Abstract
The spinicaudatan clam shrimp is a group of branchiopod crustaceans that has existed as far back as the Devonian and well-preserved fossils are known. Evidence of monophyly exists for only one family (Limnadiidae), which has a worldwide distribution and morphological conservatism. The evolutionary relationships among genera and diversification mechanisms are not deeply resolved as well as origin of the genus Eulimnadia. To address these issues, we constructed phylogenies of limnadiid clam shrimp, with both Bayesian inference and maximum likelihood methods to infer limnadiid evolutionary relationships. We then performed dated phylogenies using a relaxed clock of the Spinicaudata using fossil calibrations. Divergence date estimates show a perfect match with the break up of the Pangaea that could explain current limnadiid distributions; however the genus Eulimnadia apparently diverged 30 Ma ago. Eulimnadia phylogeography suggests an American origin and ecological patterns were analyzed to propose hypotheses on its origin and spread. This genus also shows a strong dispersive capacity, which could be explained by its reproduction modalities (androdioecy). This study and this first phylogeny with fossil calibration date the current distribution of Spinicaudata and reveal congruence with continental drift, except for Eulimnadia.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Spinicaudata is a suborder of branchiopod crustaceans (4–30 mm long) with a folded carapace and laterally compressed bodies. These continental organisms are typically found in temporary pond ecosystems in various climates. To survive in this type of biotope, especially during the dry season, many branchiopods produce resting eggs (Brendonck et al., 2008; Rogers, 2014). In addition, resting eggs disperse passively, which allows the colonization of new ponds. The taxonomy of this group and the understanding of its evolution are still controversial and have recently been challenged by molecular studies (Hoeh et al., 2006; Weeks et al., 2009). Only three families are currently recognized: Limnadiidae (Burmeister, 1843), Leptestheriidae, and Cyzicidae (Stebbing, 1902) but monophyly is supported only for the Limnadiidae (Hoeh et al., 2006). Nine genera belong to the Limnadiidae: Australimnadia, Calalimnadia, Eulimnadia, Gondwanalimnadia, Imnadia, Limnadia, Limnadopsis, Metalimnadia, and Paralimnadia (Rogers et al., 2012; Timms & Schwentner, 2012; Rogers et al., 2016). The limnadiids are known to be variable in their reproduction modes: while most genera are gonochoric, others are hermaphroditic with rare males (Limnadia lenticularis) or without males (Calalimnadia mahei). In Eulimnadia, androdioecy (populations comprised a mixture of males and hermaphrodites) seems to be the general rule even if some populations are only composed of hermaphrodites. It is also interesting to note that the limnadiids show a worldwide distribution (excluding Antarctica). Limnadia and Eulimnadia both have a particularly vast distribution area as opposed to all other limnadiids genera. These different distributions are not yet understood and, until now, the only assumption justifying the large distribution of Eulimnadia is the hypothetical old age of this genus (Weeks et al., 2006).
One way to understand the global distribution of limnadiids is to construct a complete phylogeny with all the lineages and infer divergence time among genera. The Spinicaudata, including the Limnadiidae, and their extensive fossil record are known from the Devonian (Tasch, 1967; Novojilov, 1970; Astrop & Hegna, 2015; Gueriau et al., 2016). Over the past 50 years, independent groups have comprehensively reviewed spinicaudatan phylogenetics (Astrop & Hegna, 2015). To our knowledge, there is no phylogenetic study using fossil calibrations to deduce the age of different Limnadiidae lineages.
The aim of this work is to explain the global distribution and evolution of the Limnadiidae. The first step was to perform a 28S ribosomal DNA phylogeny of the Limnadiidae. Then, we investigated spinicaudatan divergence times with fossil calibrations. Finally, we conducted a phylogeography of Eulimnadia. In order to explain the distribution of Eulimnadia we examined ecological characteristics, such as life cycle diversity and experimental breeding.
Materials and methods
Sample
A unique sample of the limnadiid sp. 1 from Bolivia was collected and substrate samples from limnadiid habitats were collected following Rabet et al. (2014), available locality data are presented in Table 1. To culture adults we placed 100 g of soil with resting eggs in 10 l milliQ water at 28°C with a permanent light. Development was checked twice daily. The adult stage was identified when resting eggs were visible under the carapace of females or hermaphrodites. The longevity was given following the maximum age reached by several individuals from the same cohort (in artificial conditions, death is regularly observed inside the cohort but some individuals seems to reach the natural limit of the species).
DNA extraction, PCR amplification, and sequencing
DNA extraction of the limnadiid sp. 1 from Bolivia was done following the Qiagen amplification DNA Mini Tissue Kit protocol from the whole specimen, stored since sampling in 96° ethanol. The 28S rRNA was amplified with the primer set D1D2fw1/D1D2rev2 (Sonnenberg et al., 2007). PCR reaction was performed with 2 µl of DNA extraction in a 20 µl final volume (0.32 µl of each primer at 10 µM, 0.8 µl of dNTP-mix at 6.6 mM, 1 µl of Bovine Serum Albumin at 1 mg/ml, 2 µl PCR buffer, and 0.12 unit Taq polymerase (Taq DNA polymerase, Qiagen). Cycler settings were conducted in a Mastercycler (Eppendorf) with an initial step of 94°C for 4 min, 45 cycles at 94°C for 20 s (denaturation), 52.5°C for 20 s (primer hybridization), 72°C for 90 s (elongation), and a final elongation at 72°C for 8 min. Successful PCRs were selected on ethidium–bromide-stained agarose gels. Sanger sequencing (both directions) was performed by a commercial company (Eurofins; http://www.eurofins.fr) using the same primers. Chromatograms in both directions were compared and a consensus sequence was assembled using Bioedit (Hall, 1999).
Phylogenetic reconstructions of Spinicaudata and Eulimnadia
An alignment of 28S ribosomal DNA sequences from 40 selected spinicaudatans (Online resource 1) was performed with Muscle (Edgar, 2004) using Geneious version 8 (http://www.geneious.com, Kearse et al., 2012). Gaps were removed using Gblocks (Castresana, 2000) carried out on the phylogeny.fr platform (Dereeper et al., 2008). Phylogenetic reconstructions were performed using both Bayesian inference (BI) and maximum likelihood (ML) from an alignment of 40 sequences of 748 bp. Evolutionary models were selected via Akaike Information Criterion or Bayesian Information Criterion using jModelTest v2.1.1 (Darriba et al., 2012). Maximum likelihood was performed using PhyML (Guindon & Gascuel, 2003) under Geneious v8 with a general time reversible model with a gamma distribution (Γ = 0.754) and a proportion of invariable sites (I = 0.578), validated with 1,000 bootstrap replicates. Bayesian analyse were carried out and done with MrBayes 3.2.3 (Ronquist & Huelsenbeck, 2003) on the CIPRES Science Gateway (Miller et al., 2010), with 4 chains of 1 × 106 generations, trees sampled every 100 generations, and burn-in value set to 20% of the sampled trees. Sequences were considered with an evolutionary model (TIM3) with a gamma distribution (Γ = 0.754) and a proportion of invariable sites (I = 0.578). We checked that standard deviation of the split frequencies fell below 0.01 and confirmed convergences of the runs to ensure convergence in tree search using the Tracer v1.6 software (http://tree.bio.ed.ac.uk/software/tracer/). Trees were visualized using FigTree v1.4.2 (http://tree.bio.ed.ac.uk/software/figtree/).
An alignment of 102 sequences (860 bp) of 28S ribosomal DNA identified as Eulimnadia in Genbank (all sequences available) was created. Protocol described above was used for the reconstruction of this Eulimnadia phylogeny. Maximum likelihood was performed with an evolutionary model (TIM2) plus a proportion of invariable sites (I = 0.88), and validated with 1,000 bootstrap replicates. Bayesian analyse were carried out with 4 chains of 2 × 106 generations, trees sampled every 200 generations, and burn-in value set to 20% of the sampled trees. Sequences were considered with an evolutionary model (TIM2) plus a proportion of invariable sites (I = 0.88).
Divergence time estimation
We constructed two datasets: the first containing 42 taxa (36 representative species of Spinicaudata and 6 notostracan outgroups) to investigate global Limnadiids emergence dates. The second one corresponds to the 36 Spinicaudatan taxa used to investigate Limnadiidae evolutionary history. Alignments were performed with Muscle and gaps were removed with Gblocks as described above. BEAST software package v2.1.3 (Bouckaert et al., 2014) was used to infer divergence time. We applied the uncorrelated lognormal relaxed clock model (Drummond et al., 2006) to account for rate heterogeneity among lineages. This model was evaluated with the CoV, where CoV values >0 were considered as evidence of non-clock evolutionary behavior. The calibrated Yule process (Heled & Drummond, 2012), an extension of the birth–death model, was selected as tree prior for the model of speciation. We applied a GTR+I+G to the complete dataset and a TN 93 +I+G to the Spinicaudata dataset, referring to evolutionary model obtained by jModelTest v2.1.1 (Darriba et al., 2012). For calibrating the phylogenetic tree we used 2 branchiopod fossils (Castracollis wilsonae (Fayers & Trewin, 2002)) and Afrolimnadia sibiriensis (Tasch, 1987) plus an estimation of the split between the genera Triops and Lepidurus with normal prior distribution. The Pragian fossil C. wilsonae was chosen for inferring the minimum age of the last common ancestor of Notostracan and Spinicaudata (Fayers & Trewin, 2002) because it shares characters from the Notostraca lineage and the Diplostraca (including Spinicaudata) (Olesen, 2009; Lagebro et al., 2015). We used the lower boundary of the Pragian stage (407.6 (±1.3) Ma) (Cohen et al., 2013) with a normal distribution prior for this node. The Notostraca order split (Triops split from Lepidurus) was estimated at 188 Ma, inferred by recent publications on molecular phylogeny and divergence time of notostracan (Korn et al., 2013; Mathers et al., 2013).
We chose the Sinemurian fossil A. sibiriensis as calibration for the Limnadiidae age estimates with a normal distribution at 190.8 (±0.5) Ma. Indeed this species has an important space without growth lines and a fragile carapace, which suggests that this species is a Limnadiidae. The space without growth lines can be explained because during the early stages, the molting of the valves is complete; the animals do not retain the external laminae of previous stages. The older laminae remain, overlaying the younger and larger plates, and forming the well-known growth lines of the typical Spinicaudatan carapace (Roessler, 1995). Our choices for spinicaudatan fossils were made in light to the lack of compatibility between biological and paleontological studies in this group. For current species, the taxonomic status is based on molecular characters associated with morphological characters such as the head or the furca and sometimes the carapace may be informative also, but never essential. In paleontology, the shape of the carapace is essential because no other details are generally found; these insufficiencies of characters imply recurrent incompatibility with modern systematics but recently a first assay of synthesis was initiated (Astrop & Hegna, 2015).
For the dataset with only Spinicaudata we used only one fossil calibration but two analyses were performed. For the first, we considered divergence times with the Sinemurian fossil but for the second we assumed a more conservative approach with a Perilimnadiidae fossil (Chen, 1975), which exhibits large carapace with limnadiform shape plus preservation of carapace gland (Zhang et al., 1976). This family was indicated to be sister group of the Limnadiidae within an attempt of clear phylogeny investigation between living and fossil spinicaudatan taxa (Astrop & Hegna, 2015). The geologic range for Perilimnadiidae was fixed to late Permian to early Paleogene; we decided to use the upper boundary with an age of 252 Ma.
For each dataset, three independent Bayesian MCMC runs were carried out for 30–50 million of generations (to obtain effective sample size values of at least 200 for each parameter), to retain a sample of 10,000 trees. Convergence of the runs was confirmed using Tracer v1.6 software (http://tree.bio.ed.ac.uk/software/tracer/). The results of the three independent runs were then combined using LogCombiner v1.8.1 (with a burn-in of 25%), and MCC trees were generated using TreeAnnotator v2.1.2 and visualized using FigTree v1.4.2 (http://tree.bio.ed.ac.uk/software/figtree/).
Results
Spinicaudatan phylogeny
Phylogenetic reconstructions produced the same topologies for 40 representative spinicaudatan specimens using either maximum likelihood or Bayesian inference. Topologies confirm the monophyly of the Limnadiidae (Fig. 1). This family is divided into three groups: Imnadia yeyetta, Limnadia lenticularis, and all other limnadiids. Within the larger group, two clades are clearly and robustly distinguished. The first one is composed of Australian limnadiids, divided into a Paralimnadia clade and a Limnadopsis + Australimnadia clade. The second is composed of African and Neotropical limnadiids + Eulimnadia species. The new genus BO sp. 1, Metalimnadia (both strict Neotropical endemism) and all Eulimnadia species form a clade suggesting that Eulimnadia has probably an American origin. Within the Eulimnadia, two different groups seem to emerge with low resolution. Percentage of nucleotide identity of BO sp. 1 with other genera are always under 96%, for example; 92–95% with Metalimnadia species, 93–96% with Eulimnadia species, 92–93% with Gondwanalimnadia, 95% with Calalimnadia mahei, 91–93% with Limnadia lenticularis, and 89–91% with Imnadia yeyetta.
Phylogenetic tree of large branchiopod computed from the partial 28S rDNA (40 sequences) by Bayesian analysis (BI) and Maximum Likelihood (ML). To simplify, only the BI tree is shown; the ML tree has the same topology. The numbers are posterior probabilities (BI) and bootstrap proportions (ML) reflecting clade support (values below 50 are indicated by dashes). Images of the new genus BO sp. 1; one male and one female (bar scale = 1 mm)
Phylogenetic analysis of Eulimnadia genus
We produced the same topologies for 101 sequences of Eulimnadia (plus one outgroup) using either maximum likelihood or Bayesian inference (Fig. 2). We observed a clear pattern with all specimens from the Americas with basal positions. Results show an important cluster regrouping of all specimens from others geographic regions as Afrotropical, Oriental, Sino-Japanese, and Australian, except four specimens in Japan (identified as E. cylindrova).
Phylogenetic tree of Eulimnadia specimens from the partial 28S rDNA (101 sequences) by Bayesian analysis (BI) and Maximum Likelihood (ML). To simplify, only the ML tree is shown; the BI tree has the same topology. The numbers are posterior probabilities (BI) and bootstrap proportions (ML) reflecting clade support (values below 50 are indicated by dashes). Color codes show the geographic distribution of each sequence. Each specimen was named with the locality (international country code) plus isolate name and accession number. Limnadiidae lineage BO sp. 1 was used as the outgroup
Divergence times
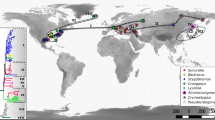
MCC trees with fossil calibration were constructed with two datasets. We illustrate only the results from the dataset containing 42 taxa (36 representatives species of Spinicaudata and six notostracan outgroups) with three calibrations (Fig. 3) but divergence dates of 17 nodes from both datasets are summarized in Table 2. In Fig. 3, we linked the MCC tree to geological stages and maps illustrating the continental drift. All analyses suggested an age between 192.9 and 295.7 Ma for the modern spinicaudatans. Pangaea was formed between the Devonian and the late Triassic, and divided into three main episodes (Scotese, 2001). The break up began in the middle Jurassic (about 180 Ma), corresponding to an initial radiation for limnadiids with a split between laurasian limnadiids (Imnadia yeyetta + Limnadia lenticularis) and gondwanian limnadiids (all others limnadiids, node 4). A second phase of the break up start in the early Cretaceous (140 Ma) with the separation of the Antarctic/Australia and the Africa/South America blocks resulting in the partition of australian limnadiids from the west gondwanian lineages (node 6). The African lineage with Gondwanalimnadia and the Neotropical limnadiids + Eulimnadia divide during the continental separation of Africa and South America (node 11). In South America, as in Australia, during the Cretaceaous to the Neogene the limnadiids diverge in several lineages (nodes 8, 9, 12, 13, 14). The expansion of Eulimnadia seems to be an exception and is not compatible with continental drift events. Emergence dates of Eulimnadia vary from 32.4, 35.7, to 47.7 Ma, clearly within the Paleogene. This genus presents an important diversity with three clades (node 14). Two of them are composed of Eulimnadia species coming from the Americas but the third seems to have colonized different continents. Estimated dates for this dispersive clade (i.e., Eulimnadia species from other geographical regions than the Americas) are 18.2, 25.2, or 45.2 Ma suggesting an American origin for them.
Time calibrated of 36 spinicaudatan species and 6 notostracan outgroups obtained in BEAST2 with 28S rDNA partial gene. Numbers at nodes identify divergence dates obtained and shown in Table 2. Gray square at nodes correspond to the calibrations given in M and M. Node bars correspond to the 95% highest posterior density (HPD) interval of each node. Maps show the continental drift at different period of history. Geographical origin of Limnadiidae were represented as Afrotropical: green Australian: purple Oriental: orange Americas: blue Palearctic: brown and Sino-Japanese: light green
Dispersal capacity of limnadiids: the particular case of Eulimnadia
Six species of Eulimnadia were discovered on eight different oceanic islands far from continental landmasses (minimum of 80 km and maximum of 3,800 km) (Table 3). We calculated the minimal distance between islands (where specimens were detected); we found a minimum gap of 20 km for E. texana and a maximum of 1,000 km for E. garretti. These islands were never connected with continental mass in the past involving that Eulimnadia colonized islands through oceans.
Life cycle of limnadiids: the short life cycle of Eulimnadia
The life cycle and the time to reach maturity (Table 4) are usually short for Eulimnadia species compared to others limnadiids, particularly in tropical conditions (>27°C) where Eulimnadia species reach maturity 4–6 days after immersion. In Weeks et al. (1997) the time of 7 days is exceptional and the average age to become adult in this experiment is only 5,1 days. In similar conditions all the other genera reach maturity slower (6–10 days) and the life cycle is longer.
Discussion
Phylogenetic and divergence date estimate for Spinicaudata
This study is the first to combine phylogenetic reconstruction and divergence time estimates within Spinicaudata using fossil calibrations. The phylogenies show well-supported topologies with the monophyly of the Limnadiidae, which is consistent with previous studies (Hoeh et al., 2006; Schwentner et al., 2009). Inside this family, results show a group of Gondwanian species, the sister position of Metalimnadia with Eulimnadia and the association of Gondwanalimnadia and Calalimnadia to west gondwanian species (Weeks et al., 2006, 2009). Nevertheless, the topology changes in the west gondwanian group: Calalimnadia is the most basal group, and not Gondwanalimnadia, as was previously thought (Weeks et al., 2009). The limnadiid Bolivian sp. 1 appears to constitute a new lineage; the percentage of identity with other populations and phylogenetic isolation suggest a new genus. Indeed, species of limnadiids have only 2–3% of difference for the 28S ribosomal DNA whereas two genera have between 5 or 6% of difference. The new limnadiid Bolivian specimen will be described as a new genus and species in a future paper. The results also demonstrate an Australian clade (Schwentner et al., 2009) composed of all Limnadopsis and Paralimnadia. The monophyly of Eulimnadia is confirmed here but the topology inside the genus is not clear (Hoeh et al., 2006; Weeks et al., 2009). A recent molecular study of 19 Eulimnadia species also confirmed the monophyly of this genus and shows a low species level phylogenetic resolution (Reed et al., 2015). Our results and previous studies underscore the great need for a revision of Eulimnadia.
Some studies tried to estimate the age of branchiopod lineages, and each time only one or two sequences were used to represent Spinicaudata, introducing a massive methodological bias. Thus, Mathers et al. (2013) suggested a common ancestor for Eulimnadia sp. and L. lenticularis around 70 millions years ago which is inconsistent with our results. Another study gave an age estimation for Spinicaudata emergence of 266.3 Ma (183.1–307), which is consistent with our results (Korn et al., 2013). We were able to estimate the age of Limnadopsis (21–25 Ma); it is a little younger than what was reported previously (Schwentner et al., 2012). In the absence of Limnadopsis fossils, Schwentner et al. (2012) used an estimation of substitution rate for a mitochondrial gene (COI) suggested by decapod crustacean pairwise divergence rates, giving divergences dates with large confidence intervals. It is difficult to compare theses studies because they did not explore the phylogeny of branchiopods at the same scale, with the same diversity or the same methods.
The particular case of Eulimnadia
Within the Limnadiidae, node ages and diversification patterns seem to perfectly fit with the Pangaea break up for most genera. This could be explained by a reduced capacity for transoceanic dispersal and a stepwise colonization via continental masses. Under this scenario, limnadiid populations, which were isolated after the continental drift, established new lineages progressively. If a limited dispersive capacity can be proposed for a majority of genera, this idea is in contradiction with the presence of Eulimnadia in oceanic islands and their distribution. Eulimnadia has the largest distribution in this family, and it appears to be an exception with a high dispersive capacity. The colonization of oceanic islands (never in direct contact with continental mass) could only be explained by the transport of eggs. Until now, it was assumed that Eulimnadia was present 180 Ma and that it was widely distributed before the disconnection of the Pangaea (Weeks et al., 2006). But this assumption was not tested with calibrated phylogeny or evolutionary rates. Our work however shows an American origin of the genus Eulimnadia and a divergence date of 30 My.
Another important outcome of the study is the age of the Eulimnadia old world clade (i.e., Eulimnadia from Australia, Japan, Africa) around 20 Ma, which colonized the rest of the world. Indeed, some populations, identified as E. cylindrova appeared in Japan suggesting that a second way of colonization of Eulimnadia could exist. In this case, the migration to Asia across the Pacific Ocean perhaps via the Galapagos Islands where the same species is also present probably occurred. However, we cannot exclude recent anthropogenic dispersal.
Eulimnadia are in part characterized by the absence of females and the presence of hermaphrodites and sometimes males, which is characteristic of androdioecy (Sassaman & Weeks, 1993; Weeks et al., 2006). Hermaphrodism seems to be a serious advantage for dispersal because only one egg we could produce one adult that will produce viable resting eggs, and a new population. Whereas gonochoric species must disperse many eggs to the same temporary pond in order to have a small probability to reproduce. Indeed, at least two eggs need to become opposite sex adults in the same time frame to support reproduction. Androdioecy presents the additional advantage that one egg can give a hermaphrodite heterozygote, which could produce genetic diversity within population by sexual recombination. These mixed descendants (hermaphrodite and male) will create diversity whereas exclusive hermaphrodites can form only clones. The resting eggs enhance the dispersive capacity of large branchiopods and two ways of dissemination are possible. First, by animals predating adults carrying the eggs (Mathias, 1936; Rogers, 2014) or by passive transport of laid eggs, for example when a pool dries out the wind spreads eggs (Graham & Wirth, 2008; Vanschoenwinkel et al., 2008). Due to the distance and the lack of biotope relay in the ocean, the long transoceanic colonization should be relatively rare.
Following our investigations on the limnadiid life cycle, tropical Eulimnadia have a short life cycle and reach maturity quickly. This ability is coupled to a high tolerance temperature, for example, Roessler (1995) indicated that tropical Eulimnadia can survive at 42°C. All these characteristics suggest that Eulimnadia is a pioneer selected to live in ephemeral and unstable ponds (<1 month). When Eulimnadia colonized other continents they compete with local limnadiids, which are characterized by a longer life cycle but they may establish in a specific ecological niche in temporary water. The monopolization hypothesis could explain this capacity of colonization and a rapid local differentiation (De Meester et al., 2002; Rogers, 2015).
Our investigation seems to confirm that most of limnadiids have a limited capacity of dispersal and most diversification in this family follows continental drift. One exception seems to be Eulimnadia, which seems to originate from the Americas and has a high dispersal capacity allowing it to colonize worldwide. Future investigation should focus on Eulimnadia with more sensitive phylogenetic markers in order to understand the initial radiation and the method of colonization.
References
Astrop, T. I. & T. A. Hegna, 2015. Phylogenetic relationships between living and fossil spinicaudatan taxa (branchiopoda Spinicaudata): reconsidering the evidence. Journal of Crustacean Biology 35: 1–16.
Belk, D., 1972. The biology and ecology of Eulimnadia antlei Mackin (Conchostraca). The Southwestern Naturalist 16: 297–305.
Benvenuto, C. & S. C. Weeks, 2011. Mate guarding behavior in clam shrimp: the influence of mating system on intersexual conflict. Behavioral Ecology and Sociobiology 65: 1899–1907.
Berry, E. W., 1926. Description and notes on the life history of a new species of Eulimnadia. American Journal of Science 11: 429–433.
Bishop, J. A., 1968. Aspects of the post-larval life history of Limnadia stanleyana King (Crustacea: Conchostraca). Australian Journal of Zoology 16: 885–895.
Bouckaert, R., J. Heled, D. Kühnert, T. Vaughan, C.-H. Wu, D. Xie, M. A. Suchard, A. Rambaut & A. J. Drummond, 2014. BEAST 2: a software platform for Bayesian evolutionary analysis. PLoS Computational Biology 10: e1003537.
Brendonck, L., A. Thiery & A. Coomans, 1990. Taxonomy and biogeography of the Galapagos branchiopod fauna (Anostraca, Notostraca, Spinicaudata). Journal of crustacean biology 10: 676–694.
Brendonck, L., Rogers, D. C., Olesen, J., S. C. Weeks & W. R. Hoeh, 2008. Global diversity of large branchiopods (Crustacea: Branchiopoda) in freshwater. In Freshwater Animal Diversity Assessment. Springer, New York: 167–176
Castresana, J., 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Molecular Biology and Evolution 17: 540–552.
Chen, P., 1975. Tertiary conchostraca from China. Scientia Sinica 6: 618-630.
Cohen, K. M., S. C. Finney, P. L. Gibbard & J. X. Fan, 2013. The ICS International Chronostratigraphic Chart, International Commission on stratigraphy.
Darriba, D., G. L. Taboada, R. Doallo & D. Posada, 2012. jModelTest 2: more models, new heuristics and parallel computing. Nature Methods 9: 772.
De Meester, L., A. Gómez, B. Okamura & K. Schwenk, 2002. The monopolization hypothesis and the dispersal-gene flow paradox in aquatic organisms. Acta Oecologica 23: 121–135.
Dereeper, A., V. Guignon, G. Blanc, S. Audic, S. Buffet, F. Chevenet, J. F. Dufayard, S. Guindon, V. Lefort, M. Lescot, J. M. Claverie & O. Gascuel, 2008. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Research 36(Web Server issue): W465–W469.
Drummond, A. J., S. Y. Ho, M. J. Phillips & A. Rambaut, 2006. Relaxed phylogenetics and dating with confidence. PLoS Biology 4: e88.
Edgar, R. C., 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research 32: 1792–1797.
Fayers, S. R. & N. H. Trewin, 2002. A new crustacean from the early Devonian Rhynie chert Aberdeenshire, Scotland. Transactions of the Royal Society of Edinburgh: Earth Sciences 93: 355–382.
Graham, T. B. & D. Wirth, 2008. Dispersal of large branchiopod cysts: potential movement by wind from potholes on the Colorado Plateau. Hydrobiologia 600: 17–27.
Giribet, G., S. Richter, G. D. Edgecombe & C. Wheeler, 2005. The position of crustaceans within Arthropoda: evidence from nine molecular loci and morphology. Crustacea and Arthropod Relationships 16: 307.
Gueriau, P., N. Rabet, G. Clément, L. Lagebro, J. Vannier, D. E. Briggs, S. Charbonnier, S. Olive & O. Béthoux, 2016. A 365-million-year-old freshwater community reveals morphological and ecological stasis in Branchiopod Crustaceans. Current Biology. doi:10.1016/j.cub.2015.12.039.
Guérin, F. E., 1837. Note monographique sur le genre limnadie, et description d’une espèce nouvelle de ce genre. Magasin de Zoologie Classe 7: 1–7.
Guindon, S. & O. Gascuel, 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Systematic Biology 52: 696–704.
Hall, T. A., 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium 41: 95–98.
Heled, J. & A. J. Drummond, 2012. Calibrated tree priors for relaxed phylogenetics and divergence time estimation. Systematic Biology 61: 138–149.
Hoeh, W. R., N. D. Smallwood, D. M. Senyo, E. G. Chapman & S. C. Weeks, 2006. Evaluating the monophyly of Eulimnadia and the Limnadiinae (Branchiopoda: Spinicaudata) using DNA sequences. Journal of Crustacean Biology 26: 182–192.
Kearse, M., R. Moir, A. Wilson, S. Stones-Havas, M. Cheung, S. Sturrock, S. Buxton, A. Cooper, S. Markowitz, C. Duran, T. Thierer, B. Ashton, P. Mentjies & A. Drummond, 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28: 1647–1649.
Korn, M., N. Rabet, H. V. Ghate, F. Marrone & A. K. Hundsdoerfer, 2013. Molecular phylogeny of the Notostraca. Molecular Phylogenetics and Evolution 69: 1159–1171.
Lagebro, L., P. Gueriau, T. A. Hegna, N. Rabet, A. D. Butler & G. E. Budd, 2015. The oldest notostracan (Upper Devonian Strud locality, Belgium). Palaeontology 58: 497–509.
MacKay, S. E. & D. D. Williams, 2011. Invertebrate colonization of the surface and deep groundwaters of a small oceanic island (Barbados, West Indies). Tropical Zoology 24: 1–48.
Mathias, P., 1936. A propos de la dissémination des Crustacés Phyllopodes. Compte Rendu de l’Association Française pour l’Avancement des Sciences 60: 335–337.
McKenzie, K. G., 1971. Entomostraca of Aldabra, with special reference to the Genus Heterocybris (Crustacea, Ostracoda). Philosophical Transactions of The Royal Society B 260: 257–297.
Mathers, T. C., R. L. Hammond, R. A. Jenner, B. Hänfling & A. Gómez, 2013. Multiple global radiations in tadpole shrimps challenge the concept of ‘living fossils’. PeerJ 1: e62.
Miller, M. A., W. Pfeiiffer & T. Schwartz, 2010 Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In Proceedings of the Gateway Computing Environments Workshop (GCE), 14 Nov 2010, New Orleans, LA: 1–8.
Nourisson, M. & P. Aguesse, 1961. Cycle annuel des Phyllopodes d’une mare temporaire de Camargue. Bulletin de la Société Zoologique de France 86: 754–762.
Novojilov, N. I., 1970. Vymershie limnadioidei (Conchostraca-Limnadioidea). Nauka, Moscow.
Olesen, J., 2009. Phylogeny of Branchiopoda (Crustacea) – character evolution and contribution of uniquely preserved fossils. Arthropod Systematics & Phylogeny 67: 3–39.
Rabet, N., D. Montero & S. Lacau, 2014. The effects of soils and soil stay on the egg morphology of Neotropical Eulimnadia (Branchiopoda: Limnadiidae). Journal of Limnology 73: 17–26.
Reed, S. K., R. J. Duff & S. C. Weeks, 2015. A systematic study of the genus Eulimnadia. Journal of Crustacean Biology 35: 379–391.
Remigio, E. A. & P. D. Hebert, 2000. Affinities among anostracan (Crustacea: Branchiopoda) families inferred from phylogenetic analyses of multiple gene sequences. Molecular Phylogenetics and Evolution 17: 117–128.
Richter, S., J. Olesen & W. C. Wheeler, 2007. Phylogeny of Branchiopoda (Crustacea) based on a combined analysis of morphological data and six molecular loci. Cladistics 23: 301–336.
Richters, F., 1882. Limnadia Garretti nov. sp. Abhandlungen der Senckenbergischen Naturforschenden Gesellschaft 12: 432–433.
Roessler, E. W., 1995. Review of Colombian conchostraca (Crustacea) – morphotaxonomic aspects. Studies on Large Branchiopod biology and aquaculture II. Springer, New York: 253–262.
Rogers, D. C., 2014. Larger hatching fractions in avian dispersed anostracan eggs (branchiopoda). Journal of Crustacean Biology 34: 135–143.
Rogers, D. C., 2015. A conceptual model for anostracan biogeography. Journal of Crustacean Biology 35: 686–699.
Rogers, D. C., N. Rabet & S. C. Weeks, 2012. Revision of the extant genera of Limnadiidae (Branchiopoda: Spinicaudata). Journal of Crustacean Biology 32: 827–842.
Rogers, D. C., N. Rabet & S. C. Weeks, 2016. Gondwanalimnadia (branchiopoda: spinicaudata) replacement name for Afrolimnadia Rogers, Rabet and Weeks, 2012 (limnadiidae), junior homonym of Afrolimnadia Tasch, 1987 (Lioestheriidae). Journal of Crustacean Biology 36: 105.
Ronquist, F. & J. P. Huelsenbeck, 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574.
Sassaman, C. & S. C. Weeks, 1993. The genetic mechanism of sex determination in the conchostracan shrimp Eulimnadia texana. American Naturalist 141: 314–328.
Scotese, C.R., 2001. Atlas of earth history: University of Texas at Arlington. Department of Geology. PALEOMAP Project
Schwentner, M., B. V. Timms, R. Bastrop & S. Richter, 2009. Phylogeny of Spinicaudata (Branchiopoda, Crustacea) based on three molecular markers - An Australian origin for Limnadopsis. Molecular Phylogenetics and Evolution 53: 716–725.
Schwentner, M., B. V. Timms & S. Richter, 2012. Flying with the birds? Recent large-area dispersal of four Australian Limnadopsis species (Crustacea: Branchiopoda: Spinicaudata). Ecology and Evolution 2: 1605–1626.
Smith, D. G. & C. D. Little, 2003. New records of and observations on Branchiopoda of St. John, United States Virgin Islands. Crustaceana 76: 631–636.
Sonnenberg, R., A. W. Nolte & D. Tautz, 2007. An evaluation of the LSU rDNA D1-D2 sequences for their use in species identification. Frontiers in Zoology 4: 6.
Stoddart, D. R. & C. A. Wright, 1967. Ecology of Aldabra Atoll. Nature 213: 1174–1177.
Tasch, P., 1967. Fossil clam shrimp distribution and its significance for the theory of continental drift. Transactions of the Kansas Academy of Science 70: 151–163.
Tasch, P., 1987. Fossil Conchostraca of the Southern Hemisphere and Continental Drift: paleontology, biostratigraphy, and dispersal. Geological Society of America Memoir 165: 1–290.
Telford, M. J., A. E. Lockyer, C. Cartwright-Finch & D. T. J. Littlewood, 2003. Combined large and small subunit ribosomal RNA phylogenies support a basal position of the acoelomorph flatworms. Proceedings of the Royal Society of London B: Biological Sciences 270: 1077–1083.
Timms, B. V. & M. Schwentner, 2012. A new genus and species of large Limnadiid clam shrimp from Australia (Spinicaudata: Limnadiidae). Journal of Crustacean Biology 32: 981–990.
Vanschoenwinkel, B., S. Gielen, M. Seaman & L. Brendonck, 2008. Any way the wind blows – frequent wind dispersal drives species sorting in ephemeral aquatic communities. Oikos 117: 125–134.
Vidrine, M. F., S. L. Sissom & R. E. McLaughlin, 1987. Eulimnadia texana Packard (Conchostraca: Limnadiidae) in rice fields in southwestern Louisiana. The Southwestern Naturalist 32: 1–4.
Weeks, S. C., V. Marcus & S. Alvarez, 1997. Notes on the life history of the clam shrimp, Eulimnadia texana. Hydrobiologia 359: 191–197.
Weeks, S. C., T. F. Sanderson, S. K. Reed, M. Zofkova, B. Knott, U. Balaraman, G. Pereira, D. M. Senyo & W. R. Hoeh, 2006. Ancient androdioecy in the freshwater crustacean Eulimnadia. Proceedings of the Royal Society B 273: 725–734.
Weeks, S. C., E. G. Chapman, D. C. Rogers, D. M. Senyo & W. R. Hoeh, 2009. Evolutionary transitions among dioecy, androdioecy and hermaphroditism in limnadiid clam shrimp (Branchiopoda: Spinicaudata). Journal of evolutionary Biology 22: 1781–1799.
Zhang, W. T., P. J. Chen & Y. B. Shen, 1976. Chinese Fossils of all Groups: Fossil Conchostraca of China. Science Press, Peking.
Zinn, D. J. & R. W. Dexter, 1962. Reappearance of Eulimnadia agassizii with notes on its biology and life history. Science 137: 676–677.
Acknowledgments
We are very grateful to Clara Lord for stimulating discussions and English corrections. We would like to thank Sébastien Lacau and Marc Pignal for soil collection in Brazil and Michaël Manuel for collecting the new Limnadiid from Bolivia. We also thank the UMS 2700 and the Service de Systématique Moléculaire (MNHN, Paris) for support in lab work.
Author information
Authors and Affiliations
Corresponding author
Additional information
Handling editor: Diego Fontaneto
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Bellec, L., Rabet, N. Dating of the Limnadiidae family suggests an American origin of Eulimnadia . Hydrobiologia 773, 149–161 (2016). https://doi.org/10.1007/s10750-016-2694-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10750-016-2694-x