Abstract
Andesiops peruvianus is widely distributed in the Andean region and has been preliminarily used as a bioindicator of good water quality. Given the morphological variations that are reported for the species, this study aimed to address whether such morphological variability in nymphs captured in tributaries of the Upper Chinchiná River Basin, Caldas-Colombia, could be connected to genetic differences, suggesting the existence of hidden, hitherto unknown taxonomic diversity. The morphological evaluation allowed confirming the presence of 73 females and 83 males belonging to what can be considered A. peruvianus sensu lato. However, Automatic Barcode Gap Discovery and Poisson Tree Processes modeling identified four different taxonomic units. The genetic distances found for specimens of A. peruvianus were higher than expected for conspecific organisms, and DNA analyses allowed to separate A. peruvianus into taxonomic units, which were further supported by morphological characters (shape and size of the abdominal gills and number of denticles of the tarsal claws). The results of this study show that A. peruvianus represents a species complex, with four putative species inferred, contributing to the growing knowledge of the existence of pseudocryptic species. These new findings could influence the conservation status of these rivers.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The Neotropical region has a great water resource richness and vast aquatic biodiversity. However, part of this biodiversity has not been thoroughly studied. Dijkstra et al. (2014) mention that nearly 100,000 species belonging to 12 orders spend one or more life stages in freshwater, yet little is known about how this remarkable diversity arose. Allopatric speciation and ecological adaptation are thought to be primary mechanisms involved, as freshwater habitats are highly susceptible to environmental change and exhibit marked ecological gradients (Gill et al., 2016). Ephemeroptera is an insect order with an aquatic nymphal stage, encompassing approximately 3,000 species that belong to 42 families and over 400 genera (Domínguez et al., 2006; Barber-James et al., 2008; Gutiérrez & Dias, 2015). Most of the species of this order are considered water quality indicators, because they do not tolerate variations in water quality and have been widely used and accepted as a biomonitoring tool (Roldán, 1999; Bonada et al., 2006; Menetrey et al., 2008; Zúñiga, 2009; Rutschmann et al., 2017).
The genus Andesiops Lugo-Ortiz & McCafferty, 1999 family Baetidae, reported exclusively in South America, was initially composed of a single species: A. peruvianus Ulmer, 1920. Currently, the genus consists of four species, A. angolinus Navás, 1933 , A. ardua Lugo-Ortiz & McCafferty, 1999, A. torrens Lugo-Ortiz & McCafferty, 1999 and A. peruvianus Lugo-Ortiz & McCafferty, 1999, which are distributed in Argentina, Peru, and Colombia (Nieto, 2004; Domínguez et al., 2006). Adults and nymphs described as A. peruvianus are distributed at high altitudes between Colombia and Argentina, along the Andean highlands (Nieto, 2004). Considering the broad geographic distribution of A. peruvianus in the Andean region and its apparent morphological variability (Finn et al., 2016; Gill et al., 2016), a molecular evaluation is warranted to investigate whether the species may actually constitute a group of closely related species, i.e., a species complex. For example, molecular data has revealed multiple cryptic species within the ephemeropteran genus Baetis (e.g., Baetis rhodani and Baetis alpinus) (Williams et al., 2006; Finn et al., 2014; Múrria et al., 2014).
Molecular tools have been widely used to discriminate aquatic insect species, mainly when morphological characters are not informative (Moore, 1995; Pereira-da-Conceicoa et al., 2012; Santos et al., 2016). Several studies (Williams et al., 2006; Virgilio et al., 2010; Gill et al., 2014; Hoyos et al., 2014; Rutschmann et al., 2014) have highlighted the existence of cryptic species in Ephemeroptera. A 658 bp fragment of the mtDNA cytochrome C oxidase subunit I (COI) gene has been standardized as the animal DNA barcode (Hebert et al., 2003).
In this study, we tested the hypothesis that A. peruvianus includes more than one species, or conversely, it is a single species with an elevated phenotypic plasticity, based on the examination of morphological features and sequence analysis of two mtDNA markers (16S rRNA and COI genes) in specimens collected from different affluents in the rural zone of the municipalities of Manizales and Villamaría, Caldas-Colombia, considering the morphological variability of A. peruvianus.
Materials and methods
Study area
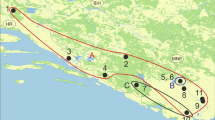
The study region included six stations located in the Chinchiná River Basin (Fig. 1). Three sampling stations were located in La Elvira stream, municipality of Manizales—Caldas (05°03′10.9″N 75°24′33.6″W; 05°03′4.4″N 75°24′33.1″W; 5°1′53″N 75°24′43.8″W) and three stations in the municipality of Villamaría—Caldas, Romerales stream (04°59′22″N 75°25′58″W), California stream (04°59′5″N 75°26′35″W), and Toldafría stream (4°59′08″N 75°26′43″W). These stations are located between 2275 and 2766 m.a.s.l. and have similar physical habitat characteristics, such as an undulating topography, the presence of riparian vegetation, and open canopy cover (0–25% of the streams are shaded). A total of six sampling events were carried out between February 2014 and February 2015; with months of high-rainfall (February, July and November 2014 and February 2015) and low-rainfall (April and September 2014). Additionally, to try to correlate environmental variables with the occurrence of hidden diversity in the studied group, the following physical and hydrobiological variables were measured in situ: water temperature, pH, conductivity, dissolved oxygen, oxygen saturation, average depth, width, and water flow velocity. Also, the following chemical variables were evaluated in the laboratory: chemical oxygen demand (COD), biological oxygen demand (BOD5), total coliforms, fecal coliforms, total suspended solids (TSS), total solids (TS), cyanide (CN), boron (B), lead (Pb), mercury (Hg), ammoniacal nitrogen (NH3-N), phosphate (PO4), sulfate (SO4), iron (Fe), chloride (Cl−), lipids and oils, nitrates (NO3), nitrites (NO2), and aluminum (Al) (Appendix 1—Supplementary Material).
Sampling stations in the Chinchiná River Basin, municipalities of Manizales and Villamaría in the department of Caldas, Colombia. (Adapted and modified from ESRI, 2014)
Two sampling stations can be considered references (areas without an evident mining impact), one located in La Elvira stream (05°03′10.9″N, 75°24′33.6″W) E1 and the other in Romerales stream (04°59′22″N, 75°25′58″W) E4. The other four sampling stations (E2, E3, E5, E6) are impacted by waste disposal generated by gold mining, according to Jiménez-Pérez et al. (2014) (Table 1). Due to the lack of normal distribution of the data, we compared the median of the physical, hydrobiological, and chemical variables between the reference and mining stations using the Wilcoxon two-sample test (W) (Zar, 1999), implemented in R version 2.15.3 (R Development Core Team, 2013).
Specimen sampling
Nymphs were collected from water using a Surber net (30.5 cm × 30.5 cm; 250 µm mesh size) and manual strainers, as well as from sediment, rocks, and fallen leaves samples. Specimens were labeled (with date, location, and coordinates), preserved in absolute ethanol, and deposited in the Entomological Collection of the Biology Program of the Universidad de Caldas (CEBUC for its initials in Spanish) in Manizales, Caldas-Colombia. This collection is registered in the Alexander von Humboldt Institute as collection number 188.
Morphological examination
Nymph morphology, sex, and aquatic stage determination was conducted using the keys and descriptions available for head, antennae, tarsal claws, labium palp, lingua, and eyes for A. peruvianus (Domínguez et al., 2006; Wilson & Kennedy, 2012; Gutiérrez & Dias, 2015). Specimens were examined under a Leica M205C stereomicroscope equipped with a MC170HD digital camera. Taxonomically important structures for the species were examined (such as labrum, dorsal surface, left and right mandible, posterior margin of terga, maxillae and maxillae palpi, dented plates distributed on tarsi, tibiae, and femora), in addition to mouthparts, tarsal claws, and abdominal gills, which were analyzed in search of genetic diversity within A. peruvianus. Euparal mountings were done, according to the procedures described by Waltz & McCafferty (1987), and were examined under a Leica DM500 microscope.
In addition, we observed 20 individuals with the help of scanning electron microscopy (SEM). For this, nymphs were subjected to five-min washes in increasing concentrations of acetone (50, 75, 90, 95, and 100%); the last concentration was repeated twice. The material was brought to critical level, mounted on stubs, and metalized with gold. Subsequently, the material was analyzed and photo-documented on a Hitachi TM3000 scanning electron microscope at the Laboratorio de Microscopia Eletrônica do Departamento de Biologia do Instituto de Biociências da UNESP de Rio Claro (Sao Paulo, Brazil).
Molecular evaluation
After the morphological identification, a total of 10 specimens from the six collection sites (six males and four females), previously identified based on shape and size of the abdominal gills and number of denticles of the tarsal claws, were individually processed for molecular analyses at the Molecular Biology Laboratory of the Department of Biological Sciences of Universidad de Caldas (Manizales, Caldas-Colombia). Tissue samples were obtained from the thorax and abdomen (the remaining body structures were preserved and mounted). Genomic DNA was extracted with the DNeasy Blood and Tissue Kit (QIAGEN®), following the standard protocol indicated by the manufacturer. DNA quality and quantity were measured using a NanoVue™ Plus spectrophotometer.
PCR amplification was performed using universal primers flanking the two target mtDNA genes widely employed in aquatic insects, including Ephemeroptera (Alexander et al., 2009; Pereira-da-Conceicoa et al., 2012; Rutschmann et al., 2014). The 16S rRNA gene was amplified using the primer pair 16Sa (F) 5′-GCCTGTTTATCAAAAACAT-3′ and 16Sb (R) 5′-CTCCGGTTTGAACTCAGATCA-3′ (Ogden & Whiting, 2005). Primers LCO1490 (F) 5′-GGTCAACAAATCATAAAGATATTGG-3′ and HCO2198 (R) 5′-TAAACTTCAGGGTGACCAAAAAATCA-3′ were used to amplify a fragment of the COI gene (Folmer et al., 1994). The final amplification reaction volume was 40 μl, which contained 21.4 μl ultrapure water, 8 μl 5× buffer, 2.4 μl MgCl2 (25 mM), 3.2 μl dNTP mix (10 mM), 0.8 μl of each primer (25 μM), 2 U of GoTaq® Flexi DNA Polymerase (Promega), and 3 μl DNA (approximately 100–130 ng of DNA). The amplifications were performed on a Techne TC-PLUS thermocycler, according to the following conditions: initial denaturation at 94°C for 2 min, followed by 7 cycles at 94°C for 30 s, 45°C for 30 s, and 72°C for 45 s, followed by 28 cycles at 94°C for 30 s, 48°C for 30 s, and 72°C for 45 s, completing the reaction with a final extension cycle at 72°C for 7 min, for the 16S rRNA gene. Initial denaturation at 95°C for 5 min, followed by 5 cycles at 94°C for 5 min, 46°C for 1 min30 s, and 72°C for 1 min 30 s, followed by 35 cycles at 94°C for 1 min, 53°C for 1 min, and 72°C for 1 min, completing the reaction with a final extension cycle at 72°C for 5 min, for the COI gene. All PCR products were visualized by horizontal electrophoresis on 1% agarose gels with 1× TBE pH 8.0 running buffer at 110 v/50 mA. Gels were stained with ethidium bromide and visualized on a GelDoc-It®2 310 Imager (UVP). PCR products were purified using the QIAquick PCR purification kit (QIAGEN®), according the manufacturer’s instructions, and sequenced at Macrogen Advancing Through Genomics—South Korea.
Sequences obtained were evaluated and edited with the programs Geneious Trial v8.14 (Drummond et al., 2009) and Sequencher 4.1 (Gene Codes Corporation, Ann Arbor, Michigan, USA). To further compare sequence divergence, we downloaded sequences of closely related taxa available in GenBank and BOLD databases. Specifically, we obtained COI gene sequences corresponding to Andesiops peruvianus (accessions KU710334–KU710343) (Finn et al., 2016) and Andesiops torrens (accessions GU175993–GU176002) (Sabando et al., 2011); while, as outgroup, we used Baetis sp. (Ephemeroptera: Leptohyphidae) (accession KR134666) (Múrria et al., 2015). There were no 16S rRNA gene sequences for Andesiops available in the public databases. The COI gene sequences were aligned using ClustalW (Thompson et al., 1997), included in the program MEGA version 7 (Tamura et al., 2013), and the 16S sequences were aligned using MAFFT (Multiple Alignment using Fast Fourier Transform), with Q-ins-i settings (Katoh et al., 2002) (http://mafft.cbrc.jp/alignment/server/).
Different taxonomic units were identified using two different approaches. First, we used Automatic Barcode Gap Discovery (ABGD; Puillandre et al., 2012), using an intraspecific prior divergence ranging from 0.001 to 0.1 and the K2P evolution model. Second, we used an evolutionary approach based on relative branch lengths. The Poisson Tree Processes (PTP) model identifies putative species boundaries on a given phylogenetic input tree (Zhan et al., 2013). The input tree was obtained by an updated version of the original maximum likelihood PTP (maximum likelihood PTP search result is part of the bPTP results), which adds Bayesian support (BS) values to species delimited on the input tree. A higher BS value on a node indicates that all descendants from this node are more likely to belong to a single species. Intraspecific nucleotide divergences between individuals and taxonomic units were estimated using the Kimura 2-Parameter distance model (K2P; Kimura, 1980), with the program MEGA. Species confirmation was carried out through a similarity analysis based on Maximum Likelihood (ML), with the K2P model and 1,000 bootstrap replications, using the program MEGA.
Results
Morphological data
A total of 156 nymphs in larval stage V were collected (Table 1). Specimens from both sexes possessed a head longer than broad, antennae three times longer than the head capsule (Fig. 2A–E); tarsal claws with an apical pair of fine setae and two rows of denticles, the first row well developed, and the second row reduced in size or in number of denticles (Figs. 2F, 3A); segment II of the labium palp with a small distomedial projection (Fig. 3C); lingua apically with a sharp-pointed projection (Fig. 3H). Finally, 73 females (Fig. 2B) and 83 males (Fig. 2C) were sexed, based on the presence of turbinate eyes in males.
Nymph, characterized as A. peruvianus: A Tarsal claw with two rows of denticles, the arrow indicates apical setae; B Labrum in ventral view (v.v.); C Labium v.v.; D Left maxilla v.v.; E Right maxilla v.v.; F Left mandible v.v.; G Right mandible v.v.; H Hypopharynx in dorsal view (d.v.), the arrow indicates the projection of the lingua (Light microscopy)
The following structures did not show variation across sampling stations: labrum longer than wide with anteromedial cleft and a small central lobe, dorsal surface with two kinds of apically bipectinate marginal setae: basally bifid near the midline of the labrum and apically bifid near the lateral setae (Figs. 3B, 4B); left mandible, prostheca robust, apically denticulate (Fig. 3F); right mandible with incisors cleft apically, prostheca bifid (Fig. 3G); posterior margin of terga serrate (Fig. 4A).
Nymphs, characterized as A. peruvianus: A Serrated margin in the posterior margin of the tergum and microsetae in the margin of the abdominal gills; B Labrum in dorsal view (d.v), the arrow indicates the clypeus; C, D Labium, the arrow points to the forked termination of the maxilla, labium palpi with short setae (Scanning Electron Microscopy—SEM)
In contrast to the characters previously reported for the species, the following two new features were recorded (with no differences observed among the A. peruvianus individuals examined): (1) top edge of the maxillae forked and setae randomly covering the short maxillae palpi (Figs. 3D–E, 4C, D); (2) dented plates distributed on tarsi, tibiae, and femora, alternated with short setae. In addition, variations in the number of denticles and abdominal gills revealed four groups: Group 1: Denticles 8/7–9 and with broadly tracheated gills; Group 2: Denticles 9/7–9 and with broadly tracheated gills (less marked than for Group 1); Group 3: Denticles 9/8–10 and very soft tracheated gills; and Group 4: Denticles 10–11/8–9 and without tracheae (Fig. 6; Table 1). There was no association between the sampling stations and the four groups of A. peruvianus (Table 1).
For the physical, hydrobiological and chemical analyses, significant differences were observed only for total suspended solids (TSS) (W = 60, P = 0.027), total solids (TS) (W = 61, P = 0.018), sulfate (SO4) (W = 63, P = 0.013), and iron (Fe) (W = 66, P = 0.003), according to the non-parametric Wilcoxon test. In addition, there were differences in organism abundance of A. peruvianus in relation to the reference and mining stations.
Molecular data
The sequences obtained in this study were deposited in GenBank and the Barcode of Life Data Systems (BOLD) under accession numbers KT625446–KT625464. Automatic Barcode Gap Discovery (ABGD) identified four different taxonomic units, based on the 16S rRNA sequences and using an intraspecific prior divergence between 0.001 and 0.0599 (Appendix 2A—Supplementary Material). Six taxonomic units were identified from the COI gene sequences, across an intraspecific prior divergence range between 0.001 and 0.1 (Appendix 2B—Supplementary Material). These taxonomic units corresponded to sequences of A. torrens and A. peruvianus, and four taxonomic units corresponding to the specimens from the Chinchiná River Basin (Caldas-Colombia), sequenced in the present study. The same taxonomic units were also recovered by the PTP model (Fig. 5).
Based on the intraspecific genetic distance of the molecular taxonomic units, A. peruvianus showed average distances between 0.0 and 0.0–0.9% for the 16S and the COI fragments, respectively (Tables 2, 3), while genetic divergence values between the taxonomic units of A. peruvianus varied between 7.3–12.4 and 17.4–24.5% for 16S and COI, respectively (Table 2, 3).
In addition, the Maximum Likelihood consensus trees obtained for the two genes clearly show a series of taxonomic units with low internal genetic divergences for the sequences of A. peruvianus. Four well-supported molecular groups are formed for A. peruvianus from the department of Caldas, based on the mtDNA 16S rDNA gene (Fig. 6), and five well-supported molecular groups are formed for A. peruvianus from the mtDNA COI gene, including the sequences deposited in GenBank (Fig. 5).
ML consensus tree with A. peruvianus samples, based on mtDNA 16S gene sequence distances. Bootstrap values are only indicated for nodes with support greater than 70%; Abdominal gills and variation in number of denticles of the tarsal claws of A. peruvianus nymphs revealed four groups: Group 1: Denticles 8/7–9 and with broadly tracheated gills; Group 2: Denticles 9/7–9 and with broadly tracheated gills (less marked than for Group 1); Group 3: Denticles 9/8–10 and very soft tracheated gills and Group 4: Denticles 10–11/8–9 and without tracheae. ML bootstrap values are shown at the nodes
Discussion
We analyzed 156 nymphs in larval stage V, collected from six stations located in the Chinchiná River Basin, that showed characteristics registered by Domínguez et al. (2006), Wilson & Kennedy (2012), Gutiérrez & Dias (2015) for A. peruvianus: head longer than broad, antennae three times longer than the head capsule (Fig. 2A–E); tarsal claws with an apical pair of fine setae and two rows of denticles, the first row well developed, and the second row reduced in size or in number of denticles (Figs. 2F, 3A); segment II of the labium palp with a small distomedial projection (Fig. 3C); lingua apically with a sharp-pointed projection (Fig. 3H), and the presence of turbinate eyes in males.
Nevertheless, we found that these morphological characteristics encompass organisms belonging to the A. peruvianus complex, based on the presence of four morphological groups, showing clear distinctions in shape and size of the abdominal gills and number of denticles of the tarsal claws (Table 1; Fig. 6). Variation in the number of denticles on the tarsal claws can be due to their function in substrate adhesion (Ditsche-Kuru et al., 2012), and the variation observed in tracheae branching relates to their importance in respiration, osmoregulation, and locomotion (Pruthi, 1927; Morgan & Grierson, 1932; Wingfield, 1939; Wichard et al., 1972; Notestine, 1994; Zhou, 2010).
The analyses show that these sampling stations have some type of anthropic impact, as evidenced by the physical, hydrobiological, and chemical analyses, where significant differences were observed only for total suspended solids (TSS) (W = 60, P = 0.027), total solids (TS) (W = 61, P = 0.018), sulfate (SO4) (W = 63, P = 0.013), and iron (Fe) (W = 66, P = 0.003), according to the non-parametric Wilcoxon test. A great amount of total and suspended solids were found, with a high content of sulfates and metals such as Fe, which are characteristic of mining disposals and can have negative effects on exposed organisms (Aduvire, 2006).
The molecular results further indicate a deep genetic structuring within A. peruvianus, showing that this species indeed constitutes a species complex, supporting the morphological results found (Table 1; Fig. 6). Four different taxonomic units were identified, using two different approaches: Automatic Barcode Gap Discovery (ABGD; Puillandre et al., 2012) for both 16S and COI, and PTP Zhan et al. (2013) for COI (Fig. 5). Accordingly, four putative species were inferred for A. peruvianus in the Chinchiná River Basin (Caldas-Colombia) (Appendix 2B—Supplementary Material), showing genetic distances between 0 and 24.5% for the COI gene, which represent higher than expected genetic distances for conspecific individuals, in comparison to the intraspecific genetic distances obtained for A. torrens (Table 3). Based on genetic divergences greater than 19% reported for B. rhodani haplogroups (Williams et al., 2006), Buckley et al. (2001), Gattolliat et al. (2015) suggested that such high divergences correspond to cryptic species. Avise (2000), Ball et al. (2009), Zhou et al. (2009) reported a 2% divergence criterion, exceeded only rarely by members of the same species and which is historically congruent with the morphological identification of aquatic insects.
Pereira-da-Conceicoa et al. (2012) suggest that Baetis harrisoni (Ephemeroptera), as is currently recognized, is not a single species with a wide geographic range and pH tolerance, but rather may encompass up to five species under the phylogenetic species concept, each with limited pH-tolerances; thus, the B. harrisoni species group is in need of taxonomic review. Further, Macher et al. (2016) studied the mayfly Deleatidium, in search of cryptic species using the COI gene, finding that Deleatidium consisted of 12 molecularly distinct clades that likely represent.
These previous findings support that the high genetic divergences obtained in A. peruvianus could correspond to confamiliar organisms belonging to a pseudocryptic species complex, as seems to occur with B. rhodani (Williams et al., 2006). Nevertheless, there was no association between the sampling stations and the four groups of A. peruvianus in the Chinchiná River Basin (Caldas-Colombia). COI gene studies in B. rhodani show that the species has at least 13 morphologically cryptic haplogroups, with local coexistence of cryptic species and their ability to adapt to different temperatures and food resources, justifying some of the differences observed in the relationship between water temperature, growth rates, and phenology documented from field studies (Lucentini et al., 2011; Rutschmann et al. 2014).
Tropical fauna is relatively unknown, therefore, comparing species richness, cryptic diversity, and altitudinal ranges of mayflies (Ephemeroptera) is crucial for understanding global biodiversity patterns (Bickford et al., 2007; Monaghan et al., 2009; Gill et al., 2014, 2016; Finn et al., 2016). Gill et al. (2016) provided evidence suggesting that the high species richness of tropical mayflies may be due not only to allopatric speciation, but also to parapatric speciation along single altitudinal gradients, because limited thermal tolerance restricts the dispersal ability of these species.
This research highlights the value of performing a detailed taxonomic classification of organisms with potential use as bioindicators of good water quality, using complementary tools such as morphology and molecular biology. The molecular data show the presence of four different taxonomic units supported by morphological characters, where the members of each group are not associated to a given study station. This agrees with reports by Finn et al. (2016); Gill et al. (2016), and with the presence of multiple cryptic o pseudocrytic species within the ephemeropteran genus Baetis (e.g., Baetis rhodani and Baetis alpinus) (Williams et al., 2006; Finn et al., 2014; Múrria et al., 2014). Nevertheless, it is necessary to continue exploring new tools and diagnostic characters, such as associations between nymphs and adults, reproductive mechanisms, mouthparts and tarsal claws in nymphs, and adult morphology such as wing venation, structures of the male genitalia with penis, forceps socket, forceps, and subgenital plate (Nieto, 2004; Domínguez et al., 2006). In addition, other molecular and biogeographic studies, as well as broader sampling efforts in the Chinchiná River basin could prove the existence of more groups within A. peruvianus s. l. (Pereira-da-Conceicoa et al., 2012; Rutschmann et al., 2014; Finn et al., 2016).
References
Aduvire, O., 2006. Drenaje acido de mina generación y tratamiento; Instituto Geológico y Minero de España 431 Dirección de Recursos Minerales y Geoambiente. España, Madrid: 1–140.
Alexander, L., M. Delion, D. J. Hawthorne & W. O. Lamp, 2009. Mitochondrial lineages DNA barcoding of closely related species in the mayfly genus Ephemerella (Ephemeroptera: Ephemerellidae). Journal of the North American Benthological Society 28: 584–595.
Avise, J. C., 2000. Phylogeography: The History and Formation of Species. Harvard University Press, Cambridge.
Ball, S. L., P. D. N. Hebert, S. K. Burian & J. M. Webb, 2009. Biological identifications of mayflies (Ephemeroptera) using DNA barcodes. Journal of the North American Benthological Society 24: 508–524.
Barber-James, H. M., J. L. Gattolliat, M. Sartori & M. D. Hubbard, 2008. Global diversity of mayflies (Ephemeroptera Insecta) in freshwater. Hydrobiologia 595: 339–350.
Bickford, D., D. J. Lohman, N. S. Sodhi, P. K. L. Ng, R. Meier, K. Winker, K. Ingram & I. Das, 2007. Cryptic species as a window on diversity and conservation. Trends in Ecology and Evolution 22: 148–155.
Bonada, N., N. Prat, V. H. Resh & B. Statzner, 2006. Developments in aquatic insect biomonitoring: a comparative analysis of recent approaches. Annual Review of Entomology 51: 495–523.
Buckley, T. R., C. Simon & G. K. Chambers, 2001. Phylogeography of the New Zealand Cicada Maoricicada campbelli based on mitochondrial DNA sequences: ancient clades associated with cenozoic environmental change. Evolution 55: 395–1407.
Dijkstra, K. D., M. T. Monaghan & S. U. Pauls, 2014. Freshwater biodiversity and aquatic insect diversification. Annual Review of Entomology 59: 143–163.
Ditsche-Kuru, P., W. Barthlott & J. H. Koop, 2012. At which surface roughness do claws cling? Investigations with larvae of the running water mayfly Epeorus assimilis (Heptageniidae Ephemeroptera). Zoology 115: 379–388.
Domínguez, E., C. Molineri, M. L. Pescador, M. D. Hubbard & C. Nieto, 2006. Diversidad Acuática en América Latina. Pensoft Publishers, Sofia.
Drummond, A. J., B. Ashton, M. Cheung, J. Heled, M. Kearse, R. Moir, H. S. Stones, T. Thierer & A. Wilson, 2009. Geneious version 8.14 [computer program]. Retrieved on June 01st 2015, http://www.geneious.com
ESRI, 2014. ArcGIS Desktop: Release 10. Environmental Systems Research Institute, Redlands.
Finn, D. S., C. Zamora-Muñoz, C. Múrria, M. Sáinz-Bariáin & J. Alba-Tercedor, 2014. Evidence from recently deglaciated mountain ranges that Baetis alpinus (Ephemeroptera) could lose significant genetic diversity as alpine glaciers disappear. Freshwater Science 33: 207–216.
Finn, D. S., A. C. Encalada & H. Hampel, 2016. Genetic isolation among mountains but not between stream types in a tropical high-altitude mayfly. Freshwater Biology 61: 702–714.
Folmer, O., M. Black, W. Hoeh, R. Lutz & R. Vrijenhoek, 1994. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Molecular Marine Biology and Biotechnology 3: 294–299.
Gattolliat, J.-L., C. Emilie, L. Vuataz & M. Sartori, 2015. DNA barcoding of Corsican mayflies (Ephemeroptera) with implications on biogeography, systematics and biodiversity. Senckenberg Gesellschaft für Naturforschung 73: 3–18.
Gene Codes Corporation, Ann Arbor, Michigan, USA. Sequencher® version 4.1 sequence analysis [computer program]. Retrieved on June 22th 2016, http://www.genecodes.com.
Gill, B. A., R. A. Harrington, B. C. Kondratieff, K. R. Zamudio, N. L. Poff & W. C. Funk, 2014. Morphological taxonomy, DNA barcoding, and species diversity in southern Rocky Mountain headwater streams. Freshwater Science 33: 288–301.
Gill, B. A., B. C. Kondratieff, K. L. Casner, A. C. Encalada, A. S. Flecker, D. G. Gannon, C. K. Ghalambor, J. M. Guayasamin, N. L. Poff, M. P. Simmons, S. A. Thomas, K. R. Zamudio & W. C. Funk, 2016. Cryptic species diversity reveals biogeographic support for the ‘mountain passes are higher in the tropics’ hypothesis. Proceedings Royal Society B 283(1832): 20160553.
Gutiérrez, Y. & L. Dias, 2015. Ephemeroptera (Insecta) de Caldas – Colombia claves taxonómicas para los géneros y notas sobre su distribución. Papéis Avulsos de Zoologia 55: 13–46.
Hebert, P. D. N., S. Ratnasingham & J. R. De Waard, 2003. Barcoding animal life: cytochrome c oxidase subunit 1 divergences among closely related species. Proceedings of the Royal Society of London, Series B 270: 596–599.
Hoyos, D., T. L. García, F. Rivera, G. López, M. D. C. Zúñiga & L. Dias, 2014. Contribución al conocimiento de las especies de Haplohyphes allen (Insecta: Ephemeroptera: Leptohyphidae) en Colombia. Caldasia 36: 164–165.
Jiménez-Pérez, P., B. Toro-Restrepo & E. Hernández-Atilano, 2014. Relación entre la comunidad de fitoperifiton y diferentes fuentes de contaminación en una quebrada de los Andes Colombianos. Relación fitoperifiton y contaminación ambiental. Boletín Científico Museo de Historia Natural 18: 49–66.
Katoh, K., K. Misawa, K. Kuma & T. Miyata, 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Research. 30: 3059–3066.
Kimura, M., 1980. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. Journal of Molecular Evolution 16: 111–120.
Lucentini, L., M. Rebora, M. E. Puletti, L. Gigliarelli, D. Fontaneto, E. Gaino & F. Panara, 2011. Geographical and seasonal evidence of cryptic diversity in the Baetis rhodani complex (Ephemeroptera, Baetidae) revealed by means of DNA taxonomy. Hydrobiologia 673: 215–228.
Lugo-Ortiz, C. R. & W. P. McCafferty, 1999. Three new genera of small minnow mayflies (Insecta: Ephemeroptera) from the Andes and Patagonia. Studies on Neotropical Fauna and Environment 34: 88–104.
Macher, J. N., R. K. Salis, K. S. Blakemorec, R. Tollriana, C. D. Matthaei & F. Leese, 2016. Multiple-stressor effects on stream invertebrates: DNA barcoding reveals contrasting responses of cryptic mayfly species. Ecological Indicators 61: 159–169.
Menetrey, N., B. Oertli, M. Sartori, A. Wagner & J. B. Lachavanne, 2008. Eutrophication: are mayflies (Ephemeroptera) good bioindicators for ponds? Hydrobiologia 597: 125–135.
Monaghan, M. T., R. Wild, M. Elliot, T. Fujisawa, M. Balke, D. J. G. Inward, D. C. Lees, R. Ranaivosolo, P. Eggleton, T. G. Barraclough & A. P. Vogler, 2009. Accelerated species inventory on Madagascar using coalescent-based models of species delineation. Systematic Biology 58: 298–311.
Moore, W. S., 1995. Inferring phylogenies from mtDNA variation: mitochondrial-gene trees versus nuclear-gene trees. Evolution 49: 718–729.
Morgan, A. H. & M. C. Grierson, 1932. The functions of the gills in burrowing mayflies (Hexagenia recurvata). Physiological Zoology 5: 230–245.
Múrria, C., M. Morante, M. Rieradevall, C. Ribera & N. Prat, 2014. Genetic diversity and species richness patterns in Baetidae (Ephemeroptera) in the Montseny Mountain range (North-East Iberian Peninsula). Limnetica 33(2): 313–326.
Múrria, C., A. T. Rugenski, M. R. Whiles & A. P. Vogler, 2015. Long-term isolation and endemicity of Neotropical aquatic insects limit the community responses to recent amphibian decline. Diversity and Distributions 21: 938–949.
Navás, L., 1933. Algunos Insectos de Chile. Revista Chilena De Historia Natural 37: 230–234.
Nieto, C., 2004. South American Baetidae (Ephemeroptera): a new generic synonymy. Studies on Neotropical Fauna and Environment 39: 95–101.
Notestine, M. K., 1994. Comparison of the respiratory currents produced by ephemeropteran nymphs with operculate gills. Journal of the Australian Entomological Society 33: 399–403.
Ogden, T. H. & M. F. Whiting, 2005. Phylogeny of Ephemeroptera (mayflies) based on molecular evidence. Molecular Phylogenetics and Evolution 37: 625–643.
Pereira-da-Conceicoa, L. L., B. W. Price, H. M. Barber-James, N. P. Barker, F. C. De Moor & M. H. Villet, 2012. Cryptic variation in an ecological indicator organism: mitochondrial and nuclear DNA sequence data confirm distinct lineages of Baetis harrisoni Barnard (Ephemeroptera: Baetidae) in southern Africa. BMC Evolutionary Biology 29: 12–26.
Puillandre, N., A. Lambert, S. Brouillet & G. Achaz, 2012. ABGD, Automatic Barcode Gap Discovery for primary species delimitation. Molecular Ecology 21: 1864–1877.
Pruthi, H. S., 1927. The ability of fishes to extract oxygen at different hydrogen ion concentrations of the medium. Journal of the Marine Biological Association UK 14: 741–748.
R Development Core Team. 2013. R: A language and environment for statistical computing. R Foundation for Statistical Computing.
Roldán, G., 1999. Los macroinvertebrados y su valor como indicadores de la calidad del agua. Revista de la Academia Colombiana de Ciencias Exactas Físicas y Naturales 23(88): 375–387.
Rutschmann, S., J. L. Gattolliat, S. L. Hughes, M. Báez, M. Sartori & M. T. Monaghan, 2014. Evolution and island endemism of morphologically cryptic Baetis and Cloeon species (Ephemeroptera Baetidae) on the Canary Islands and Madeira. Freshwater Biology 59: 2516–2527.
Rutschmann, S., H. Detering, S. Simon, D. H. Funk, J. L. Gattolliat, S. J. Hughes, P. M. Raposeiro, R. DeSalle, M. Sartori & M. T. Monaghan, 2017. Colonization and diversification of aquatic insects on three Macaronesian archipelagos using 59 nuclear loci derived from a draft Genome. Molecular Phylogenetics and Evolution 107: 27–38.
Sabando, M. C., I. Vila, R. Peñaloza & D. Véliz, 2011. Contrasting population genetic structure of two widespread aquatic insects in the Chilean high-slope rivers. Marine and Freshwater Research 62: 1–10.
Santos, A. P. M., D. M. Takiya & J. L. Nessimian, 2016. Integrative taxonomy of Metrichia Ross (Trichoptera: Hydroptilidae: Ochrotrichiinae) microcaddisflies from Brazil: descriptions of twenty new species. PeerJ 4: e2009.
Tamura, K., G. Stecher, D. Peterson, A. Filipski & S. Kumar, 2013. MEGA 6: molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution 30: 2725–2729.
Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin & D. G. Higgins, 1997. The CLUSTAL X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research 25: 4876–4882.
Ulmer, G., 1920. Neue ephemeropteren. Archiv für Naturgeschichte 85: 1–80.
Virgilio, M., M. T. Backeljau, B. Nevado & M. De Meyer, 2010. Comparative performances of DNA barcoding across insect orders. BMC Bioinformatic 11: 1–10.
Waltz, R. D. & W. P. McCafferty, 1987. Revision of the genus Cloeodes Traver (Ephemeroptera: Baetidae). Annals of the Entomological Society of America 80: 191–207.
Wichard, W., H. Komnick & J. H. Abel Jr., 1972. Typology of ephemerid chloride cells. Zeitschrift für Zellforschung und Mikroskopische Anatomie 132: 533–551.
Williams, H. C., S. J. Ormerod & M. W. Bruford, 2006. Molecular systematics and phylogeography of the cryptic species complex Baetis rhodani (Ephemeroptera Baetidae). Molecular Phylogenetics and Evolution 40: 370–382.
Wilson, R. & J. Kennedy, 2012. Life histories of Edwardsina sp. and Andesiops ardua from Navarino Island Chile. Retrieved on August 01st 2015, http://digital.library.unt.edu/ark:/67531/metadc86770/.
Wingfield, C. A., 1939. The function of the gills of mayfly nymphs from different habitats. Journal of Experimental Biology 16: 363–373.
Zar, J. H., 1999. Biostatistical analysis, 4th ed. Prentice Hall, Upper Saddle River.
Zhan, J., P. Kapli, P. Pavlidis & A. Stamatakis, 2013. A general species delimitation method with applications to phylogenetic placements. Bionformatics 29(2): 2869–2876.
Zhou, C.-F., 2010. Accessory gills in mayflies (Ephemeroptera). Stuttgarter Beiträge zur Naturkunde, Serie A Biologie 3: 79–84.
Zhou, X., S. J. Adamowicz, L. M. Jacobus, R. E. De Walt & P. D. N. Hebert, 2009. Towards a comprehensive barcode library for arctic life – Ephemeroptera, Plecoptera, and Trichoptera of Churchill, Manitoba, Canada. Frontiers in Zoology 6: 1–9.
Zúñiga, M. D. C., 2009. Bioindicadores de calidad de agua y caudal ambiental. In Cantera, J. R. K., Y. E. Carvajal & L. M. H. Castro (eds), El caudal ambiental: conceptos experiencias y desafíos. Programa Editorial Universidad del Valle, Valle: 176–206.
Acknowledgements
To the Research Group GEBIOME, the IIES Institute, Laboratory of Microbiology, and the Colección Entomológica del Programa de Biología de la Universidad de Caldas (CEBUC), especially Dra. Lucimar Gomes Dias for the taxonomic confirmation of the species. To Carlos Augusto Gil for the illustrations. To COLCIENCIAS for financing the project “Evaluation of mining, agriculture, and livestock impact through ecological and genetic responses of aquatic macroinvertebrates”.
Author information
Authors and Affiliations
Corresponding author
Additional information
Handling editor: Diego Fontaneto
Electronic supplementary material
Below is the link to the electronic supplementary material.
Appendix 1
Physical, hydrobiological, and chemical characteristics of the creeks assessed. The values correspond to the mean values of the parameters measured in each sampling station. E1, E2 and E3: La Elvira streams, E4: Romerales stream, E5: California stream, E6: Toldafría stream. Supplementary material 1 (DOCX 19 kb)
Appendix 2
Automatic partitioning of the ABGD method Kimura two – Parameters; A) mtDNA 16SrDNA gene; B) mtDNA COI gene. Supplementary material 2 (TIFF 12039 kb)
Rights and permissions
About this article
Cite this article
Ossa-López, P.A., Camargo-Mathias, M.I. & Rivera-Páez, F.A. Andesiops peruvianus (Ephemeroptera: Baetidae): a species complex based on molecular markers and morphology. Hydrobiologia 805, 351–364 (2018). https://doi.org/10.1007/s10750-017-3321-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10750-017-3321-1