Abstract
Branchinecta lindahli is a generalist fairy shrimp that has a widespread distribution in North America. We initiated a study to better understand the geographic structure of genetic variation within this species and to test if cryptic species are present in B. lindahli. Additionally, we examined four other species of Branchinecta to test if there were any generalities in the levels of genetic divergence within and among species. Genetic data from the mitochondrial cytochrome c oxidase I gene revealed two distinct clades within B. lindahli: one found in the Central Valley and Mojave Desert of California, USA and one found in Baja California, Mexico. The remaining haplotypes were widespread throughout the remainder of North America. Further examination of the distribution within clades revealed no additional phylogeographic structure. The amount of intraspecific divergence observed for B. lindahli and B. hiberna was high compared to B. mackini and B. sandiegonensis. However, maximal intraspecific divergences were less than what was observed among Branchinecta species. We argue that the amount of intraspecific divergence observed in B. lindahli is not consistent with the presence of cryptic species and that caution should be taken when attempting to delimit cryptic species within this group.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Phylogeographic studies on passively dispersed zooplankton have revealed that complex non-equilibrium processes drive both regional and localized genetic differentiation (Boileau et al., 1992; Hebert et al., 2003; Gomez et al., 2007; Schwentner et al., 2015). The dogma states that widespread species have high dispersal capabilities and should have limited genetic structure (Boileau et al., 1992; De Meester et al., 2002; but see Bohonak and Jenkins, 2003). However, a large body of work has shown that this is not always the case and that in fact many widespread aquatic zooplankton species are composed of cryptic genetic lineages, often with complex geographic patterns (Penton et al., 2004; Muñoz et al., 2008; Meusel & Schwentner, 2016).
Attempting to elucidate the forces that drive phylogeographic patterning in aquatic zooplankton is contingent upon evaluating many complex processes. Strong founder (or priority) effects are thought to be extremely important and can create non-equilibrium patterns of genetic differentiation (De Meester et al., 2002). Post-Pleistocene expansions can also generate non-equilibrium patterns of high gene flow among recently founded populations (Boileau et al., 1992; De Gelas & De Meester, 2005; Millette et al., 2011). Many studies do find geographically differentiated phylogroups and, in some instances, cryptic species that are associated with major barriers (Hebert et al., 2003; Xu et al., 2009). Complex geographic associations and regional endemism of clades have been observed within freshwater crustaceans (Aguilar, 2011; Ketmaier et al., 2012; Schwentner et al., 2014).
Fairy shrimp of the genus Branchinecta, generally considered cool water organisms, have a Holarctic and New World distribution (Belk & Brtek, 1995, 1997). In the New World, they occur from Canada to the Patagonia/Antarctic regions and inhabit a wide variety of temporary aquatic habitats (Rogers, 2009). They can be found in deserts, arid/prairie grasslands, and mountain regions. The fairy shrimp Branchinecta lindahli Packard, 1883 is the most widespread North American Branchinecta species. It was originally described from an ephemeral pool near Wallace, Wallace County, Kansas, USA; however, due to confusion surrounding the original description, a neotype locality was designated by Lynch (1964) which is 16 km east of Garden City, Finney County, Kansas, USA. Branchinecta lindahli is reported from Canada (Alberta), USA (Arizona, California, Colorado, Iowa, Kansas, Montana, Nebraska, Nevada, New Mexico, North Dakota, Oklahoma, Oregon, Texas, Utah, Washington, and Wyoming), and Mexico (Baja California (Norte), and Baja California (Sur)) (Lynch, 1964; Hartland-Rowe, 1965; Belk, 1975, 1977, 1983; Belk and Lindberg, 1979; Belk & Brtek, 1995; Eng et al., 1990; Eriksen and Belk, 1999; Maeda-Martínez et al., 2002; Obregón-Barboza et al., 2002, 2015a, b). This species utilizes a large range of temporary aquatic habitats from seasonally arid regions, including: alkaline vernal pools, prairie potholes, and slightly saline pools and playas (Hartland-Rowe, 1965; Horne, 1967, 1971; Belk, 1977, 1983; Eng et al., 1990; Eriksen & Belk, 1999; Maeda-Martínez et al., 2002). Horne (1967, 1971) reported that B. lindahli is remarkably tolerant of a wide range of dissolved salts types and concentrations, and Eng et al. (1990) recorded it from both turbid and clear water habitats. Rogers (2014a) examined geochemical parameters from 23 populations across the species distribution and found that B. lindahli occurs in sites with 0–11% gypsum, 0–30% CaCO3, 0–24 mS/cm salinity, and always in locations where sodium and calcium salts were present. It is commonly encountered in disturbed habitats, such as roadside ditches and railroad bed toe drains, co-occurring with fifteen other fairy shrimp species from five genera (Lynch, 1964; Hartland-Rowe, 1965; Belk, 1977, 1983; Eng et al., 1990; Eriksen and Belk, 1999; Maeda-Martínez, 1991; Maeda-Martínez et al., 1997, 2002). The fact that B. lindahli has been observed in such diverse habitats indicates a high potential for gene flow in this species.
Schwentner et al. (2011) outlined the utility of an integrated taxonomic approach to delineating species in the Branchiopoda. They utilized multiple species concepts to better define species limits within the clam shrimp genus Limnadopsis. The vast majority of work in the Branchinectidae has applied a pure morphological approach to delineating and describing species (Rogers, 2013). In our initial assessment of genetic variation within the species of Branchinecta, we rely on morphological characteristics to identify and delineate species. We then apply genetic approaches under a phylogenetic species concept to establish the limits of intra and inter-specific divergence in selected Branchinecta. The identification of cryptic lineages could then be further investigated under additional conceptual frameworks to verify their status.
The goals of this study were to examine the extent of genetic divergence within B. lindahli throughout most its geographical range and discuss these results in light of the potential existence of cryptic species in this morphological entity. If B. lindahli is truly a generalist and has the potential for high gene flow among regions, we would expect to observe limited genetic differentiation across its range. However, given that genetic structure is often high among passively dispersed zooplankton, we expect to observe cryptic lineages within B. lindahli. We also address the presence of cryptic species within the Branchinectidae through a comparative analysis of within and among species divergence. This information will be useful for future work that attempts to assess species limits in this genus.
Materials and methods
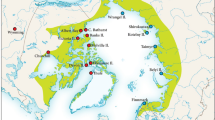
Branchinecta lindahli populations were opportunistically sampled from various habitat types across the species range (Fig. 1). Fairy shrimp were obtained directly from the natural habitats or from cultures derived from wild collected eggs by one of two methods: outdoor cultures with fiber glass tanks of 1200 l using commercial drinking water (total dissolved solids <0.3 g/l) and 20 kg of dry soil containing B. lindahli eggs as substrate, and laboratory cultures with plastic containers using 10 l of water and 100 g of soil containing B. lindahli eggs. In the field, individuals were collected using dip nets. All specimens were stored in 95% ethanol and taxonomically identified in the laboratory. Samples of three additional species (Branchinecta hiberna Rogers and Fugate, 2001, B. mackini Dexter, 1956, and B. lutulenta Rogers and Hill, 2013) were taken from the collections of D.C. Rogers.
Genetic analysis
Genetic analysis was performed on 231 Branchinecta lindahli individuals from 14 sites, 16 B. hiberna individuals from two sites, 19 B. mackini individuals from four sites, and three B. lutulenta individuals from a single site (Table 1). DNA was extracted from whole specimens using either the Qiagen DNA easy extraction kit (Qiagen Inc.) or with a Chelex extraction protocol. For the Chelex extraction, a small portion of the abdomen was placed in 5% Chelex and 2 µl of 20 mg/ml proteinase K. The sample was then incubated for 1 h at 60°C, followed by 5 min at 95°C, then cooled on ice. Samples were then centrifuged and the supernatant removed and used directly or in a 1:10 dilution for subsequent amplifications. Qiagen extractions were done following the manufacturer’s protocols. A portion of the mitochondrial cytochrome c oxidase I gene was amplified using the primers LCO1490 (5′-GGTCAACAAATCATAAAGATATTG-3′) and HCO2198 (5′-TAAACTTCAGGGTGACCAAAAAATCA-3′) (Folmer et al., 1994). Each reaction contained 2µl of 1:10 diluted template DNA, 3µl of 10× PCR buffer from Applied Biosystems Inc. (ABI, Foster City, CA), 2.1 mM of MgCl2, 0.4 µM of dNTPs, 0.4 µM of each primer, 5 ng of BSA, and 0.4 units of Taq polymerase in a 30 µl reaction. Reactions were run on an ABI 2720 thermocycler under the following conditions: initial denaturation 3 min at 94°C, 35 cycles of 94°C for 30 s, 48°C for 30 s, and 72°C for 45 s followed by a final extension for 5 min at 72°C. PCR products from the COI gene were directly sequenced in both directions on an Applied Biosystems (ABI) 3130xl automated sequencer using BigDye (v3.1) chemistry. Sequences were deposited in GenBank (Table 1). For this study, we included GenBank sequences of Branchinecta sandiegonensis Fugate, 1993 (Accession #’s: FJ439689-FJ439743) in the ingroup, and Streptocephalus sealii Ryder, 1879 (AY519832) and Thamnocephalus platyurus Packard, 1877 (AF209066) as outgroups.
DNA sequences were aligned using MUSCLE and checked for proper reading frame manually. They were imported to jMODELTEST (Posada, 2008) for DNA sequence model evaluation using the Bayesian Information Criterion. This approach found that the GTR + G model of sequence evolution was most appropriate for our alignment. Phylogenetic analysis of the COI sequences was conducted in a Bayesian MCMC framework (Ronquist & Huelsenbeck, 2003). Bayesian analyses involved two independent chains per run and were run for 10,000,000 generations sampling every 1000 generations on MrBayes (Ronquist & Huelsenbeck, 2003). The first 25% of recorded runs were discarded as burn-in. Consensus trees were visualized in FigTree (beast.bio.ed.ac.uk/FigTree). Haplotype networks were constructed independently for each group that was revealed in the phylogenetic analysis using the median-joining algorithm (Bandelt et al., 1999) in PopART (Leigh & Bryant, 2015). Pairwise estimates of sequence divergence, using the K2P model of sequence evolution, were estimated among all possible pairwise sequence comparisons using MEGA4 (Tamura et al., 2007). The K2P model is commonly used in ‘DNA Barcoding’ studies (Lefébure et al., 2006; Costa et al., 2007) and we applied it so that we could make appropriate comparisons with the previously published work. This information was then used to explore genetic divergence within B. lindahli and across the genus Branchinecta. We also assessed various measures of diversity for the COI data (number of segregating sites, haplotype diversity, and nucleotide diversity) with DnaSP v5 (Librado & Rozas, 2009).
Results
The COI gene fragment (670 bp) was sequenced from 269 individuals (Haplotype information in Supplementary Data). Bayesian phylogenetic reconstruction and neighbor-net network of all unique COI haplotypes of B. lindahli revealed two well-supported groups (Clade A and B) and a basal set of haplotypes (Group C) (Fig. 2). Clade A is found in the Great Central Valley and Mojave Desert of California, and clade B is found in Baja California. Group C is composed of a basal group of paraphyletic haplotypes within B. lindahli and is widespread, found in southern California, Colorado, and Wyoming. Pairwise comparisons across all samples of B. lindahli ranged from 0 to 7.4% sequence divergence. We observed a mean sequence divergence of 1.2% (range 0.1–3.5%) within clade A, 0.5% (range 0.1–1.1%) divergence within clade B, and 1.4% (range 0.1–3.3%) divergence within group C. Mean divergences between clades A–B, B–C, and A–C were 4.0, 5.1, and 4.9%, respectively. Haplotype and nucleotide diversity varied throughout the sample sites (Table 1); however, sites in Mexico had the lowest levels of variation.
Phylogenetic analysis
Phylogenetic analysis of all five species with the COI sequence revealed a tree where all species of Branchinecta were well resolved (Fig. 2). There was also strong support for a monophyletic Branchinecta (Fig. 2). Internal nodes had little statistical support and any inference on the relationships among species based on COI gene variation cannot be made at this point.
Haplotype networks
Networks from all clades showed partitioning of haplotypes over various geographic distances; however, clear geographic patterns are not evident across the three clades. Clade A (Fig. 3A) showed little sharing of haplotypes across sampling sites, with the most divergent haplotypes at Edwards Airforce Base. The samples from northern Baja California shared haplotypes among sites (Fig. 3B); however, these sites were in much closer geographic proximity to one another than the most other sampled sites in this study. Group C contained the most complex haplotype network (Fig. 3C). There were only three haplotypes shared among the sampled sites and the network shows little to no geographic association of haplotypes.
Haplotype networks generated with the median-joining algorithm for the three major clades from Fig. 1. A Clade A; B Clade B; C Group C. Tick marks indicate the number of changes between haplotypes and the size of the circles is proportional to the frequency of the haplotype in our sample
Sequence divergence
Branchinecta lindahli and B. hiberna had the highest levels of within-species divergence (Fig. 4). Within the B. hiberna samples, there was a mean sequence divergence of 3.9% (range 0.1–6.6%). Sequence divergence within B. lindahli was 7.4%. Branchinecta mackini (range 0.2–2.8%) and B. sandiegonensis (range 0.2–3.2%) contained much lower levels of within-species divergence (Fig. 4). Most levels of pairwise sequence divergence between all recognized species, with the exception of B. lindahli-B. lutulenta, were greater than 7% (Fig. 4).
Box plots of between and within Branchinecta species divergence at the COI gene based on the Kimura 2-parameter model. Dashed lines indicate maximal and minimal values within 75 and 25% quantiles and dots indicate outlier values outside this range. Intraspecific comparisons below the dashed lines and inter-specific comparisons are above the dashed line
Discussion
We uncovered two divergent clades within B. lindahli. The amount of intraspecific genetic divergence, at a common barcoding gene (COI), was consistent with what is observed in other species. These results are similar to what is found in other passively dispersed aquatic zooplankton (Schwentner et al., 2014; Schwentner et al., 2015; Lindholm et al., 2016); however, they do not support the designation of cryptic species within B. lindahli given their divergence is below what is observed between species of Branchinecta.
Phylogenetic patterns
The phylogenetic patterns within B. lindahli cannot be attributed to any simple geographic hypotheses. Our three clades/groups cover overlapping geographic regions. Clade A is geographically restricted to sites in the Mojave Desert and California’s Great Central Valley. Clade B was restricted to a region of northern Baja California. Group C was widely distributed across western North America, including sites in Colorado and Wyoming, and multiple sites in California. Clades A and B are sister to one another and their relationship has strong statistical support (posterior probability = 1.0; maximum likelihood bootstrap support = 0.92).
Haplotype networks suggests a high degree of haplotype endemism, with the exception of the Baja California sites. The pattern of shared haplotypes, with little geographic structure, may be due to the close proximity of sites sampled in this region. However, the patterns in clade A and group C are more indicative of what is observed in other passively dispersed aquatic zooplankton, with little sharing of haplotypes across sites with complex geographic patterns. Such patterns are attributed to the stochastic nature of dispersal and colonization for these organisms. Clearly, denser sampling across western North America would be useful in delineating the geographic boundaries of these clades and possibly identifying additional clades.
Intraspecific divergence in the Branchinectidae
Intraspecific variation of four species of Branchinecta shows that two of the species (B. hiberna and B. lindahli) contain high intraspecific genetic divergences when compared to the other two species. There is no clear pattern with regards to species geographic distribution. Branchinecta sandiegonensis has intraspecific variation and is restricted to southern California and northern Baja California (Fugate, 1993). Branchinecta mackini is widely distributed in North America west of the continental divide and also has low levels of within-species divergence. Branchinecta hiberna has been found in the Great Basin regions of northern California, eastern Oregon and Nevada, as well as southern Idaho (Rogers and Fugate, 2001; Rogers et al., 2006; Rogers pers. obs.) and contains intraspecific divergences similar to B. lindahli. If a wide geographic distribution was predictive of high intraspecific divergences, we would expect to observe high K2P distances within B. mackini; however, the species-level divergences for B. mackini are similar to that of B. sandiegonensis. This may be due to the fact that B. mackini occurs in very large temporary lakes and their surrounding wetlands. These larger habitats would attract larger numbers of birds, thus dispersal vectors are stronger and more predictable (Rogers, 2014b, 2015).
The two species with the highest intraspecific divergences are B. hiberna and B. lindahli. Maximal intraspecific divergences for these two species are near what has been proposed as a ‘species-level cutoff’ for crustaceans based on the COI gene (Lefébure et al., 2006; Costa et al., 2007). While it would be tempting to suggest that cryptic species exist within these two taxa given the presence of well-supported clades at the intraspecific level, we feel it is premature to utilize this designation at this time. This is predominately due to the observed levels of pairwise genetic divergence among the different species and how this compares to the levels of intraspecific divergence (see below).
Intra-species genetic distances in the COI gene of Branchinecta were reported to be 0–3% (Vandergast et al. (2009), 2.5–5.3 in Chirocephalus (Ketmaier et al., 2012), 2.1–5.0 in Tanymastix (Ketmaier et al., 2005), and 0.1–2.5 in Thamnocephalus (Obregón-Barboza et al., 2015a). The observed pairwise sequence divergence within B. hiberna (0.1–6.6%), B. lindahli (0–7.4%), B. mackini (0.2–2.8%), and B. sandiegonensis (0.2–3.2%) is more consistent with deep within-species divergences rather than the presence of cryptic species. Lefébure et al. (2006) found a maximal level of within-species sequence divergence in the COI gene for a variety of crustacean taxa to be ~7.0%. Costa et al. (2007) found that in a large-scale survey of crustaceans, within-species divergence for the COI gene ranged from 0.0 to 5.0%, while inter-specific comparisons within genera showed divergences of 9.0–31.0%. Both of these studies used the K2P distance and a similar region of the COI gene that we present here. Therefore, our intraspecific divergences for B. lindahli (0–7.4%) and B. hiberna (0.1–6.6%) are comparable to intraspecific variation from other studies.
The lowest between-species divergence was observed in the comparison of B. lindahli and B. lutulenta. Divergences between these two species range from 7 to 10%, while pairwise divergences between other species comparisons range from 8 to 17%. It would be difficult and arbitrary to establish a level of divergence that one could use to delimit species within the Branchinectidae based on COI sequence data. Therefore, comparative analysis, like the one presented here, is useful in establishing a baseline for this type of discussion.
Our data and analysis are not without limitations. Hybridization would easily obscure any genetic signal that could be used to establish species boundaries, as hybridization would lead to sharing of haplotypes across species (Hebert et al., 2004). Our data do not suggest hybridization between species or even among distinct lineages within B. lindahli; however, this could pose problems in applying an approach similar to ours. Lastly, the inclusion of genomic data would greatly improve one’s ability to correctly delimit species (in a purely genetic context). Therefore future work should move away from solely mitochondrial loci and utilize more information from the nuclear genome.
Conclusion
The deep genetic divergences and distinct clades found within B. lindahli in this study might lead one to infer that cryptic species exist for this taxon; however, our results from a comparison of intraspecific COI divergences in this and other Branchinecta suggest that cryptic taxa are not present in the studied taxon. Additional data (e.g., nuclear DNA) may improve our ability to assess reproductive isolation among these lineages. Therefore, it is premature to suggest that speciation has or is occurring in B. lindahli. Our comparison of intra/inter-specific divergence at the COI locus does suggest that intraspecific divergences for B. lindahli are well below what is observed among other Branchinecta species. Further study of the clades with more restricted geographic distributions (A and B) will provide a more detailed analysis of genetic, reproductive, and ecological divergence in this widespread generalist crustacean.
References
Aguilar, A., 2011. Weak phylogeographic structure in the endemic western North American fairy shrimp Branchinecta lynchi (Eng, Belk and Eriksen 1990). Aquatic Sciences 73: 15–20.
Bandelt, H. J., P. Forster & A. Röhl, 1999. Median-joining networks for inferring intraspecific phylogenies. Molecular Biology and Evolution 16: 37–48.
Belk, D., 1975. Key to the Anostraca (fairy shrimps) of North America. The Southwestern Naturalist 20: 91–103.
Belk, D., 1977. Zoogeography of the Arizona fairy shrimps (Crustacea: Anostraca). Arizona Academy of Sciences 12: 70–78.
Belk, D., 1983. New fairy shrimp distribution records among collections at the California Academy of Sciences. The Southwestern Naturalist 28: 380–381.
Belk, D. & J. Brtek, 1995. Checklist of the Anostraca. Hydrobiologia 298: 315–353.
Belk, D. & J. Brtek, 1997. Supplement to ‘Checklist of the Anostraca’. Hydrobiologia 359: 243–245.
Belk, D. & D. R. Lindberg, 1979. First freshwater animal reported for Isla de Guadalupe represents a southern range extension for Branchinecta lindahli. (Crustacea: Anostraca). The Southwestern Naturalist 24: 390–391.
Bohonak, A. & D. Jenkins, 2003. Ecological and evolutionary significant of dispersal by freshwater invertebrates. Ecology Letters 6: 783–796.
Boileau, M. G., P. N. D. Hebert & S. S. Schwartz, 1992. Nonequilibrium gene frequency divergence – persistent founder effects in natural populations. Journal of Evolutionary Biology 5: 25–39.
Costa, F. O., J. R. de Waard, J. Boutillier, S. Ratnasingham, R. T. Dooh, M. Hajibabae & P. D. N. Hebert, 2007. Biological identifications through DNA barcodes: the case of the Crustacea. Canadian Journal of Fisheries and Aquatic Sciences 64: 272–295.
De Gelas, K. & L. De Meester, 2005. Phylogeography of Daphnia magna in Europe. Molecular Ecology 14: 753–763.
De Meester, L., A. Gomez, B. Okamura & K. Schwenk, 2002. The monopolization hypothesis and the dispersal-gene flow paradox in aquatic organisms. Acta Oecologia 23: 121–135.
Dexter, R. W., 1956. A new fairy shrimp from western United States, with notes on other North American species. Journal of the Washington Academy of Sciences 46: 159–165.
Eng, L. L., D. Belk & C. H. Eriksen, 1990. Californian Anostraca: Distribution, habitat, and status. Journal of Crustacean Biology 10: 247–277.
Eriksen, C. H. & D. Belk, 1999. The fairy shrimps of California’s pools, puddles, and playas. Mad River Press, Eureka: 196.
Folmer, O., M. Black, W. Hoeh, R. Lutz & R. Vrijenhoek, 1994. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Molecular Marine Biology and Biotechnology 3: 294–299.
Fugate, M., 1993. Branchinecta sandiegonensis, a new species of fairy shrimp (Crustacea: Anostraca) from western North America. Proceedings of the Biological Society of Washington 106: 296–304.
Gomez, A., J. Montero-Pau, D. H. Lunt, M. Serra & S. Campillo, 2007. Persistent genetic signatures of colonization in Branchionus manjavacas rotifers in the Iberian Peninsula. Molecular Ecology 16: 3228–3240.
Hartland-Rowe, R., 1965. The Anostraca and Notostraca of Canada with some new distribution records. The Canadian Field Naturalist 79: 185–189.
Hebert, P. D. N., J. D. S. Witt & S. J. Adamowicz, 2003. Phylogeographical patterning in Daphnia ambigua: Regional divergence and intercontinental cohesion. Limnology and Oceanography 48: 261–268.
Hebert, P. D., M. Y. Stoeckle, T. S. Zemlak & C. M. Francis, 2004. Identification of birds through DNA barcodes. PLoS Biology 2: e312.
Horne, F. R., 1967. Effects of physical-chemical factors on the distribution and occurrence of some southeastern Wyoming phyllopods. Ecology 48: 472–477.
Horne, F. R., 1971. Some effects of temperature and oxygen concentration on phyllopod ecology. Ecology 52: 343–347.
Ketmaier, V., R. Mandatori, E. DeMatthaeis & G. Mura, 2005. Molecular systematics and phylogeography in the fairy shrimp Tanymastix stagnalis based on mitochondrial DNA. Journal of Zoology 206: 401–410.
Ketmaier, V., F. Marrone, G. Alfonso, K. Paulus, A. Wiemann, R. Tiedemann & G. Mura, 2012. Mitochondrial DNA regionalism and historical demography in the extant populations of Chirocephalus kerkyrensis (Branchiopoda: Anostraca). PLoS ONE 7: e30082.
Lefébure, T., C. J. Douady, M. Gouy & J. Gibert, 2006. Relationship between morphological taxonomy and molecular divergence within Crustacea: proposal of a molecular threshold to help species delimitation. Molecular Phylogenetics and Evolution 40: 435–447.
Leigh, J. W. & D. Bryant, 2015. POPART: full-feature software for haplotype network construction. Methods in Ecology and Evolution 6: 1110–1116.
Librado, P. & J. Rozas, 2009. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25: 1451–1452.
Lindholm, M., M. A. D’auriac, J. Thaulow & A. Hobæk, 2016. Dancing around the pole: Holarctic phylogeography of the Arctic fairy shrimp Branchinecta paludosa (Anostraca, Branchiopoda). Hydrobiologia 772: 189–205.
Lynch, J. E., 1964. Packard’s and Pearse’s species of Branchinecta: Analysis of a nomenclatural involvement. Amercan Midland Naturalist 71: 466–488.
Maeda-Martínez, A. M., 1991. Distribution of species of Anostraca, Notostraca, Spinicaudata, and Laevicaudata in Mexico. Hydrobiologia 212: 209–212.
Maeda-Martínez, A. M., H. Obregón-Barboza & H. García-Velazco, 1997. New records of large branchiopods (Branchiopoda: Anostraca, Notostraca, and Spinicaudata) in Mexico. Hydrobiologia 359: 63–68.
Maeda-Martínez, A. M., H. Obregon-Barboza, H. García-Velazco, & M. A. Prieto-Salazar. 2002. Branchiopoda: Anostraca, In: Bousquets, J. L., and J. J. Morrone (eds) Biodiversidad, Taxonomía y Biogeografía de Artrópodos de México: hacia una síntesis de su conocimiento, Vol. III. Universidad Nacional Autonóma de México, pp. 305–322.
Millette, K. L., S. Xu, J. D. Witt & M. E. Cristescu, 2011. Pleistocene-driven diversification in freshwater zooplankton: Genetic patterns of refugial isolation and postglacial recolonization in Leptodora kindtii (Crustacea, Cladocera). Limnology and Oceanography 56: 1725–1736.
Meusel, F. & M. Schwentner, 2016. Molecular and morphological delimitation of Australian Triops species (Crustacea: Branchiopoda: Notostraca)—large diversity and little morphological differentiation. Organisms Diversity & Evolution 17: 1–20.
Muñoz, J., A. Gomez, A. J. Green, J. Figuerola, F. Amat & C. Rico, 2008. Phylogeography and local endemism of the native Mediterranean brine shrimp Artemia salina (Branchiopoda: Anostraca). Molecular Ecology 17: 3160–3177.
Obregón-Barboza, H., A. M. Maeda-Martínez, H. García-Velazco & H. J. Dumont, 2002. Branchinecta oterosanvicentei n. sp. (Branchiopoda: Anostraca), a new fairy shrimp from the Chihuahuan desert, with a proposal for the conservation of the Branchinectidae of Mexico. Hydrobiologia 467: 45–56.
Obregón-Barboza, H., G. Murugan, H. García-Velazco & A. M. Maeda-Martínez, 2015a. A systematic review of mexican populations of the fairy shrimp genus Thamnocephalus (Branchiopoda: Anostraca). Journal of Crustacean Biology 35: 407–432.
Obregón-Barboza, H., G. Murugan, H. García-Velazco & A. M. Maeda-Martínez, 2015b. A review of the Branchinecta (Branchiopoda: Anostraca) from the Baja California Peninsula: First record of the giant fairy shrimp B. gigas Lynch, 1937 from Mexico. Journal of Crustacean Biology 35: 433–440.
Packard, A. S., 1877. Description of new phyllopod Crustacea from the West. Bulletin of the United States Geological and Geographical Survey of the Territories 3: 171–179.
Packard, A. S., 1883. A monograph of the phyllopod Crustacea of North America, with remarks on the order Phyllocarida. Twelfth annual report of the United States Geological and Geographical Survey of the Territories: a report of progress of the exploration in Wyoming and Idaho for the year 1878, F. V. Hayden, Part 1, section 2: 295–593.
Penton, E. H., P. D. Hebert & T. J. Crease, 2004. Mitochondrial DNA variation in North American populations of Daphnia obtusa: continentalism or cryptic endemism? Molecular Ecology 13: 97–107.
Posada, D., 2008. jModelTest: phylogenetic model averaging. Molecular Biology and Evolution 25: 1253–1256.
Rogers, D. C. 2009. Branchiopoda (Anostraca, Notostraca, Laevicaudata, Spinicaudata, Cyclestherida). In: Likens, G. F. (editor) Encyclopedia of Inland Waters, vol. 2, pp. 242–249.
Rogers, D. C., 2013. Anostraca catalogus (Crustacea: Branchiopoda). The Raffles Bulletin of Zoology. 61: 525–546.
Rogers, D. C. & M. A. Hill, 2013. Annotated Checklist of the large branchiopod crustaceans of Idaho, Oregon and Washington, USA, with the “rediscovery” of a new species of Branchinecta (Anostraca: Branchinectidae). Zootaxa 3694: 249–261.
Rogers, D. C., 2014a. Anostracan (Crustacea: Branchiopoda) zoogeography II. Relating distribution to geochemical substrate properties in the USA. Zootaxa 3865: 1–49.
Rogers, D. C., 2014b. Larger hatching fractions in avian dispersed anostracan eggs (Branchiopoda). Journal of Crustacean Biology 34: 135–143.
Rogers, D. C., 2015. A conceptual model for anostracan biogeography. Journal of Crustacean Biology 35: 686–699.
Rogers, D. C. & M. Fugate, 2001. Branchinecta hiberna, a new species of fairy shrimp (Crustacea: Anostraca) from western North America. Western North American Naturalist 61: 11–18.
Rogers, D. C., D. Quinney, J. Weaver & J. Olesen, 2006. A new giant species of predatory fairy shrimp from Idaho (Branchiopoda: Anostraca). Journal of Crustacean Biology 26: 1–16.
Ronquist, F. & J. P. Huelsenbeck, 2003. MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574.
Ryder, J. A., 1879. Description of a new branchiopod. Proceedings of the Academy of Natural Sciences of Philadelphia 1879: 200–202.
Schwentner, M., B. V. Timms & S. Richter, 2011. An integrative approach to species delineation incorporating different species concepts: a case study of Limnadopsis (Branchiopoda: Spinicaudata). Biological Journal of the Linnean Society 104: 575–599.
Schwentner, M., B. V. Timms & S. Richter, 2014. Evolutionary systematics of the Australian Eocyzicus fauna (Crustacea: Branchiopoda: Spinicaudata) reveals hidden diversity and phylogeographic structure. Journal of Zoological Systematics and Evolutionary Research 52: 15–31.
Schwentner, M., B. V. Timms & S. Richter, 2015. Spinicaudata (Branchiopoda: Diplostraca) in Australia’s arid zone: unparalleled diversity at regional scales and within water bodies. Journal of Crustacean Biology 35: 366–378.
Tamura, K., J. Dudley, M. Nei & S. Kumar, 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Molecular Biology and Evolution 24: 1596–1599.
Vandergast, A. G., D. A. Wood, M. A. Simovich & A. J. Bohonak, 2009. Identification of co-occurring Branchinecta fairy shrimp species from encysted embryos using multiplex polymerase chain reaction. Molecular Ecology Resources 9: 767–770.
Xu, S., P. D. N. Hebert, A. A. Kotov & M. E. Cristescu, 2009. The noncosmopolitanism paradigm of freshwater zooplankton: insights from the global phylogeography of the predatory cladoceran Polyphemus pediculus (Linnaeus, 1761) (Crustacea, Onychopoda). Molecular Ecology 18: 5161–5179.
Acknowledgements
We thank Mr. Humberto García-Velazco and Mr. José Ángel Camacho for helping during field collection in Baja California, Mexico. Mr. J. Lopez and Mr. J. Ulloa assisted in the generation of genetic data. This study was supported by a grant from the UC MEXUS-CONACYT program to AA and AMMM (Project No. CN08). Collections made by AA and AMMM were done so under scientific collecting permits SCP-17338 (CDFW) and SGPA/DGVS/08331/10, respectively. We also thank Dr. M. Schwentner and two anonymous reviewers for comments on an earlier version of this manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Guest editors: Federico Marrone, D. Christopher Rogers, Paola Zarattini & Luigi Naselli-Flores / New Challenges in Anostracan Research: a Tribute to Graziella Mura
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Aguilar, A., Maeda-Martínez, A.M., Murugan, G. et al. High intraspecific genetic divergence in the versatile fairy shrimp Branchinecta lindahli with a comment on cryptic species in the genus Branchinecta (Crustacea: Anostraca). Hydrobiologia 801, 59–69 (2017). https://doi.org/10.1007/s10750-017-3283-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10750-017-3283-3