Abstract
Environmental changes can trigger endoplasmic reticulum (ER) stress and misfolded protein accumulation, potentially leading to pre-eclampsia (PE). Amyloid-β (Aβ) is a crucial misfolded protein that can overactivate autophagy. Our study assessed the expression of Aβ1−42 and autophagic activity in PE placental tissues and trophoblasts under ER stress. Placental tissues were surgically collected from normal pregnant women (NP) and pregnant women with late-onset PE (LOPE) delivering through cesarean section. The expression levels of Aβ1−42 were detected in both PE and NP placental tissues, as well as in tunicamycin (TM)-induced HTR-8/SVneo cells. Autophagy-related proteins, such as Beclin-1, the ratio of LC3-II to LC3-I, ATG5, and SQSTM1/p62 in the placental tissues and HTR-8/SVneo cells were measured by Western blot. The number and morphology of autophagosomes were observed using transmission electron microscopy (TEM). Potential targets associated with the unfolded protein response (UPR) in the placental tissues of NP and PE cases were screened using PCR Arrays. The misfolded protein was significantly upregulated in the PE group. In both PE placental tissues and TM-induced HTR-8/SVneo cells, not only was Aβ1−42 upregulated, but also Beclin-1, ATG5, and LC3BII/I were significantly increased, accompanied by an increase in autophagosome count, while SQSTM1/P62 was downregulated. A total of 17 differentially expressed genes (DEGs) associated with the UPR were identified, among which elevated calnexin (CANX) was validated in the placenta from both PE and TM-induced HTR-8/SVneo cells. Autophagy is significantly upregulated in PE cases due to ER stress-induced Aβ1−42 accumulation, likely mediated by autophagy-related proteins involved in the UPR.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Pre-eclampsia (PE), a severe human-specific complication of pregnancy with an incidence of 3–5% (Bijl et al. 2022), is characterized by a new onset of maternal hypertension diagnosed after 20 weeks of gestation and significant end-organ damage (Espinoza et al. 2019). Depending on whether it occurs prior to or after the 34th week of gestation, PE is typically categorized into early-onset PE (EOPE) and late-onset PE (LOPE). LOPE is observed at a significantly higher frequency compared to EOPE. Although great strides have been made in exploring the pathogenesis of LOPE, its pathogenic molecular mechanisms remain unclear. Abnormal placentation is widely considered the primary etiological factor of PE. Insufficient trophoblast invasion in the maternal uterine spiral arteries triggers imbalanced angiogenesis and subsequent local ischemia/hypoxia, endoplasmic reticulum (ER) stress, oxidative stress, and endothelial dysfunction. Recent research reveals that the placenta is likely dysfunctional in LOPE (Staff 2019), potentially indicating a global deficiency in trophoblasts development and deficient invasion of EVT cells (Yang et al. 2023).
Recent evidence has shown that PE is linked with the aggregation of misfolded proteins, which may accumulate in the urine, placenta, and serum of PE patients (Millen et al. 2018; Rood et al. 2019; Sergeeva et al. 2020). Irina A et al. found that the pathophysiological characteristics of PE were similar to those of the recognized protein misfolding disorders that involve Aβ (Buhimschi et al. 2014). Aβ is produced by the amyloid precursor protein (APP), abundant in the human placenta. Aβ peptides are produced by APP through hydrolysis and cleavage via α-secretase, β-secretase, and γ-secretase. Aβ is highly oligomeric and self-assembling (Cheng et al. 2016), and its formation may be related to the imbalanced hydrolysis of APP. Interestingly, misfolded proteins in PE patients share similar characteristics with those of Aβ aggregation (Kouza et al. 2017). Aβ can polymerize into many forms, among which the Aβ1−42 oligomer has the strongest toxicity.
Autophagy, an evolutionarily conserved lysosome-dependent cell event in eukaryotes, regulates protein metabolism. Damaged organelles and misfolded proteins are degraded by autophagosomes and then recycled to maintain protein homeostasis (Zhang et al. 2021). APP and the four subunits of the γ-secretase complex are located in autophagosomes, suggesting that some Aβ polypeptides are produced via the autophagy signaling pathways. Moreover, autophagy is essential to the secretion and clearance of Aβ (Zhang et al. 2021).
Although PE is linked to misfolded proteins, research on the specific expression of β-amyloid protein, especially Aβ1−42, in the placenta of PE patients is limited.This study aims to explore the potential involvement of Aβ1−42 expression and its association with autophagy in PE. Our findings indicate a correlation between PE and endoplasmic reticulum (ER)-induced autophagy and protein misfolding. Aβ1−42 was significantly upregulated, and autophagy was enhanced in the placental tissues of PE patients and tunicamycin (TM)-induced HTR-8/SVneo cells. These findings indicate that Aβ1−42 may serve as a potential novel diagnostic marker and therapeutic target for PE.
Materials and methods
Study design
It was a case-control study involving pregnant women with LOPE and normal pregnant women (NP) admitted to the Wusong Central Hospital from May 2021 to December 2022. The study was approved by the Wusong Central Hospital Human Research Ethics Committee. Written informed consent was obtained before recruitment.
PE was diagnosed based on the following criteria: new-onset of hypertension defined as systolic blood pressure ≥ 140 mmHg and/or diastolic blood pressure ≥ 90 mmHg after 20 weeks of gestation and new-onset of proteinuria or in the absence of` proteinuria, new-onset hypertension accompanied by at least one of the following new-onset features: impaired liver function, thrombocytopenia, renal insufficiency, cerebral or visual disturbances, pulmonary edema or uteroplacental dysfunction including fetal growth restriction and/or abnormal uteroplacental circulation. Given the rarity of early-onset PE cases, our study focused exclusively on LOPE in pregnant women. Eligible singleton pregnancies aged 18–55 years were recruited. Pregnant women with a history of chronic hypertension, renal disease, diabetes mellitus, neurological disease, respiratory diseases, and heart diseases, and those who did not sign the written informed consent were excluded. The clinical features of recruited subjects are listed in Table 1.
Collection of placental tissues
Placental tissues sized 1*1*1 cm were collected from the center of the maternal surface within 1 h after delivery in pregnant women of each group (hemorrhagic, infarcted, and calcified tissues were avoided) and rinsed three times in sterile PBS. Each piece of placental tissue sample was prepared into two parts: one was rapidly frozen in liquid nitrogen and stored at -80℃ for protein and RNA extraction, and the other was fixed in 4% paraformaldehyde or 2.5% glutaraldehyde and prepared for histological analysis. All procedures were performed on ice as quickly as possible.
Preparation of Aβ1–42 oligomer
The synthetic peptide Aβ1−42 (GL Biochem Ltd, Shanghai, China) was dissolved in chilled hexafluoroisopropanol (HFIP). The peptide-HFIP solution was placed on ice for 10 min and then evaporated at room temperature in a fume hood to dry overnight. Aβ1−42 peptide was added to fresh anhydrous 100% DMSO to form the peptide membrane. It was then diluted with phenol red-free F12 culture medium to a final concentration of 1 µmol/L and incubated at 4℃ for 24 h before further utilization.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) assay
Total RNA was extracted from tissues and cells using the TRIzol kit (Invitrogen, Carlsbad, CA, USA). cDNA was synthesized using the reverse transcription kit (Takara, Dalian, China) and then subjected to the thermal reaction. Thermal cycling was performed using ABI7500(Applied Biosystems, Foster) at 95 °C for 30 s, followed by 40 cycles of 95 °C for 1 s and 60 °C for 34 s. All primers were synthesized by Wuhan Service Bio Technology Co., Ltd. Table 2 gives the primer sequences that were used. GAPDH was used as an internal control. The mRNA expression was calculated using the 2 −△△Cq method. All samples were detected in duplicate.
Immunohistochemical staining (IHC)
Three placental tissue samples from PE or NP groups were subjected to IHC. Briefly, placental tissues were paraffin-embedded and sectioned (thickness 10 μm). Then, the sections were incubated in 3% hydrogen peroxide, followed by pre-treatment with 5% bovine serum albumin (BSA) for 30 min. Then, the sections were incubated with the rabbit anti-human Aβ1−42 antibody (1:1000, ab180956, Abcam) at 4℃ overnight and the corresponding secondary antibody (1:2000, ab150077, Abcam) at room temperature for 30 min. After counterstaining with hematoxylin, the sections were visualized, and the images were captured using a microscope (Nikon, E100).
Thioflavin-S fluorescence assay
Deparaffinized sections were successively incubated with xylene (15 min×2), pure ethanol (5 min×2), 85% ethanol (5 min×1), and 75% ethanol (5 min×1). After rinsing with water, sections were stained with 0.3% Thioflavin-S(Thio-S)(Sigma), filtered, incubated at room temperature for 8 min, and counterstained with DAPI. Finally, the sections were rinsed with PBS thrice and mounted with coverslips. Images were captured by fluorescent microscopy. The nuclei were labeled blue by DAPI, and Thio-S positive cells were labeled green. The positive rate of fluorescence was calculated based on the recorded data by dividing the number of fluorescent positive cells by the total number of cells and multiplying by 100%.
Western blot
Tissues and cells were processed to extract total protein. Tissues underwent cutting into small pieces, homogenization using Tissue Grinders (Potter‑Elvehjem) on ice, and lysis in RIPA buffer (Sigma-Aldrich; Merck KGaA) containing 1 mM PMSF. Total protein extraction from HTR-8/SVneo cells utilized RIPA lysis buffer. Subsequently, total protein was quantified through a BCA assay (ab207002, abcam, Cambridge). Prepared protein samples (20 µg/lane) were separated by 10% SDS-PAGE and transferred onto PVDF membranes (Millipore Sigma). Following blockage by 5% skim milk for 1 h at room temperature, the membranes were incubated with primary antibodies against Aβ1−42 (1:500, cat. no. ab201060), anti-LC3B antibody (1:500, cat. no. ab221794), anti-Beclin-1 antibody (1:2000, cat. no. ab207612), anti-ATG5 antibody (1:2000, cat. no. A11427), anti-SQSTM1/P62 antibody (1:1000, cat. no. A19700) and GAPDH (1:2500, cat. no. ab181602) overnight at 4℃, and homologous secondary antibodies [rabbit anti-mouse IgG H&L (HRP), 1:2500, cat. no. ab3728; and goat anti-rabbit IgG H&L (HRP), 1:2500, cat. no. ab6721] for 2 h at room temperature. Images were captured using ChemiScope 6000 (CLINX). Protein expression was quantified using ImageJ software (National Institutes of Health).
Cell culture and treatments
The human trophoblast cell line HTR8/SVneo was purchased from Shanghai Zhong Qiao Xin Zhou Biotechnology Co., Ltd., comprising a diverse mix of trophoblast and mesenchymal cells (Abou-Kheir et al. 2017). HTR8/SVneo was cultured in RPMI 1640 medium (Invitrogen Ltd, Paisley, UK) supplemented with 10% fetal bovine serum (FBS, Invitrogen) and 1% penicillin in a humidified incubator containing 5% CO2 at 37℃. HTR8/SVneo cells were seeded in a 6-well plate with 2 ml cell suspension (2 × 105 cells/ml per well). After culturing for 48 h, the cells were washed in serum-free medium twice and induced with 0.078 µg/ml TM (ab120296, Abcam) for 24 h. The cells treated with DMSO were considered as the controls. Cells were also treated with or without 3-Methyladenine (1mM, Macklin) or Rapamycin (100 nM, Macklin) for 24 h.
Enzyme-linked immunosorbent assay (ELISA)
Aβ1−42 peptides secreted in the supernatant of TM-induced HTR8/SVneo cells were quantitatively measured using a Human Amyloid Beta1-42 ELISA Kit (Colorimetric, NBP2-69913) according to the manufacturer’s protocol. Briefly, the cells were gently washed with moderate pre-cooled PBS and dissociated using trypsin. The cell suspension was centrifuged for 5 min at 1000×g. The supernatant was discarded, and the precipitant was washed three times with pre-cooled PBS and then resuspended in pre-cooled PBS to yield a concentration of 4–6 × 106 cells/ml. The freeze-thaw process was repeated several times until the cells were fully lysed. After 10 min of centrifugation at 1500×g at 4℃, the supernatant was collected for measuring Aβ1−42 levels by determining the optical density (OD value) in each well at 450 nm using a microplate reader.
PCR array analysis
Total RNA was extracted as mentioned above. A similar amount of RNA (750–880 NG) was added to each reverse transcriptase reaction to produce a similar amount of cDNA. The cDNA was stored immediately or at -20℃ for 2–3 h and then subjected to PCR Arrays and gene expression analysis on 84 unfolded protein response (UPR) associated genes using the RT2 Profiler PCR array (pahs-089z, Qiagen). Thermal cycling was performed using ABI-9700 Fast (Applied Biosystems, Foster), with an initial denaturation at 95℃ for 10 min, followed by 40 cycles at 95℃ for 15 s, and at 60℃ for 1 min. The signal was collected at 60℃ for each period. Cycle threshold (CT) values were obtained to calculate fold changes in mRNA abundance using the 2−∆∆Ct method. Results were normalized to the housekeeping gene.
Transmission electron microscopy (TEM)
Placental tissues and cells were washed three times in PBS, fixed in ice-cold 2.5% glutaraldehyde at 4 °C overnight, and osmic acid for 2 h at room temperature. After araldite-embedding, the sections were prepared and stained with uranyl acetate and lead citrate. Representative TEM images were captured, and the number of autophagosomes in 20 randomly selected visual fields for each group was quantified.
Trophoblast invasion assay
HTR8/SVneo cells were subjected to a Transwell invasion assay. The cells were suspended in a serum-free medium, and the concentration was adjusted to 2 × 105/ml. In the assay, 600 µl of culture medium containing 10% serum was added to the lower chamber, while 100 µl of the cell suspension was added to the upper chamber. The cells were then incubated in the incubator for 24 h. After incubation, the medium from the upper chamber was removed. The lower surface of the insert was fixed by immersing it in a 4% methanol solution for 30 min. Subsequently, the cells were stained with crystal violet (diluted 4% in PBS) for 20 min. The number of cells below the PET membrane was determined by examining them under a microscope (OLYMPUS, CKX41). Five fields were counted in the center and periphery for each sample, and the average value was calculated.
Statistical analysis
All statistical analyses were performed using IBM SPSS 26.0. Each experiment was performed in triplicate. Quantitative data were expressed as mean ± standard deviation if appropriate and compared using unpaired Student’s t-test for normally distributed data. Categorical variables were reported as frequencies and percentages, and Fisher’s exact test was employed for comparisons. P < 0.05 was considered statistically significant.
Results
Clinical characteristics
There were no significant differences in maternal age, parity, birth weight percentile, and Apgar score between the PE and NP groups, while significant differences were observed in the body mass index, systolic and diastolic blood pressure, and gestational age (all P < 0.05) (Table 1).
Expression levels of Aβ1−42 in the placental tissues of PE
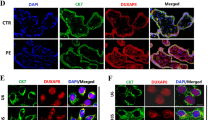
Thio-S fluorescence assay showed that the positive rate of Thio-S in the PE group was significantly higher than that of the NP group (Fig. 1a). The percentage of stained plaque in PE was higher than in the NP group (Fig. 1b). As an essential β-sheet domain in misfolding proteins, IHC staining showed that Aβ1−42 was mainly located in the cell membrane and cytoplasm of trophoblast cells, especially in the EVT and syncytiotrophoblast (ST) (Fig. 1c). Consistently, the protein level of Aβ1−42 was significantly upregulated in the PE group than in the NP group (Fig. 1d, e).
Expression levels of Aβ1−42 in placenta tissues of PE. (a) Thio-S staining of placental tissue from NP and PE. The nuclei were labeled blue by DAPI, and Thio-S positive cells were labeled green. n = 15 per group; scale bar = 50 μm. (b) The percentage of stained plaque was analyzed quantitatively. n = 15 per group. ***P < 0.001 vs. normal. (c) IHC staining of placenta tissues. Aβ1−42 was mainly located in the cell membrane and cytoplasm of EVT and ST (black frame). Scale bar = 20 μm. (d, e) The protein level of Aβ1−42 in the placenta tissues of PE and NP was detected by Western blot. n = 15 per group. **P < 0.01 vs. normal
Enhanced autophagy in the placental tissues of PE
Autophagy is a process in which cytoplasmic components are delivered to be digested by lysosomes. Overactivated autophagy is involved in the development of PE (Gao et al. 2015). We detected expression levels of autophagy-related proteins in NP and PE groups. The ratio of LC3‑II to LC3‑I, along with Beclin-1 and ATG5, were significantly upregulated, while P62 was downregulated in the PE group compared to the NP group, as demonstrated in Fig. 2a, b. TEM images revealed abundant and regular microvilli in the trophoblast membrane, many intact subcellular organelles in the cytoplasm, and massive endoplasmic reticulum and mitochondria with well-arranged mitochondrial cristae in the NP group (Fig. 2c, d). However, a small number of irregular microvilli in the trophoblast membrane, much fewer subcellular organelles in the cytoplasm, and disordered mitochondrial cristae were observed in the PE group. In addition, typical autophagosomes were detectable in the PE group (Fig. 2e, f), whose number was significantly higher in the PE group than in the NP group (Fig. 2g).
Autophagy in placental tissues of PE. (a, b) Expression levels of autophagy-relate proteins (LC3II/I, ATG5, P62, and Beclin-1) in placental tissues of NP and PE groups. n = 10 per group. (c, d, e, f) Representative TEM images of placental tissues in NP (c, d) and PE groups (e, f). d and f (scale bar = 1.0 μm) are the magnified images of red boxes in c and e (scale bar = 5.0 μm) to visualize autophagic vacuoles, respectively. Representative TEM images of the formation of autophagosomes (blue arrow) and mitochondria (red arrow). g. Semi-quantitative analysis on the number of autophagosomes in NP and PE groups. Twenty randomly selected fields were selected for each sample. *P < 0.05, **P < 0.01 and ***P < 0.001 vs. normal
Cell autophagy and upregulation of Aβ1−42 induced by ER stress
HTR-8/SVneo cells present many similar characteristics to those of primary human trophoblasts. TM is a classical ER stress inducer (Jiang et al. 2015). HTR-8/SVneo cells were induced with DMSO and 0.078 µg/ml TM, respectively (Lee et al. 2019; Zhang et al. 2022). ELISA revealed that the expression level of Aβ1−42 was dose-dependently enhanced in TM-induced HTR-8/SVneo cells (Fig. 3a). The ratio of LC3‑II to LC3‑I, along with Beclin-1 and ATG5, were significantly upregulated, while P62 was downregulated after 24 h of TM induction, suggesting that the autophagic activity of HTR-8/SVneo cells was significantly enhanced under ER stress (Fig. 3b, c). The morphological changes of TM-induced HTR-8/SVneo cells were noticeable under TEM. As shown in Fig. 3d, the structure of mitochondrial cristae was disordered following the induction of TM. During the induction of the UPR, massive autophagosomes accumulated in TM-induced HTR-8/SVneo cells. The significantly increased number of autophagosomes in the trophoblasts and decreased number of undegraded substrates indicated the enhancement of autophagic activity (Fig. 3f). Cell invasion is an essential characteristic of PE development. In the Transwell experiment, the number of invading cells significantly decreased in TM-treated cells (Fig. 3e, g).
Cell autophagy and upregulation of Aβ1−42 induced by ER stress. (a) Total expression of Aβ1−42 was measured in HTR-8/SVneo cells induced with DMSO, 0.078 µg/mL TM by ELISA. (b, c) Expression levels of autophagy-relate proteins (LC3II/I, ATG5, P62, and Beclin-1) were measured in HTR-8/SVneo cells induced with DMSO, 0.078 µg/mL TM. n = 5 per group. (d) Representative TEM images of the formation of autophagosomes (blue arrow) and mitochondria (red arrow) in HTR-8/SVneo cells induced with DMSO and 0.078 µg/mL TM. Magnification=×7000. (e, g) Assessment of the invasion ability using Transwell; (f) The number of autopfagosomes was measured in HTR-8/SVneo cells induced with DMSO, 0.078 µg/mL TM; 20 randomly selected fields were selected for each sample. Scale bar = 2 μm. *P < 0.05, **P < 0.01 and ***P < 0.001 vs. DMSO
Aβ1−42 oligomer treatment induced autophagy in HTR-8/SVneo cells
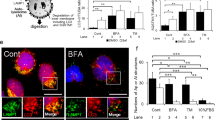
To evaluate whether Aβ1–42 oligomer induced autophagy in HTR-8/SVneo cells, we determined the autophagosomes and autophagy-related proteins, The number of autophagosomes, The ratio of LC3-II to LC3-I, along with Beclin-1 and ATG5, were significantly upregulated, while P62 was downregulated in Aβ1−42 treatment significantly increased (Fig. 4a, b,c, d). However, no significant differences in the concentration of Aβ1−42 were observed in HTR-8/SVneo cells treated with 3-MA or rapamycin (Fig. 4e).
Aβ induced autophagy in HTR-8/SVneo cells. (a) Representative TEM images of the formation of autophagosomes (blue arrow) and mitochondria (red arrow) in HTR-8/SVneo cells induced with control and Aβ1−42 oligomer. (b) The number of autophagosomes was measured in HTR-8/SVneo cells induced with Aβ1−42 oligomer, 20 randomly selected fields were selected for each sample. Scale bar = 2 μm. (c, d) Expression levels of autophagy-relate proteins (LC3II/I, ATG5, P62, and Beclin-1) were determined by western blotting after both cell lines were treated with Aβ1−42 for the indicated time. GAPDH was used as a loading control. The results of three independent experiments were expressed as mean ± SEM, ∗P < 0.05 vs. control. (e) Total expression of Aβ1−42 was measured in HTR-8/SVneo cells induced with Aβ1−42 oligomer, 3-MA, or rapamycin by ELISA. *P < 0.05 vs. control
Identification of DEGs associated with the UPR in PE and their validation
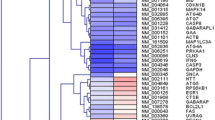
Here, we screened DEGs associated with the UPR in the placental tissues of PE and NP and visualized them in the volcano plots and heatmap. Sixteen significantly upregulated genes associated with the UPR and one significantly downregulated gene were identified (Fig. 5a, b). According to the literature, Adrenomedullin2 (ADM2), activating transcription factor 6 (ATF6), CANX, Endoplasmic reticulum degradation-enhancing α-mannosidase-like protein 1 (EDEM1), Mesencephalic astrocyte-derived neurotrophic factor (MANF), and Ubiquitin-specific protease 14 (USP14) are closely linked with PE or Aβ(Paquet et al. 2005; Lian et al. 2011; Chauhan et al. 2016; Nowakowska-Gołacka et al. 2021; Wang et al. 2021; Zhao and Zong 2021), which further validated their expression levels in our study. Consistently, they were significantly differentially expressed in placental tissues of NP and PE, as well as in HTR-8/SVneo cells induced with or without TM (Fig. 5c, d). Among them, CANX showed the most prominent difference in expression.
DEGs associated with the UPR in PE. (a, b) Volcano plots and heatmap visualizing the DEGs associated with the UPR in placental tissues of NP and PE identified by PCR arrays. Significantly upregulated and downregulated genes were labeled red and blue, respectively. (c, d) The mRNA levels of ADM2, ATF6, CANX, EDEM1, MANF, and USP14 in placental tissues of NP and PE (c) and HTR-8/SVneo cells induced with or without TM (d). n = 5 per group. *P < 0.05, **P < 0.01 and ***P < 0.001vs. control
Discussion
Protein misfolding may lead to pathological conditions when generating higher-level structures. PE is associated with the formation and aggregation of misfolded proteins, which are detectable in the urine, serum, and placenta of pregnant women with PE (Gerasimova et al. 2019). Dysregulated proteins during PE, including Aβ, contribute to the toxic deposition of amyloid-like aggregates in the placenta and body fluids. We primarily collected placentas from LOPE cases for research because most.
PE occurs after 34 weeks of pregnancy, which leads to a higher risk to pregnant women. Thio-S staining, a technique specific for amyloid fibrils with β-pleated sheet conformation (McCarthy et al. 2016), revealed a significant difference between PE and NP groups. In our study, placental tissues from PE patients exhibited strong Thio-S fluorescence, indicating an abundance of these fibrils within amyloid plaques (Bussière et al. 2004). This finding aligns with the observed upregulation of Aβ1−42, a protein known to form β-pleated sheet aggregates, and strengthens the link between protein misfolding and PE.
Aβ is formed by APP through amyloid metabolism. Misfolded proteins have been found to match Aβ signatures, including various polymeric forms of monomers, oligomers, and fibers. Aβ1−42 oligomers are the most toxic among them. Our results showed that Aβ1−42 was significantly upregulated in the placental tissues of PE, especially in the membrane and cytoplasm of trophoblast cells like EVT and ST. This provides evidence that the features of protein misfolding in PE and Aβ aggregation are overlapped.
Autophagy, an evolutionarily conserved process to maintain cell homeostasis, is of great significance in embryogenesis, implantation, and maintenance of pregnancy. Through autophagy, large protein aggregates or damaged organelles are degraded by lysosomes, and the impairment of the autophagy-lysosomal system is a pivotal factor for accumulating aggregated proteins. The role of autophagy in the human placenta remains controversial, and some believe that autophagy can prevent protein aggregation in trophoblast cells (Nakashima et al. 2019). Autophagy is activated in the placenta of pregnant women with PE, and excessive autophagy exacerbates PE(Veerbeek et al. 2015). Beclin-1 is upregulated during autophagy and participates in the recruitment of corresponding proteins in autophagosome formation (Mizushima et al. 2002). The expression of autophagy-related proteins, including ATG5 and LC3-I/LC3-II, increased, and the expression of P62 was reduced after Tm treatment (Zheng et al. 2019). The ratio of LC3‑II to LC3‑I is upregulated in pregnant women with severe PE placental tissues. It is speculated that elevated autophagic activity in placental tissues (mainly reflected by upregulated LC3B) is involved in the onset of PE (Oh et al. 2008).
In this study, an increased number of autophagosomes and upregulated Beclin-1, ATG5, the ratio of LC3‑II to LC3‑I protein expression, and reduced P62 were consistently found in the placental tissues of PE. Increased autophagic activity is closely related to Aβ production and secretion. Autophagy enhancement may serve as a strategy to counteract the overproduction of neurotoxic Aβ (Luo et al. 2020). In this study, HTR8/SVneo cells were stimulated with Aβ1−42 oligomer. We observed that the number of autophagosomes increased, along with the upregulation of the ratio of LC3‑II to LC3‑I, ATG5, Beclin-1 expression, and downregulation of P62. This suggests a close association between autophagy and the accumulation of Aβ aggregates, potentially playing a pathological role in PE. Because Aβ1−42 is the main form of intracellular Aβ and is more prone to oligomerization, extracellular exposure to Aβ1−42 can induce a robust autophagic response. Aβ-induced autophagy is a harmful reaction. However, further research is needed to explore the potential mechanisms underlying Aβ1−42-induced autophagy and the role of autophagy.
However, the interaction between Aβ and autophagy is highly complex. In patients with Alzheimer’s disease (AD), Aβ secretion may increase if the high number of autophagosomes is caused by increased autophagy. Meanwhile, the intracellular Aβ level also ascends with the accumulation of autophagosomes in the last stage of autophagy due to impaired autophagosome clearance (Nilsson et al. 2013). Rapamycin is an mTOR inhibitor known to activate the autophagy process. 3-MA is a classical autophagy inhibitor that primarily inhibits the formation and development of autophagosomes. We treated HTR8/SVneo cells with rapamycin and 3-MA separately and found no significant difference in the expression of Aβ1−42. Based on the analysis, it can be concluded that ER stress in HTR8/SVneo cells increases Aβ1−42 content, thereby enhancing autophagic activity. The autophagy activity in trophoblast cells affects their invasive function. Excessive autophagy activity may be one factor involved in the occurrence and development of PE (Gao et al. 2015).
ER is an essential organelle for protein synthesis, folding, post-translational modification, and secretion. ER stress rises upon pathological stimuli that disrupt protein folding processes, accumulating misfolded proteins in the ER. Accumulating evidence has proven the close association between ER stress and PE (Cheng et al. 2019; Castro et al. 2022). To clear misfolded and aggregated proteins, a panel of signaling pathways is activated to maintain the balance of the ER stress, a phenomenon known as UPR. In pregnancy-related diseases, the accumulation of unfolded or misfolded proteins in the ER lumen promotes ER stress. UPR is activated to alleviate it and restore protein folding capacity (Li et al. 2015). We hypothesized that sustained ER stress impairs UPR and converts it from protective into destructive to promote protein misfolding and accumulation of aggregated proteins. In the present study, ER stress in HTR8/SVneo cells was induced by TM. Our results revealed that upregulation of Aβ1−42 and autophagy-related proteins (LC3II/I, ATG5, and Beclin-1) and an increased number of autophagosomes were detected in TM-induced HTR8/SVneo cells, suggesting that ER stress increased the autophagic activity. Autophagosomes are formed at the mitochondria-ER contact sites. Autophagy can be induced by intrinsic stimuli like aging or misfolded proteins, suggesting its involvement in the occurrence and development of PE. Autophagy can be induced by ER stress, indicating that misfolded proteins have already accumulated in ER but cannot be eliminated in PE.
PCR arrays were performed in the present study to screen DEGs associated with the UPR in the placental tissues of NP and PE. A total of 17 DEGs were identified. Some were validated in placental tissues of NP and PE and HTR8/SVneo cells induced with or without TM. Among them, CANX was the most significant DEG. It is the most abundant lectin chaperone in the ER that participates in quality control in the ER. Incompletely folded or misfolded glycoproteins of CANX were retained within the ER (Paquet et al. 2005). Proteomic analysis of extracellular vesicles in syncytial trophoblasts from patients with early-onset severe PE using isobaric tags for relative and absolute quantification and LC-MS/MS showed that CANX is upregulated in the syncytiotrophoblast extracellular vesicles. It cannot rule out the possibility that these proteins may be autophagy triggers, and they may be able to turn on or exaggerate the action of key effector molecules (Li et al. 2015). In AD patients, CANX positively regulates Aβ1−42, which may involve the autophagy-lysosomal (mitochondrial autophagy) system (Gerber et al. 2019). It is suggested that CANX may be implicated in regulating APP metabolism and Aβ production. As an autophagy co-receptor, CANX targets misfolded proteins for autophagy and also, as an ER chaperone, assists in the initiation of autophagy (Fan and Simmen 2019).
Our data revealed that CANX was significantly upregulated in the placental tissues of PE and TM-induced HTR8/SVneo cells. We speculate that CANX may serve as an ER marker that causes PE by inducing autophagy in the placenta via Aβ. While our study indicates that Aβ1−42 enhances autophagy activity in PE and placental tissue, we have not delved into the specific mechanism of this effect. Further investigation is required to elucidate the potential role of CANX in Aβ production and the etiology of PE.
Our findings suggest that modulation of Aβ1−42 production or autophagic activity may hold promise for the prevention or alleviation of PE symptoms. Future studies will be directed towards elucidating the molecular mechanisms underlying the interplay between Aβ1−42 and autophagy.To further validate the potential of Aβ1−42 as a novel diagnostic marker for PE, we plan to expand our investigation to include patients with EOPE. Additionally, our future plan aims to collect and detect Aβ1−42 in blood and urine samples from PE patients at different stages of pregnancy. This comprehensive dataset will facilitate a deeper understanding of the relationship between Aβ1−42, autophagy and the development and progression of PE.
Data availability
No datasets were generated or analysed during the current study.
References
Abou-Kheir W, Barrak J, Hadadeh O, Daoud G (2017) HTR-8/SVneo cell line contains a mixed population of cells. Placenta 50:1–7. https://doi.org/10.1016/j.placenta.2016.12.007
Bijl RC, Bangert SE, Shree R, Brewer AN, Abrenica-Keffer N, Tsigas EZ, Koster MP, Seely EW (2022) Patient journey during and after a pre-eclampsia-complicated pregnancy: a cross-sectional patient registry study. BMJ Open 12(3):e057795. https://doi.org/10.1136/bmjopen-2021-057795
Buhimschi IA, Nayeri UA, Zhao G, Shook LL, Pensalfini A, Funai EF, Bernstein IM, Glabe CG, Buhimschi CS (2014) Protein misfolding, congophilia, oligomerization, and defective amyloid processing in preeclampsia. Sci Transl Med 6(245):245. https://doi.org/10.1126/scitranslmed.3008808
Bussière T, Bard F, Barbour R, Grajeda H, Guido T, Khan K, Schenk D, Games D, Seubert P, Buttini M (2004) Morphological characterization of Thioflavin-S-positive amyloid plaques in transgenic Alzheimer mice and effect of passive Aβ immunotherapy on their clearance. Am J Pathol 165(3):987–995. https://doi.org/10.1016/S0002-9440(10)63360-3
Castro KR, Prado KM, Lorenzon AR, Hoshida MS, Alves EA, Francisco RP, Zugaib M, Marques AL, Silva EC, Fonseca EJ (2022) Serum from preeclamptic women triggers endoplasmic reticulum stress pathway and expression of angiogenic factors in trophoblast cells. Front Physiol 12:799653. https://doi.org/10.3389/fphys.2021.799653
Chauhan M, Balakrishnan M, Vidaeff A, Yallampalli U, Lugo F, Fox K, Belfort M, Yallampalli C (2016) Adrenomedullin2 (ADM2)/Intermedin (IMD): a potential role in the pathophysiology of preeclampsia. J Clin Endocrinol Metabolism 101(11):4478–4488. https://doi.org/10.1210/jc.2016-1333
Cheng SB, Nakashima A, Sharma S (2016) Understanding pre-eclampsia using Alzheimer’s etiology: an intriguing viewpoint. Am J Reprod Immunol 75(3):372–381. https://doi.org/10.1111/aji.12446
Cheng S-B, Nakashima A, Huber WJ, Davis S, Banerjee S, Huang Z, Saito S, Sadovsky Y, Sharma S (2019) Pyroptosis is a critical inflammatory pathway in the placenta from early onset preeclampsia and in human trophoblasts exposed to hypoxia and endoplasmic reticulum stressors. Cell Death Dis 10(12):927. https://doi.org/10.1038/s41419-019-2162-4
Espinoza J, Vidaeff A, Pettker C, Simhan H (2019) ACOG practice bulletin 202: gestational hypertension and preeclampsia. Obstet Gynecol 133(1):e1–25. https://doi.org/10.1097/AOG.0000000000003018
Fan Y, Simmen T (2019) Mechanistic connections between endoplasmic reticulum (ER) redox control and mitochondrial metabolism. Cells 8(9):1071. https://doi.org/10.3390/cells8091071
Gao L, Qi H-B, Kamana K, Zhang X-M, Zhang H, Baker PN (2015) Excessive autophagy induces the failure of trophoblast invasion and vasculature: possible relevance to the pathogenesis of preeclampsia. J Hypertens 33(1):106–117. https://doi.org/10.1097/HJH.0000000000000366
Gerasimova EM, Fedotov SA, Kachkin DV, Vashukova ES, Glotov AS, Chernoff YO, Rubel AA (2019) Protein misfolding during pregnancy: new approaches to preeclampsia diagnostics. Int J Mol Sci 20(24):6183. https://doi.org/10.3390/ijms20246183
Gerber H, Mosser S, Boury-Jamot B, Stumpe M, Piersigilli A, Goepfert C, Dengjel J, Albrecht U, Magara F, Fraering PC (2019) The APMAP interactome reveals new modulators of APP processing and beta-amyloid production that are altered in Alzheimer’s disease. Acta Neuropathol Commun 7:1–18. https://doi.org/10.1186/s40478-019-0660-3
Jiang H, Zou J, Zhang H, Fu W, Zeng T, Huang H, Zhou F, Hou J (2015) Unfolded protein response inducers tunicamycin and dithiothreitol promote myeloma cell differentiation mediated by XBP-1. Clin Experimental Med 15:85–96. https://doi.org/10.1007/s10238-013-0269-y
Kouza M, Banerji A, Kolinski A, Buhimschi IA, Kloczkowski A (2017) Oligomerization of FVFLM peptides and their ability to inhibit beta amyloid peptides aggregation: consideration as a possible model. Phys Chem Chem Phys 19(4):2990–2999. https://doi.org/10.1039/c6cp07145g
Lee C-L, Veerbeek JH, Rana TK, van Rijn BB, Burton GJ, Yung HW (2019) Role of endoplasmic reticulum stress in proinflammatory cytokine–mediated inhibition of trophoblast invasion in placenta-related complications of pregnancy. Am J Pathol 189(2):467–478. https://doi.org/10.1016/j.ajpath.2018.10.015
Li H, Han L, Yang Z, Huang W, Zhang X, Gu Y, Li Y, Liu X, Zhou L, Hu J (2015) Differential proteomic analysis of syncytiotrophoblast extracellular vesicles from early-onset severe preeclampsia, using 8-Plex iTRAQ labeling coupled with 2D Nano LC-MS/MS. Cell Physiol Biochem 36(3):1116–1130. https://doi.org/10.1159/000430283
Lian I, Løset M, Mundal S, Fenstad M, Johnson M, Eide I, Bjørge L, Freed K, Moses E, Austgulen R (2011) Increased endoplasmic reticulum stress in decidual tissue from pregnancies complicated by fetal growth restriction with and without pre-eclampsia. Placenta 32(11):823–829. https://doi.org/10.1016/j.placenta.2011.08.005
Luo R, Su L-Y, Li G, Yang J, Liu Q, Yang L-X, Zhang D-F, Zhou H, Xu M, Fan Y (2020) Activation of PPARA-mediated autophagy reduces Alzheimer disease-like pathology and cognitive decline in a murine model. Autophagy 16(1):52–69. https://doi.org/10.1080/15548627.2019.1596488
McCarthy FP, Adetoba A, Gill C, Bramham K, Bertolaccini M, Burton GJ, Girardi G, Seed PT, Poston L, Chappell LC (2016) Urinary congophilia in women with hypertensive disorders of pregnancy and preexisting proteinuria or hypertension. Am J Obstet Gynecol 215(4):464. https://doi.org/10.1016/j.ajog.2016.04.041
Millen KR, Buhimschi CS, Zhao G, Rood KM, Tabbah S, Buhimschi IA (2018) Serum and urine thioflavin-T–enhanced fluorescence in severe preeclampsia. Hypertension 71(6):1185–1192. https://doi.org/10.1161/HYPERTENSIONAHA.118.11034
Mizushima N, Ohsumi Y, Yoshimori T (2002) Autophagosome formation in mammalian cells. Cell Struct Funct 27(6):421–429. https://doi.org/10.1247/csf.27.421
Nakashima A, Tsuda S, Kusabiraki T, Aoki A, Ushijima A, Shima T, Cheng S-B, Sharma S, Saito S (2019) Current understanding of autophagy in pregnancy. Int J Mol Sci 20(9):2342. https://doi.org/10.3390/ijms20092342
Nilsson P, Loganathan K, Sekiguchi M, Matsuba Y, Hui K, Tsubuki S, Tanaka M, Iwata N, Saito T, Saido TC (2013) Aβ secretion and plaque formation depend on autophagy. Cell Rep 5(1):61–69. https://doi.org/10.1016/j.celrep.2013.08.042
Nowakowska-Gołacka J, Czapiewska J, Sominka H, Sowa-Rogozińska N, Słomińska-Wojewódzka M (2021) EDEM1 regulates amyloid precursor protein (APP) metabolism and Amyloid-β production. Int J Mol Sci 23(1):117. https://doi.org/10.3390/ijms23010117
Oh S-Y, Choi S-J, Kim KH, Cho EY, Kim J-H, Roh C-R (2008) Autophagy-related proteins, LC3 and Beclin-1, in placentas from pregnancies complicated by preeclampsia. Reproductive Sci 15(9):912–920. https://doi.org/10.1177/1933719108319159
Paquet M-E, Leach MR, Williams DB (2005) In vitro and in vivo assays to assess the functions of calnexin and calreticulin in ER protein folding and quality control. Methods 35(4):338–347. https://doi.org/10.1016/j.ymeth.2004.10.005
Rood KM, Buhimschi CS, Dible T, Webster S, Zhao G, Samuels P, Buhimschi IA (2019) Congo red dot paper test for antenatal triage and rapid identification of preeclampsia. EClinicalMedicine 8:47–56. https://doi.org/10.1016/j.eclinm.2019.02.004
Sergeeva VA, Zakharova NV, Bugrova AE, Starodubtseva NL, Indeykina MI, Kononikhin AS, Frankevich VE, Nikolaev EN (2020) The high-resolution mass spectrometry study of the protein composition of amyloid-like urine aggregates associated with preeclampsia. Eur J Mass Spectrom 26(2):158–161. https://doi.org/10.1177/1469066719860076
Staff AC (2019) The two-stage placental model of preeclampsia: an update. J Reprod Immunol 134:1–10. https://doi.org/10.1016/j.jri.2019.07.004
Veerbeek JH, Van Patot MT, Burton GJ, Yung H-W (2015) Endoplasmic reticulum stress is induced in the human placenta during labour. Placenta 36(1):88–92. https://doi.org/10.1016/j.placenta.2014.11.005
Wang Y, Wen W, Li H, Clementino M, Xu H, Xu M, Ma M, Frank J, Luo J (2021) MANF is neuroprotective against ethanol-induced neurodegeneration through ameliorating ER stress. Neurobiol Dis 148:105216. https://doi.org/10.1016/j.nbd.2020.105216
Yang J, Gong L, Liu Q, Zhao H, Wang Z, Li X, Tian W, Zhou Q (2023) Single-cell RNA-seq reveals developmental deficiencies in both the placentation and the decidualization in women with late-onset preeclampsia. Front Immunol 14:1142273. https://doi.org/10.3389/fimmu.2023.1142273
Zhang Z, Yang X, Song Y-Q, Tu J (2021) Autophagy in Alzheimer’s disease pathogenesis: therapeutic potential and future perspectives. Ageing Res Rev 72:101464. https://doi.org/10.1016/j.arr.2021.101464
Zhang L, Ding H, Shi Y, Zhang D, Yang X (2022) CTRP9 decreases high glucose–induced trophoblast cell damage by reducing endoplasmic reticulum stress. Mol Med Rep 25(5):1–8. https://doi.org/10.3892/mmr.2022.12701
Zhao Y, Zong F (2021) Inhibiting USP14 ameliorates inflammatory responses in trophoblast cells by suppressing MAPK/NF-κB signaling. Immun Inflamm Dis 9(3):1016–1024. https://doi.org/10.1002/iid3.465
Zheng W, Xie W, Yin D, Luo R, Liu M, Guo F (2019) ATG5 and ATG7 induced autophagy interplays with UPR via PERK signaling. Cell Communication Signal 17:1–16. https://doi.org/10.1186/s12964-019-0353-3
Acknowledgements
Not applicable.
Funding
This study was funded by the Baoshan District Science and Technology Committee (Grant No 20-E-10), the Baoshan District Science and Technology Committee (Grant No 21-E-36), the National Natural Science Foundation of China (Grant No 81701468), and Shanghai Baoshan District Health Commission Key Subject Construction Project (BSZK-2023-A18).
Author information
Authors and Affiliations
Contributions
QG and KC made the most substantial contributions to the paper regarding the conception and design of the experiments; YD, LC, YL, and ZN were involved with data acquisition, analysis, and interpretation. QL was responsible for drafting and revising the article for important intellectual content. All authors read and approved the final manuscript.QG and KC confirm the authenticity of all the raw data.All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
All procedures performed in studies involving human participants were per the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards. Specifically, the present study was approved by the Wusong Central Hospital Human Research Ethics Committee (approval number 2021-Y-08), and informed consent from all participants was obtained.
Competing interests
The authors declare no competing interests.
Patient consent for publication
Not applicable.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Gao, Q., Cheng, K., Cai, L. et al. Aβ1−42 stimulates an increase in autophagic activity through tunicamycin-induced endoplasmic reticulum stress in HTR-8/SVneo cells and late-onset pre-eclampsia. J Mol Histol 55, 513–525 (2024). https://doi.org/10.1007/s10735-024-10203-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10735-024-10203-7