted with its absence in the Llobregat River basin, being the main responsible for the high genetic differentiation between native gene pools in the studied area. Such pattern suggests a long-time isolation of trout inhabiting the Cardener streams, a situation very similar to that described for the endemic Duero (DU) lineage haplotypes in the Miño-Sil drainage (Vera et al., 2015). Estimated time of divergence between the endemic MEcs23 haplogroup and the ancestral MEcs1 haplogroup was of 120,000–290,000 years, a range including the Riss-Würm interglacial (130,000 to 115,000 years before present) ~ 8 ± 4°C warmer than the average of the last 1000 years before present (Dahl-Jensen et al., 2013). Increased temperatures during that interglacial period likely confined trout populations to the uppermost regions. Fossil data in other regional basins have suggested a downstream expansion of trout populations during the subsequent glacial period (Muñoz & Casadevall, 1997), colonizing those basins at altitudes lower than that of the Cardener and Llobregat confluence (260 m.a.s.l.). However, trout populations in the Cardener River likely remained isolated, avoiding the expansion of the MEcs23 haplogroup into the mainstream of the Llobregat River. The detection of endemic trout groups in the Southern European peninsulas is not surprising because they are hot spots of genetic diversity for S. trutta reflecting different evolutionary processes, including secondary contacts among lineages, drift and local adaptation (Sanz et al., 2002; Meraner et al., 2007; Bouza et al., 2008; Snoj et al., 2008). Several haplotype lineages and brown trout morphs described in Italian and Balkan peninsulas are currently recognized as different species (Jadan et al., 2015; Maric et al., 2017; Fabiani et al., 2018), but the taxonomic revision of Iberian trout endemisms is still pending (García-Marín et al., 2017).
The second interesting evolutionary feature is the absence of the AD lineage in the Llobregat drainage. This situation is rather surprising because the AD lineage expanded in all the other regional rivers (i.e. Ter and Ebro rivers) surrounding the Llobregat, likely between 77,000 and 155,000 years before present (Cortey et al., 2004). The lack of the AD lineage might be explained by different causes, such as insufficient survey, selective forces against this lineage in the whole Llobregat basin, demographic events producing strong genetic drift, or the existence of barriers preventing the colonization of the Llobregat River basin during the expansion of the AD lineage. Sampling bias seems unlikely due to the important number of analysed individuals (295). For example, Cortey et al. (2004) analysed 264 individuals from 19 localities distributed along the main Eastern Pyrenees drainages (including Riutort location) and the global frequency of AD lineage was 0.598. Selective pressures have been suggested to explain the absence of the AT brown trout lineage in the inner section of some Atlantic Iberian basins inhabited by the DU lineage (Vera et al., 2010). However, selection looks unlikely in our case since the AD lineage has been reported in all other Mediterranean basins of the region where the ME lineage (Cortey et al., 2004). Demographic decline determining loss of diversity by genetic drift seems a plausible explanation for the absence of AD lineage on the Llobregat River. For instance, Maric et al. (2016) suggested that the rapid post-glacial colonization of Middle Volga and Ural drainages would explain the reduced mtDNA diversity observed in these regions. Certainly, genetic drift is more severe over mtDNA markers than on nuclear markers, because of their smaller effective population size (Ne = ¼ of nuclear; Moore, 1995), leading to a faster loss of haplotypes within populations. The impact of genetic drift on Western Mediterranean trout populations has been previously reported (Sanz et al., 2002; Vera et al., 2013) and it is confirmed in our study by the low native haplotype diversity (see Table 2) and the loss of intermediate haplotypes within MEcs23 haplogroup. However, a surprising observation is that both lineages seem to have a similar abundance in the other regional river basins (average 0.598 AD lineage, 0.402 ME lineage). Consequently, diversity loss should have affected both lineages equally if they had been present in the Llobregat basin and, therefore, the exclusive presence of the AD lineage would be expected in some localities of the Llobregat. However, this result has not been observed and suggests the existence of natural barriers preventing the colonization of the Llobregat basin by the AD lineage. The Cardener River and the Llobregat River at their confluence flow over upper Eocene evaporites (sulphates and halides). The geochemical signature of streams in this area is dominated by the weathering of these sediments (Marce et al., 2012). To what extent these dissolved materials might represent a barrier to trout dispersal in the past is unknown (trout is not currently present at that altitude, 260 m a.s.l.), but salinity is high in the Cardener River (conductivity range from 480 to 2260 µS/cm; Soler et al., 2002). Furthermore, the quality index of macroinvertebrate fauna at the closest sections to the confluence of both rivers is lower than in upstream stretches, despite the similar investment performed on sewage treatment (Prat & Rieradevall, 2006).
The maintenance of connectivity among populations contributes to the long-term conservation of populations for freshwater species. Connectivity promotes gene flow and limits the effect of genetic drift expected from reduction of the effective population size (Ne) in fragmented and isolated populations (Fumagalli et al., 2002). River fragmentation by anthropological activities (i.e. dam buildings, water over-extraction) has caused significant decrease of the effective size in fish populations that compromised the population persistence (McDermid et al., 2014). In fact, recovery of fluvial connectivity has been one of the main tasks carried out by the ACA (Catalan Water Agency) during the last ten years. Recent reviews on the river connectivity index (ACA, 2017) listed the headstreams of the Llobregat River as “very good” or “good”, meanwhile the Cardener River is classified as “very bad” due to the presence of impassable barriers (Fig. 1). Similarly, “very bad” results are obtained at the confluence of both rivers. This information along with the unsuitable environmental conditions for trout in the lower Cardener river course may have facilitated the preservation of the MEcs23 haplogroup into this river. Nevertheless, connectivity among Cardener locations would be also desirable to increase local effective sizes to prevent genetic drift. Otherwise, two of the studied locations in the Llobregat River (AJ02 and GRE17) were almost composed of naturalized hatchery fish and their dispersion might have contributed to increase introgression in other places of the drainage. So, the maintenance of some barriers may facilitate the eradication of these alien populations as well as prevent their expansion to areas inhabited by native fish. Under the ongoing global climate warming, the preservation of some artificial barriers may also prevent trout areas of being populated by downstream fish communities mostly composed by exotic species.
The singularity of the Cardener River basin has promoted the inclusion of some of their streams (i.e. Aiguadora) into the NATURA 2000 Network of the EU. In addition, river stretches at the Aiguadevalls and Aiguadora streams (both tributaries of the Cardener River) have been declared as genetic refuges for brown trout since 2008. These areas are thought to maintain native genetic pools and prevent introgression from foreign stocks (reviewed in García-Marín et al., 2017). In 2017, all analysed locations were under a “catch-and-release” status, except Riutort and Gressolet which were “unfished”. However, according to our results, the fishery status for some locations should be likely revised, especially those with a high introgression impact (e.g. Arija and Gressolet), because they are mainly reservoirs of alien gene pools. Allowing the extractive fishery at such locations (i.e. changing their status to “fished” areas) could help to eradicate exogenous variants in the territory.
The present study is the widest performed to date on S. trutta populations within the Llobregat River basin, a drainage basin with high anthropological pressure (Gonzalez et al., 2012). Our study has enabled the detection of a new ME endemic brown trout haplogroup of high conservation interest. Thus, wider surveys including unexplored locations both within Llobregat and other neighbouring drainages should be performed to ascertain the current distribution of this new MEcs23 haplogroup. The use of highly variable nuclear markers (i.e. microsatellites) would also help to define limits among population groups. Finally, temporal genetic monitoring and management actions balancing conservation and socio-economic interests (e.g. increasing the number of “catch-and-release” sections, avoid stocking with hatchery fish, definition of new genetic refuges, minimizing anthropological disturbances…) are suggested to reach a self-sustained recreational fisheries with native fish in the region.
References
ACA, 2017. Connectivitat longitudinal als rius de les Conques Internes de Catalunya. Estructures presents als rius i actuacions de millora realitzades. Agència Catalana de l’Aigua, Departament de Territori i Sostenibilitat, Generalitat de Catalunya, Barcelona.
Allendorf, F. W., R. F. Leary, N. P. Hitt, K. L. Knudsen, L. L. Lundquist & P. Spruell, 2004. Intercrosses and the US Endangered Species Act: should hybridized populations be included as Westslope cutthroat trout? Conservation Biology 18: 1203–1213.
Almodovar, A., G. G. Nicola, B. Elvira & J. L. García-Marín, 2006. Introgression variability among Iberian brown trout Evolutionary Significant Units: the influence of local management and environmental features. Freshwater Biology 51: 1175–1187.
Almodovar, A., G. G. Nicola, D. Ayllon & B. Elvira, 2012. Global warming threatens the persistence of Mediterranean brown trout. Global Change Biology 18: 1549–1560.
Aparicio, E., E. Garcia-Berthou, R. M. Araguas, P. Martinez & J. L. García-Marín, 2005. Body pigmentation pattern to assess introgression by hatchery stocks in native Salmo trutta from Mediterranean streams. Journal of Fish Biology 67: 931–949.
Araguas, R. M., N. Sanz, R. Fernandez, F. M. Utter, C. Pla & J. L. García-Marín, 2008. Genetic refuges for a self-sustained fishery: experience in wild brown trout populations in the eastern Pyrenees. Ecology of Freshwater Fish 17: 610–616.
Araguas, R. M., N. Sanz, R. Fernandez-Cebrian, F. M. Utter, C. Pla & J. L. García-Marín, 2009. Role of genetic refuges in the restoration of native gene pools of brown trout. Conservation Biology 23: 871–878.
Araguas, R. M., M. Vera, E. Aparicio, N. Sanz, R. Fernandez-Cebrian, C. Marchante & J. L. García-Marín, 2017. Current status of the brown trout (Salmo trutta) populations within eastern Pyrenees genetic refuges. Ecology of Freshwater Fish 26: 120–132.
Arthington, A. H., N. K. Dulvy, W. Gladstone & I. J. Winfield, 2016. Fish conservation in freshwater and marine realms: status, threats and management. Aquatic Conservation: Marine and Freshwater Ecosystems 26: 838–857.
Bandelt, H. J., P. Forster & A. Rohl, 1999. Median-joining networks for inferring intraspecific phylogenies. Molecular Biology and Evolution 16: 37–48.
Bernas, R., A. Burzynski, P. Debowski, A. Pocwierz-Kotus & R. Wenne, 2014. Genetic diversity within sea trout population from an intensively stocked southern Baltic river, based on microsatellite DNA analysis. Fisheries Management and Ecology 21: 398–409.
Bernatchez, L., 2001. The evolutionary history of brown trout (Salmo trutta L.) inferred from phylogeographic, nested clade, and mismatch analyses of mitochondrial DNA variation. Evolution 55: 351–379.
Berrebi, P., D. Jesenek & A. Crivelli, 2017. Natural and domestic introgressions in the marble trout population of Soa River (Slovenia). Hydrobiologia 785: 277–291.
Bouza, C., R. Vilas, J. Castro & P. Martínez, 2008. Mitochondrial haplotype variability of brown trout populations from Northwestern Iberian Peninsula, a secondary contact area between lineages. Conservation Genetics 9: 917–920.
Cortey, M. & J. L. García-Marín, 2002. Evidence for phylogeographically informative sequence variation in the mitochondrial control region of Atlantic brown trout. Journal of Fish Biology 60: 1058–1063.
Cortey, M., C. Pla & J. L. García-Marín, 2004. Historical biogeography of Mediterranean trout. Molecular Phylogenetics and Evolution 33: 831–844.
Cortey, M., M. Vera, C. Pla & J. L. García-Marín, 2009. Northern and Southern expansions of Atlantic brown trout (Salmo trutta) populations during the Pleistocene. Biological Journal of the Linnean Society 97: 904–917.
Dahl-Jensen, D., M. R. Albert, A. Aldahan, N. Azuma, D. Balslev-Clausen, M. Baumgartner, A. M. Berggren, M. Bigler, T. Binder, T. Blunier, J. C. Bourgeois, E. J. Brook, S. L. Buchardt, C. Buizert, E. Capron, J. Chappellaz, J. Chung, H. B. Clausen, I. Cvijanovic, S. M. Davies, P. Ditlevsen, O. Eicher, H. Fischer, D. A. Fisher, L. G. Fleet, G. Gfeller, V. Gkinis, S. Gogineni, K. Goto-Azuma, A. Grinsted, H. Gudlaugsdottir, M. Guillevic, S. B. Hansen, M. Hansson, M. Hirabayashi, S. Hong, S. D. Hur, P. Huybrechts, C. S. Hvidberg, Y. Iizuka, T. Jenk, S. J. Johnsen, T. R. Jones, J. Jouzel, N. B. Karlsson, K. Kawamura, K. Keegan, E. Kettner, S. Kipfstuhl, H. A. Kjaer, M. Koutnik, T. Kuramoto, P. Koehler, T. Laepple, A. Landais, P. L. Langen, L. B. Larsen, D. Leuenberger, M. Leuenberger, C. Leuschen, J. Li, V. Lipenkov, P. Martinerie, O. J. Maselli, V. Masson-Delmotte, J. R. McConnell, H. Miller, O. Mini, A. Miyamoto, M. Montagnat-Rentier, R. Mulvaney, R. Muscheler, A. J. Orsi, J. Paden, C. Panton, F. Pattyn, J. R. Petit, K. Pol, T. Popp, G. Possnert, F. Prie, M. Prokopiou, A. Quiquet, S. O. Rasmussen, D. Raynaud, J. Ren, C. Reutenauer, C. Ritz, T. Rockmann, J. L. Rosen, M. Rubino, O. Rybak, D. Samyn, C. J. Sapart, A. Schilt, A. M. Z. Schmidt, J. Schwander, S. Schuepbach, I. Seierstad, J. P. Severinghaus, et al., 2013. Eemian interglacial reconstructed from a Greenland folded ice core. Nature 493: 489–494.
Excoffier, L., P. E. Smouse & J. M. Quattro, 1992. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131: 479–491.
Excoffier, L., G. Laval & S. Schneider, 2005. Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evolutionary Bioinformatics 1: 47–50.
Fabiani, A., P. Gratton, I. A. Zappes, M. Seminara, A. D’Orsi, V. Sbordoni & G. Allegrucci, 2018. Investigating the genetic structure of trout from the Garden of Ninfa (central Italy): suggestions for conservation and management. Fisheries Management and Ecology 25: 1–11.
Fernandez-Cebrian, R., R. M. Araguas, N. Sanz & J. L. García-Marín, 2014. Genetic risks of supplementing trout populations with native stocks: a simulation case study from current Pyrenean populations. Canadian Journal of Fisheries and Aquatic Sciences 71: 1243–1255.
Frankham, R., J. D. Ballou & D. A. Briscoe, 2002. Introduction to conservation genetics. Cambridge University Press, Cambridge.
Fumagalli, L., A. Snoj, D. Jesensek, F. Balloux, T. Jug, O. Duron, F. Brossier, A. J. Crivelli & P. Berrebi, 2002. Extreme genetic differentiation among the remnant populations of marble trout (Salmo marmoratus) in Slovenia. Molecular Ecology 11: 2711–2716.
García-Marín, J. L., P. E. Jorde, N. Ryman, F. Utter & C. Pla, 1991. Management implications of genetic differentiation between native and hatchery populations of brown trout (Salmo trutta) in Spain. Aquaculture 95: 235–249.
García-Marín, J. L., R. M. Araguas, M. Vera & N. Sanz, 2017. Understanding the brown trout population genetic structure in the Iberian Peninsula. In Lobón-Cerviá, J. & N. Sanz (eds), Brown Trout: biology, ecology and management. Wiley, Chichester: 103–126.
Gonzalez, S., R. Lopez-Roldan & J. L. Cortina, 2012. Presence and biological effects of emerging contaminants in Llobregat River basin: a review. Environmental Pollution 161: 83–92.
Hansen, M. M., D. Bekkevold, L. F. Jensen, K. L. D. Mensberg & E. E. Nielsen, 2006. Genetic restoration of a stocked brown trout Salmo trutta population using microsatellite DNA analysis of historical and contemporary samples. Journal of Applied Ecology 43: 669–679.
Hansen, M. M., D. J. Fraser, K. Meier & K. L. D. Mensberg, 2009. Sixty years of anthropogenic pressure: a spatio-temporal genetic analysis of brown trout populations subject to stocking and population declines. Molecular Ecology 18: 2549–2562.
Jadan, M., I. Strunjak-Perovic, N. Topic-Popovic & R. Coz-Rakovac, 2015. Three major phylogenetic lineages of brown trout (Salmo trutta Linnaeus, 1758) in the Krka River system (Croatia) revealed by complete mitochondrial DNA control region sequencing. Journal of Applied Ichthyology 31: 192–196.
Kohout, J., I. Jaskova, I. Papousek, A. Sediva & V. Slechta, 2012. Effects of stocking on the genetic structure of brown trout, Salmo trutta, in Central Europe inferred from mitochondrial and nuclear DNA markers. Fisheries Management and Ecology 19: 252–263.
Kumar, S., G. Stecher & K. Tamura, 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution 33: 1870–1874.
Marce, R., J. Honey-Roses, A. Manzano, L. Moragas, B. Catllar & S. Sabater, 2012. The Llobregat River basin: a paradigm of impaired rivers under climate change threats. The Handbook of Environmental Chemistry 21: 1–26.
Maric, S., O. Askeyev, A. Askeyev, S. Monakhov, N. Yanybaev, I. Askeyev, D. Galimova & A. Snoj, 2016. Lack of mtDNA variation among remote middle Volga and upper Ural brown trout suggests recent and rapid recolonization. Journal of Applied Ichthyology 32: 948–953.
Maric, S., S. S. Bajec, J. Schoffmann, V. Kostov & A. Snoj, 2017. Phylogeography of stream-dwelling trout in the Republic of Macedonia and a molecular genetic basis for revision of the taxonomy proposed by S. Karaman. Hydrobiologia 785: 249–260.
McDermid, J. L., S. Nienhuis, M. Al-Shamlih, T. J. Haxton & C. C. Wilson, 2014. Evaluating the genetic consequences of river fragmentation in lake sturgeon (Acipenser fulvescens Rafinesque, 1817) populations. Journal of Applied Ichthyology 30: 1514–1523.
McMeel, O. M., E. M. Hoey & A. Ferguson, 2001. Partial nucleotide sequences, and routine typing by polymerase chain reaction-restriction fragment length polymorphism, of the brown trout (Salmo trutta) lactate dehydrogenase, LDH-C1*90 and*100 alleles. Molecular Ecology 10: 29–34.
Meraner, A., S. Baric, B. Pelster & J. Dalla Via, 2007. Trout (Salmo trutta) mitochondrial DNA polymorphism in the centre of the marble trout distribution area. Hydrobiologia 579: 337–349.
Moore, W. S., 1995. Inferring phylogenies from mtDNA variation mitochondrial-gene trees versus nuclear-gene trees. Evolution 49: 718–726.
Muñoz, M. & M. Casadevall, 1997. Fish remains from Arbreda Cave (Serinyà Girona), northeast Spain, and their palaeoecological significance. Journal of Quaternary Science 12: 111–115.
Nei, M. & F. Tajima, 1981. DNA polymorphism detectable by restriction endonucleases. Genetics 97: 145–163.
Nonnis Marzano, F., N. Corradi, R. Papa, J. Tagliavini, F. N. Marzano & G. Gandolfi, 2003. Molecular evidence for introgression and loss of genetic variability in Salmo (trutta) macrostigma as a result of massive restocking of Apennine populations (Northern and Central Italy). Environmental Biology of Fishes 68: 349–356.
Ormerod, S. J., 2009. Climate change, river conservation and the adaptation challenge. Aquatic Conservation: Marine and Freshwater Ecosystems 19: 609–613.
Pfenninger, M. & D. Posada, 2002. Phylogeographic history of the land snail Candidula unifasciata (Helicellinae, Stylommatophora): fragmentation, corridor migration, and secondary contact. Evolution 56: 1776–1788.
Prat, N. & M. Rieradevall, 2006. 25-years of biomonitoring in two Mediterranean streams (Llobregat and Besòs basins, NE Spain). Limnetica 25: 541–550.
Rasmussen, G., 1986. The population dynamics of brown trout (Salmo trutta L.) in relation to year-class size. Polskie Archiwum Hydrobiologii 33: 489–508.
Rousset, F., 2008. GENEPOP ‘ 007: a complete re-implementation of the GENEPOP software for Windows and Linux. Molecular Ecology Resources 8: 103–106.
Ryman, N., F. Utter & L. Laikre, 1995. Protection of intraspecific biodiversity of exploited fishes. Reviews in Fish Biology and Fisheries 5: 417–446.
Sambrook, J., E. F. Fritsch & T. Maniatis, 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, New York.
Sanz, N., J. L. García-Marín & C. Pla, 2002. Managing fish populations under mosaic relationships. The case of brown trout (Salmo trutta) in peripheral Mediterranean populations. Conservation Genetics 3: 385–400.
Sanz, N., M. Cortey, C. Pla & J. L. García-Marín, 2006. Hatchery introgression blurs ancient hybridization between brown trout (Salmo trutta) lineages as indicated by complementary allozymes and mtDNA markers. Biological Conservation 130: 278–289.
Saura, M. & R. Faria, 2011. Genetic tools for restoration of fish populations. Journal of Applied Ichthyology 27: 5–15.
Schenekar, T., E. Lerceteau-Koehler & S. Weiss, 2014. Fine-scale phylogeographic contact zone in Austrian brown trout Salmo trutta reveals multiple waves of post-glacial colonization and a pre-dominance of natural versus anthropogenic admixture. Conservation Genetics 15: 561–572.
Snoj, A., I. Bogut & S. Susnik, 2008. Evidence of a genetically distinct population of Vrljika softmouth trout Salmo obtusirostris Heckel evolved by vicariance. Journal of Fish Biology 72: 1945–1959.
Soler, A., A. Canals, S. L. Goldstein, N. Otero, N. Antich & J. Spangenberg, 2002. Sulfur and strontium isotope composition of the Llobregat river (NE Spain): tracers of natural and anthropogenic chemicals in streams waters. Water, Air, and Soil Pollution 136: 207–224.
Sonstebo, J. H., R. Borgstrom & M. Heun, 2008. High genetic introgression in alpine brown trout (Salmo trutta L.) populations from Hardangervidda, Norway. Ecology of Freshwater Fish 17: 174–183.
Sostoa, A., N. M. Caiola, F. Casals, E. Garcia-Berthou, C. Alcaraz, L. Benejam, A. Maceda, C. Solà & A. Munné, 2010. Ajust de l’Index d’Integritat Biològica (IBICAT) basat en l’ús dels peixos com a indicadors de la qualitat ambiental als rius de Catalunya. Agència Catalana de l’Aigua, Departament de Medi Ambient i Habitatge, Generalitat de Catalunya, Barcelona.
Splendiani, A., M. Giovannotti, P. N. Cerioni, M. L. Caniglia & V. Caputo, 2006. Phylogeographic inferences on the native brown trout mtDNA variation in central Italy. Italian Journal of Zoology 73: 179–189.
Splendiani, A., P. Ruggeri, M. Giovannotti & V. Caputo, 2013. Role of environmental factors in the spread of domestic trout in Mediterranean streams. Freshwater Biology 58: 2089–2101.
Splendiani, A., P. Ruggeri, M. Giovannotti, S. Pesaresi, G. Occhipinti, T. Fioravanti, M. Lorenzoni, P. Nisi Cerioni & V. Caputo Barucchi, 2016. Alien brown trout invasion of the Italian peninsula: the role of geological, climate and anthropogenic factors. Biological Invasions 18: 2029–2044.
Suarez, J., J. M. Bautista, A. Almodovar & A. Machordom, 2001. Evolution of the mitochondrial control region in Palaearctic brown trout (Salmo trutta) populations: the biogeographical role of the Iberian Peninsula. Heredity 87: 198–206.
Susnik, S., J. Schoffmann & S. Weiss, 2005. Genetic verification of native brown trout from the Persian Gulf (Catak Cay River, Tigris basin). Journal of Fish Biology 67: 879–884.
Tamura, K. & M. Nei, 1993. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Molecular Biology and Evolution 10: 512–526.
Trigo, R. M., D. Pozo-Vazquez, T. J. Osborn, Y. Castro-Diez, S. Gamiz-Fortis & M. J. Esteban-Parra, 2004. North Atlantic oscillation influence on precipitation, river flow and water resources in the Iberian Peninsula. International Journal of Climatology 24: 925–944.
Vera, M., M. Cortey, N. Sanz & J. L. García-Marín, 2010. Maintenance of an endemic lineage of brown trout (Salmo trutta) within the Duero river basin. Journal of Zoological Systematics and Evolutionary Research 48: 181–187.
Vera, M., J. L. García-Marín, P. Martínez, R. M. Araguas & C. Bouza, 2013. Identification and conservation of remnant genetic resources of brown trout in relict populations from Western Mediterranean streams. Hydrobiologia 707: 29–45.
Vera, M., J. L. García-Marín, P. Martínez & C. Bouza, 2015. Phylogenetic diversity within the endemic brown trout Duero lineage: implications for conservation and management. Marine and Freshwater Research 66: 1066–1071.
Vera, M., P. Martínez & C. Bouza, 2018. Stocking impact, population structure and conservation of wild brown trout populations in inner Galicia (NW Spain), an unstable hydrologic region. Aquatic Conservation: Marine and Freshwater Ecosystems 28: 435–443.
Walsh, P. S., D. A. Metzger & R. Higuchi, 1991. CHELEX-100 as a medium for simple extraction of dna for pcr-based typing from forensic material. Biotechniques 10: 506–513.
Weiss, S. & S. Schmutz, 1999. Performance of hatchery-reared brown trout and their effects on wild fish in two small Austrian streams. Transactions of the American Fisheries Society 128: 302–316.
WWF, 2016. Living Planet Report 2016. Risk and resilience in a new era. WWF International, Gland.
Acknowledgements
Authors wish to thank M. López, S. Sánchez-Darriba and S. Gómez for their laboratory support; E. Aparicio for his fieldwork; and J. Ruiz, C. Marchante and J. Capdevila from the Inland Fisheries Service of the Generalitat de Catalunya Autonomous Government (Spain) for their technical advice. Authors are also indebted with Prof. M. Power and two anonymous reviewers for their constructive comments on the earlier version of this manuscript. A. Casanova was funded by a predoctoral reasearch fellowship from Xunta de Galicia Autonomous Government (Spain). This study has been supported by Spanish Science and Technology Ministry projects (REN2000-0740-C02-01; REN2003-05931/GLO), University of Girona project (MPCUdG2016/060), Generalitat de Catalunya (Spain) contracts (AMB/acs/itt 2001-3; AG-2017-617) and Xunta de Galicia project (GRC2014/010).
Author information
Authors and Affiliations
Corresponding author
Additional information
Handling editor: Michael Power
Rights and permissions
About this article
Cite this article
Vera, M., Bouza, C., Casanova, A. et al. Identification of an endemic Mediterranean brown trout mtDNA group within a highly perturbed aquatic system, the Llobregat River (NE Spain). Hydrobiologia 827, 277–291 (2019). https://doi.org/10.1007/s10750-018-3775-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10750-018-3775-9