Abstract
Although pregnancy is initiated and maintained through highly complex mechanisms, it is essential to understand the events that occur before and during early pregnancy to understand a healthy implantation process. The Notch signal, thought to be involved in this process, is frequently the subject of research with its different aspects. To better understand the role of Notch signaling in the peri-implantation period of the mouse uterus, we investigated the state of expression and localization of Notch 3, Notch 4, Rbp-J, Hes1, Hes7, Hey2, HeyL, and Fbw7 in the uterus and implantation sites in early pregnancy. Balb/C mice were divided into groups D1, D4, D5, D6, and D8. For D5 and D6 groups, implantation sites were identified by intravenous injection of Chicago blue. IHC, WB, and QRT-PCR methods were used. Notch 3 was very strong positive on the 4th day of pregnancy. Notch 4 was highly expressed on days 4, 5, 6, and 8 of pregnancy when P4 levels were high. Hes 1 level was at the lowest on the 4th day of pregnancy. Hes 7 protein expression gradually increased from D1 to D8 in the uteri and implantation sites. Hey 2 expression was at the highest level on the 1st and 4th days. Hey L expression was on the apical of the glands. Fbxw7 that expression was high on the 1st and 4th days of pregnancy. Notch signaling may play an essential role in regulating endometrial receptivity. In addition, our Hes7 results are new to the literature.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Pregnancy, a complicated process, comprises fertilization, implantation, decidualization, placentation, and parturition (Cha et al. 2012). Each process affects the other sequentially, and the trouble in any of these events is an obstacle to a healthy birth. (Dey 2010). The fusion of sperm and oocyte is called fertilization. The fertilized oocyte, called a zygote, undergoes several mitotic divisions and forms the blastocyst. Implantation is the attachment and invasion of the endometrium by the blastocyst. Estrogen and progesterone (P4) are two crucial ovarian steroid hormones that prepare the uterus for embryo development and implantation. During the first two days of pregnancy in mice, preovulatory ovarian estrogen triggers the proliferation of the luminal and glandular epithelial cells. On the 3rd day of pregnancy, P4 secretion from corpora lutea inducts stromal cell proliferation. On the 4th day of pregnancy, known as the day of implantation, proliferation is superimposed by preimplantation ovarian estrogen secretion. On this pregnancy day, epithelial cells do not proliferate anymore and become differentiated, and endometrial capillary permeability increases at the site of the blastocyst (Tan et al. 1999).
Implantation occurs within a precise and transient time known as “the implantation window.“ A reciprocal interaction between the blastocyst and receptive uterus is essential for implantation. Synchronization of embryonic development until the blastocyst stage with the uterine receptive state is critical to successful implantation (Wang and Dey 2006). Uterine sensitivity to the process of implantation of the blastocyst into the uterus is divided into three phases in mice: pre-receptive (days 1–3), receptive (day 4), and non-receptive (refractory; day 5 onward). Embryos can only implant during the receptive phase (Cha et al. 2012). Decidualization is the transformation of stromal cells into morphologically and functionally distinct cells. In mice, it is triggered by blastocyst attachment to the uterus endometrium (Wang and Dey 2006; Dey 2010). Decidualization is initiated on the anti-mesometrial side, then extends to the mesometrial side, the presumed site of placentation (Wang and Dey 2006). Decidua formation is essential for coordinated trophoblast invasion, placental growth, and formation (Afshar et al. 2012a).
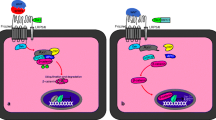
The Notch signaling pathway is pivotal throughout development by controlling numerous cell fate decisions. It is evolutionarily conserved and has been shown in many organisms, from sea urchins to humans (Cormier et al. 2004).
Notch receptors which are ligand-dependent transmembrane proteins, are expressed by one cell and interact with the ligand present in a neighboring cell (Gasperowicz and Otto 2008; Afshar et al. 2012a). In mammals, Notch family members consist of Notch1, 2, 3, and 4 receptors; which are activated by Jagged 1, 2, and Delta-like ligands 1, 3, and 4 (Dll 1, 3, and 4), which are homologs of Serrate and Delta in Drosophila, respectively. After the ligand binds, the Notch receptor undergoes two proteolytic cleavages. Proteolytic processing of the Notch receptor creates two domains that are non-covalently bound to each other. The Notch protein-intracellular domain (NICD) is transferred to the nucleus and cooperates with a transcription factor complex to express Notch target genes. (Gasperowicz and Otto 2008; Sahin et al. 2011; Cormier et al. 2004).
After the NICD translocates to the nucleus, it communicates with DNA binding protein, recombining binding protein suppressor of hairless (RBPJ, also known as CBF1). If there is no NICD, RBPJ serves as a transcriptional repressor and inhibits gene expression via transcription repressors. When NICD associates with RBPJ, it transforms it into an activator, leading to the transcription of target genes. The best-defined Notch targets are the basic inhibitory helix-loop-helix (bHLH) class transcription factors of the Hes and Hey families (Gasperowicz and Otto 2008; Rozenberg et al. 2018). After RBPJ stimulation, ubiquitin-dependent proteolysis moderated by Fbw7 cause the NICD to be destroyed (Tetzlaff et al. 2004).
Cormier et al. (2004) indicated the expressions of Notch1, 2, Jag1-2, Dll-3, and Rbpj transcripts from unfertilized oocyte to late blastocyst stage during preimplantation mouse embryo development using RT-PCR. They showed the expression of all the genes mentioned above in embryonic and trophoblast stem cells. They supposed the activation of the Notch pathway during mouse preimplantation development (Cormier et al. 2004). Chu et al. (2011) inhibited the generation of Notch1’s intracellular domain using γ-secretase inhibitor (DAPT) in the embryo culture medium. They suggested that Notch 1 is essential for the competency of embryo implantation (Chu et al. 2011).
In a study by Gasperowicz et al. (2013), they analyzed mRNA expressions of all Notch receptors-Notch1,2,3,4- and ligands -Jagged1,2; Dll1,3,4- in all trophoblast cell types of developing mouse placenta from the embryonic day 7.5 to E12.5. They showed that Notch receptors and ligands were precisely and dynamically expressed in multiple cell layers of the developing placenta. They concluded that Notch signaling might play diverse roles during mouse placenta development (Gasperowicz and Otto 2008).
In their study investigating the role of Notch1 during early pregnancy using Notch1 uterus of conditional knock-out mice, Afshar et al. (2012a, b) showed that Notch1 has a significant physiological role in endometrial stromal cell differentiation and regulates decidualization by preventing stromal fibroblast apoptosis (Afshar et al. 2012a).
Although some Notch receptors, ligands, and downstream effectors have been studied in the mouse embryo and placenta development and endometrial stromal cell differentiation, the role of Notch signaling hasn’t been investigated during the peri-implantation period in mouse uterus. To better understand the role of Notch signaling in the peri-implantation period of mouse uterus, we examined some Notch signaling members’ expression and localization patterns. We investigated the expression and localization of Notch3, Notch4, Rbp-J, Hes1, Hes7, Hey2, HeyL, and Fbw7 on uterus and implantation sites during early pregnancy.
Materials and methods
Animals
The present study was conducted with unpaired, 6 to 8 weeks-old female (n = 80) and 12 to 18 weeks old male (n = 20) Balb/C mice. Mice had not been mated and used in any experiment before and were procured from the Experimental Animals Unit of Akdeniz University. The mice used in the study were subjected to a 12-h light/dark cycle without any restrictions on water and feed. Two or three female mice were kept with a male mouse overnight for mating. The female mice were checked for vaginal plugs the next day. Female mice observed to have vaginal plugs were admitted on the 1st day of pregnancy, thereby forming the experimental groups; D1: 1st day of pregnancy (n = 10), D4: 4th day of pregnancy (n = 10), D5: 5th day of pregnancy (n = 10); D6: 6th day of pregnancy (n = 10); D8: 8th day of pregnancy (n = 10). Mice were sacrificed by cervical dislocation following anesthesia (Ketamine-Xylazine, 0.1 ml/10 g, #K113 Sigma Aldrich, St. Louis, MO, USA) on determined days, and uteri or implantation sites were collected. For the D1 and D4 groups, pregnancy was confirmed by recovering embryos from the oviducts and uterus, respectively. For D5 and D6 groups, implantation sites were identified by intravenous injection of 0.1 ml 1% Chicago blue (#C8679, Sigma Aldrich, St. Louis, MO, USA) in saline, and discrete blue bands demarcated implantation sites, for the D8 group implantation sites were visible without Chicago blue injection. Some uteri and implantation sites of sacrificed mice were preserved in liquid nitrogen for Western blotting and QRT-PCR. The others were fixed in 10% formalin (#15,512, Merck, Darmstadt, Germany) for routine tissue processing and subsequent immunohistochemical staining. Approval for the experimental protocols was received from the Animal Care and Usage Committee of Akdeniz University. The protocols mentioned above were consistent with the declaration of Helsinki and the International Association for the study of pain guidelines. The local ethics committee approved the experimental protocol for animal experiments at Akdeniz University (approval number:2013.10.04).
Tissue processing
Uteri and implantation sites were fixed in 10% formalin at room temperature for approximately 24 h. Formalin was removed by washing under tap water for 3 h. This was followed by dehydration, immersion in 70%, 80%, and 90% ethanol (#100,986, Merck, Darmstadt, Germany) for 24 h each, and 100% ethanol for 4 h. After dehydration, tissues were cleared with submersion in xylene (#16,446, Sigma Aldrich, St. Louis, MO, USA) for approximately 4 min and embedded in paraffin wax (#107,337, Merck, Darmstadt, Germany).
Immunohistochemistry
Formalin-fixed paraffin-embedded samples were cut into 5 μm sections and placed on electrostatically charged slides (#J1800AMNZ, Thermo Scientific, Braunschweig, Germany). Slides were placed in an oven and incubated at 56 °C overnight the day before immunostaining. For the immunohistochemical procedure, sections were deparaffinized in xylene and rehydrated through a graded ethanol series. To unmask antigens, an antigen retrieval procedure was performed by treating the samples in 10 mM citrate buffer (#100,242, Merck, Darmstadt, Germany), pH6.0, in a microwave oven at 750 W for 5 min, three times. After cooling for 20 min at room temperature, the sections were washed in phosphate-buffered saline (PBS; pH7.4) for 5 min. Endogenous peroxidase activity was blocked by incubation in methanol containing 3% H2O2 (#108,597, Merck, Darmstadt, Germany) for 30 min and washed with PBS three times. Afterward, sections were incubated in a blocking solution (Ultra UVBlock, #TA-125UB, LabVision Corporation, Fremont, CA, USA) for 7 min at room temperature to block non-specific binding. Excess serum was drained, and sections were incubated with antibodies shown in Table 1 at 4 °C overnight. The primary antibodies were substituted with the normal goat IgG (#sc-2028 Santa Cruz Biotechnology, Santa Cruz, CA, USA) or with the normal rabbit IgG (#sc-2027; Santa Cruz Biotechnology, Santa Cruz, CA, USA) in the same dilutions as Notch 3 and other antibodies, respectively. The next day, after washing out the primary antibody, slides were incubated with biotinylated secondary antibodies for Notch 3 (Biotinylated rabbit anti-goat Antibody, #BA-5000, Vector Laboratories, Burlingame, USA) and other antibodies (Biotinylated goat anti-rabbit Antibody, #BA-1000, Vector Laboratories, Burlingame, USA), respectively at 1:500 dilution for 45 min at room temperature, followed by incubation with horseradish peroxidase-conjugated streptavidin (#TS125HR, Thermo Scientific, Fremont, CA, USA) for 30 min at room temperature. All incubation steps were performed in a humidified chamber to avoid dehydration of the slides. After washing the sections with PBS, antibody binding was visualized using the 3.3 di Amino Benzidine (DAB) chromogen (#D4168, Sigma Aldrich, St. Louis, MO, USA). Mayer’s Haematoxylin (#109,249, Merck, Darmstadt, Germany) was utilized for the counterstaining of sections, and they were mounted with Kaiser’s glycerin gelatin (#1.09242.0100, Merck, Darmstadt, Germany) and investigated using Zeiss Axioplan 100 light microscope (Zeiss, Germany), and their photographs were taken.
The evaluation of the immunohistochemical staining of Notch3, Notch4, Rbp-J, Hes1, Hes7, Hey2, HeyL, and Fbw7 in peri-implantation period mouse uteri and implantation sites was determined semi-quantitatively: ø= negative; (+) = weak positive; += positive; ++= strong positive; +++= very strong positive.
Quantitative real-time PCR (QRT-PCR)
RNA isolation and cDNA synthesis
Total RNA was isolated from uteri and implantation sites using Trizol Reagent (#15596-018; Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. RNA concentration was quantified by measuring the absorbance at 260 and 280 nm. To eliminate DNA contamination, isolated RNA was treated with DNase I (#AM1906, Ambion, Buckinghamshire, UK). Preparation of cDNA was prepared from 2 µg of total RNA using the SuperScript III First-Strand Synthesis System for RT-PCR (#11,904,018, Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions.
PCR amplification
Quantitative real-time PCR with SYBR Green I detection (Applied Biosystems, Warrington, U.K.) was performed in a MyiQ Single-Color Real-Time PCRDetection System (Bio-Rad Laboratories, Hercules, CA). Primers were designed using Primer Express Software (Applied Biosystems) and synthesized by Integrated DNA Technologies, Inc (Coralville, IA). Reactions were performed in a total volume of 25 µl, including 12.5 µl 2 × Power SYBR Green PCR Master Mix (#4,368,706, Qiagen, Valencia, CA, USA), 1 µl of 0.4 µM primer, and 1 µl of the previously reverse-transcribed cDNA template and PCR cycling was performed on 96-well iCycler iQ PCR plates (#2,239,441, Bio-Rad, Hercules, CA, USA). PCR amplification for Notch3, Notch4, Hey2, HeyL, Hes7, Fbw7, and Rbp-J were performed for 40 cycles in which the initial 3 min denaturation was at 95 °C followed by a program consisting of denaturation at 92 °C for 20 s, annealing at 55 °C for 15 s and elongation at 72 °C for 30 s. PCR amplification for Hes1 was performed for 40 cycles in which the initial 3 min denaturation was at 95 °C, followed by a program consisting of denaturation at 92 °C for 20 s, annealing at 58 °C for 15 s, and elongation at 72 °C for 30 s. Beta-actin was used as an internal control housekeeping gene to normalize the expression of genes. Sequences of primers are listed in Table 2. The transcripts’ relative expression profiles were calculated using the 2−ΔΔCt (cycle threshold) method and reported as fold changes. A melting curve analysis confirmed the specificity of the genes and Beta-actin qRT-PCR products.
Western blotting
Uteri or implantation sites were weighed and put into a homogenization buffer supplemented with a complete protease inhibitor cocktail (#11,697,498,001, Merck, Mannheim, Germany). After homogenization, samples were centrifuged at 10,000 g for 10 min. Supernatants were collected and stored at − 80 °C. Protein concentrations were determined using a standard bicinchoninic acid assay (Bicinchoninic Acid solution (#B9643) and Copper (II) sulfate solution (#C2284), Sigma Aldrich, St. Louis, MO, USA). Before electrophoresis, samples were heated for 5 min at 95 °C, and 50 µg protein was applied per lane. Samples subjected to SDS polyacrylamide gel electrophoresis under standard conditions were transferred onto polyvinylidene difluoride (PVDF) membrane (#1,620,177, Biorad, USA) in a buffer containing 0.2 mol/L glycine (#1,610,718, Biorad, USA), 25 mM Tris (#108,387, Merck, Darmstadt, Germany) and 20% methanol, overnight at 4 °C. After blocking with 5% non-fat dry milk (#70,166, Sigma Aldrich, St. Louis, MO, USA) membrane was incubated with antibodies listed in Table 1 at shown dilutions overnight at 4oC. After washing, membranes were incubated for one h at room temperature with horseradish peroxidase-conjugated secondary anti-rabbit IgG diluted by 1:2000 (#PI-1000, Vector Laboratories, Burlingame, USA). The reaction was visualized using a chemiluminescence-based Super Signal CL HRP Substrate System (#34,080, Thermo Fisher Scientific, Rockford, USA). The membranes were exposed to Hyperfilm (#28-9068-37 Amersham Hyperfilm ECL, GE Healthcare, Buckinghamshire, UK). The film was processed through a developer (#1,757,855, Ilford, England) and fixative (#1,984,565, Ilford, England), followed by washing with distilled water and drying. An identical protocol also labeled membranes for binding a 1:3000 dilution of a rabbit monoclonal beta-actin antibody (#4970, Cell Signaling) as an internal control to confirm the equal loading of the samples. The bands were quantified using NIH image analysis software (Image J Version 1.36b, National Institutes of Health, USA).
Statistical analysis
Statistical analyses were performed by One-Way ANOVA tests followed by post hoc Tukey tests. Data are presented as the mean ± SD. Probability values of less than 0.05 were considered significant. All data were analyzed using GraphPad Prism (version 6.01, San Diego, USA).
Results
Notch 3 mRNA and protein expressions were high on day 4 of pregnancy, especially in stromal cells
Notch 3 mRNA expression was similar on pregnancy days D1, D5, D6, and D8. It was higher on pregnancy D4 than on other days, but there was no statistically significant difference (Fig. 1a). We weren’t able to get any results for NOTCH 3 Western blotting. According to our immunohistochemistry results, Notch 3 was cytoplasmic and membranous. On day 1 of pregnancy, Notch 3 was strongly positive in the luminal-glandular epithelia, stroma, and myometrium. On day 4 of pregnancy, Notch 3 expression was weakly positive in the glandular epithelium, positive in the luminal epithelium and myometrium, and strongly positive in stromal cells. On day 5, expression was very strong positive in the glandular epithelium, strong positive in stroma and myometrium, and positive in the luminal epithelium. On day 6, interestingly, Notch 3 expression was weakly positive in PDZ, strong positive in SDZ, and very intense in some regions of SDZ. Also, on day 6, it was weakly positive in the luminal epithelium and myometrium and strongly positive in the glandular epithelium. On day 8 of pregnancy, Notch 3 was positive in SDZ and weakly positive in the myometrium; also embryo was strong positive (Fig. 1b, c).
Notch 4 mRNA and protein expressions were high on day 8 of pregnancy when P4 levels were high
Notch 4 mRNA expression was lowest on D5 and statistically significant compared to D6 and D8 (p < 0.05). It was highest on D8 and statistically significant compared to D1 and D5 (p < 0.05) (Fig. 2a). Our Western blotting results showed that NOTCH 4 protein expression was lowest on D1 and then increased through D8. Expression patterns on D4, D5, D6, and D8 were similar and expression on these days of pregnancy was higher than on D1 with a statistically significant difference (p < 0.05). NOTCH 4 expressions on D8 were also higher than on D5 and D6, with a statistically significant difference (p < 0.05) (Fig. 2b). Comparing our QRT-PCR and Western blot results for Notch 4, we conclude that Notch 4 mRNA transcripts were partially translated or degraded on days D6 and D8. According to our immunohistochemistry results, Notch 4 was mostly cytoplasmic and membranous, occasionally nuclear. On day 1 of pregnancy, Notch 4 was positive in the luminal and glandular epithelia, stroma, and myometrium. On day 4, expression was increased. It was strongly positive in the luminal epithelium, stroma, and myometrium; very strong positive in glandular epithelium. On day 5, Notch 4 expression was strongly positive in the luminal and glandular epithelia and myometrium and very strong positive in the stroma. On day 6 of pregnancy, Notch 4 was strongly positive in all examined zones. On day 8, Notch 4 was a strong positive in myometrium and; a very strong positive in SDZ. Also, the embryo was very strong positive on day 8 (Fig. 2c, d).
Rbpj mRNA and protein expressions were highest on day 6 of pregnancy and were strongly expressed by decidual cells
Rbpj mRNA expression was similar on D1, D4, D5, and D8. It was highest on D6 and statistically significant compared to D1, D4, D5, and D8 (p < 0.05) (Fig. 3a). Our Western blotting results showed that RBPJ protein expression was highest on D6 compared to other pregnancy days, and it was statistically significant (p < 0.05) (Fig. 3b). RBPJ protein expression was compatible with Rbpj mRNA expression. According to our immunohistochemistry results Rbpj was mainly nuclear. On day 1 of pregnancy, Rbpj was weakly positive in the glandular epithelium, positive in the luminal epithelium, and strongly positive in stroma and myometrium. On days 4, 5, and 6 of pregnancy; there was no Rbpj expression in the luminal and glandular epithelia. On day 4, Rbpj was strong positive in myometrium and strong positive in the stroma. On day 5, it was very strong positive in the stroma and positive in the myometrium. On day 6 of pregnancy, Rbpj expression was positive in myometrium and strongly positive in PDZ and SDZ. On day 8, Rbpj was weakly positive in myometrium and very strong positive in SDZ (Fig. 3c, d).
Hes 1 mRNA and protein expressions were high on days 6 and 8 of pregnancy and were strongly expressed by decidual cells
Hes 1 mRNA expression was similar on D1 and D8; on D4 and D5, it was highest on D6. There was no statistically significant difference between groups (p < 0.05) (Fig. 4a). Western blotting showed that HES 1 protein expression was lowest on D4 as Hes 1 mRNA expression, and it was statistically significant compared to D1, D5, D6, and D8 groups (p < 0.05). After D4, it showed an increase. An increase in D8 was statistically significant compared to D1 (p < 0.05) (Fig. 4b). Since HES 1 protein expressions on D1, D4, and D6 were higher than Hes1 mRNA expressions, it is possible that mRNAs were degraded or weren’t translated to protein. According to our immunohistochemistry results, Hes 1 was nuclear. On day 1 of pregnancy, it was positive on the apical side of the luminal epithelium and myometrium. On day 4 of pregnancy, it was weakly positive in the luminal epithelium and myometrium, positive in the stroma, and strongly positive in the glandular epithelium. On day 5 of pregnancy, it was strongly positive in the luminal-glandular epithelium and myometrium, very strong in the stroma. Interestingly, Hes 1 was only localized to the anti-mesometrial side of the luminal epithelium, close to the embryo. On day 6 of pregnancy, Hes 1 was positive in the luminal and glandular epithelium and strongly positive in the myometrium, PDZ, and SDZ. On day 8 of pregnancy, Hes 1 expression was positive in SDZ and strongly positive in the myometrium (Fig. 4c, d).
Hes 7 expression gradually increased and was strongly expressed by decidual cells
Hes 7 mRNA expression was lowest on D1 and was statistically significant compared to D4 and D8. Hes 7 mRNA expression was highest on D4, and it was statistically significant compared to D6 (p < 0.05) (Fig. 5a). Our Western blotting results indicated that HES 7 protein expression increased from D1 to D8. HES 7 expression was lowest on D1 as Hes7 mRNA expression. It was highest on D8, and the difference was statistically significant compared to D1 and D4 (p < 0.05) (Fig. 5b). Hes 7 mRNA and HES 7 protein expressions generally showed a similar pattern, except on D4 and D5. These days, not all mRNA transcripts are translated into protein, or mRNAs are degraded. According to our immunohistochemistry results, Hes 7 was nuclear. On days 1, 4, and 5 of pregnancy, it was strongly positive in the luminal-glandular epithelium, stroma, and myometrium. On day 6 of pregnancy, Hes 7 expression was strongly positive in the luminal-glandular epithelium and myometrium and very strong positive in PDZ and SDZ. On day 8 of pregnancy, it was a strong positive in myometrium and a very strong positive in SDZ and embryo (Fig. 5c, d).
Hey 2 expression showed an increase after day 4 of pregnancy and was strongly expressed by decidual cells
Hey 2 mRNA expression was similar to Hes 1 mRNA expression. It was highest on D4 and statistically significant compared to D1, D5, and D6 (p < 0.05) (Fig. 6a). Our Western blotting results showed that HEY 2 protein expression was highest on D1 and decreased from D1. It was lowest on D4 and then increased through D8. HEY 2 expression on D4, D5, and D6 was lower than D1 with a statistically significant difference (p < 0.05). HEY 2 expression on D6 was higher than on D4, and expression on D8 was higher than on D4 and D5 (p < 0.05) (Fig. 6b). On D4, although Hey 2 mRNA transcripts were high, HEY 2 protein amount wasn’t so high. We concluded that not all Hey 2 mRNA transcripts were translated into HEY 2 protein. According to our immunohistochemistry results, Hey 2 was both nuclear and cytoplasmic. On day 1 of pregnancy, it was generally cytoplasmic and strongly positive in the luminal and glandular epithelium, stroma, and myometrium. On day 4 of pregnancy, it was cytoplasmic and weaker compared to day 1; it was positive in the luminal epithelium and weakly in the glandular epithelium, stroma, and myometrium. Hey 2 expression in the endothelial cells was prominent. On day 5 of pregnancy, Hey 2 expression was nuclear and cytoplasmic. It was strongly positive in the luminal epithelium and stroma and positive in glandular epithelium and myometrium. On day 6 of pregnancy, Hey 2 expression was strongly positive in the luminal and glandular epithelium, myometrium, PDZ, and SDZ. On day 8 of pregnancy, Hey 2 expression was strongly positive in SDZ and weakly positive in the myometrium (Fig. 6c, d).
Hey L expression was similar between experimental groups and exclusively expressed at the apical of the glands on day 4 of pregnancy
Hey L mRNA expression on D4, D5, and D6 was higher than on D1 and D8. But there wasn’t a statistically significant difference between the groups (Fig. 7a). According to our Western blotting results, HEY L protein expression showed a similar pattern on D1, D4, D5, and D8. It decreased on D8 compared to other pregnancy days, and it was statistically significantly lower compared to D6 (p < 0.05) (Fig. 7b). Our Hey L mRNA and HEY L protein expression results were compatible. According to our immunohistochemistry results Hey L showed both nuclear and cytoplasmic localization. On day 1 of pregnancy, it was generally cytoplasmic and strongly positive in the luminal epithelium, stroma, and myometrium. On day 4 of pregnancy, it was cytoplasmic and strongly positive in the luminal epithelium, stroma, and myometrium and very strong positive in glandular epithelium. Hey L expression at the apical of the glands on this pregnancy day was remarkable. On day 5 of pregnancy, Hey L expression was nuclear. It was positive in the luminal and glandular epithelium and strongly positive in stroma and myometrium. On day 5 of pregnancy, Hey L expression was cytoplasmic. It was weakly positive in the luminal epithelium, positive in glandular epithelium and myometrium, and strongly positive in PDZ and SDZ. On day 8 of pregnancy, Hey L expression was cytoplasmic, and it was positive in myometrium and very strong in SDZ (Fig. 7c, d).
Fbxw 7 expression gradually decreased and was weakly expressed by decidual cells
Fbxw 7 mRNA expression on pregnancy days 5 (D5) and 6 (D6) was similar and lower than D1 and D4. Fbxw 7 mRNA expression was highest on D8, and it was statistically significant compared to the D1, D5, and D6 groups (p < 0.05) (Fig. 8a). According to our Western blotting results FBXW 7 protein showed a similar expression pattern to Fbxw 7 mRNA for days 1, 4, 5, and 6 of pregnancy. It was highest in D4 and statistically significant compared to D5, D6, and D8 groups (p < 0.05). FBXW 7 expression on D1 was higher than on D5, D6, and D8, with a statistically significant difference (p < 0.05) (Fig. 8b). Although Fbxw 7 mRNA expression was highest on D8, FBXW 7 protein was not as high as mRNA. So it made us think that mRNA was degraded or wasn’t translated to protein at all. According to our immunohistochemistry results Fbxw 7 showed both cytoplasmic and nuclear staining. On day 1 of pregnancy, Fbxw 7 expression was strong on the apical side of the luminal epithelium and myometrium; weak on the glandular epithelium. On day 4 of pregnancy, it was strong on the apical side of the luminal epithelium, glandular epithelium, and endothelial cells in the stroma and myometrium. Interestingly, on day 5 of pregnancy, expression decreased periphery of the luminal epithelium but an increased neighborhood of myometrium and was membranous (Fig. 8c, d).
Discussion
For a successful pregnancy in mammalian species, the uterus must be receptive to the embryo’s healthy development and differentiation into the blastocyst stage. In mice, uterine receptivity is divided into 3 phases: pre-receptive, receptive, and non-receptive (Tu et al. 2014). The uterus becomes receptive only on the night of the 4th day of pregnancy. On the 5th day, the uterus becomes non-receptive and does not allow the blastocyst implantation. In mice, implantation occurs at the anti-mesometrial side of the luminal epithelium, the opposite side of the blood vessel-rich mesometrium (Ye 2020). Implantation is divided into three stages: apposition, adhesion, and penetration. During these stages, trophoblast cells converge, attach, and invade the luminal epithelium. As a result of embryo attachment to the luminal epithelium, the stromal vascular permeability increases at the site of the blastocyst. Proliferation, differentiation, migration, and remodeling occur in cells of both the embryo and the uterus throughout these phases (Enders and Schlafke 1969). One of the most important causes of infertility is implantation failure (Koot et al. 2011). The interactions between the blastocyst and endometrium are critical for implantation, and there is little information about these interactions for humans due to ethical constraints. Therefore, information obtained from the studies in rodents such as mice and rats is quite valuable.
One of the most highly conserved signaling cascades-the Notch pathways, is involved in regulating various cellular processes such as cell proliferation, invasion, adhesion, survival, apoptosis, and differentiation (Massimiani et al. 2019). Although there are studies about the expression of Notch signaling pathway members in normal and pathologic human placenta (De Falco et al. 2007; Cobellis et al. 2008; Mikhailik et al. 2009; Herr et al. 2011) and preimplantation period embryo development in mice, (Cormier et al. 2004; Chu et al. 2011) there are limited studies about their role during implantation and decidualization (Afshar et al. 2012a, b; Wu et al. 2021). For these reasons, in our study, we aimed to examine the expression of some members of the Notch signaling pathway at the mRNA and protein level on different days during early pregnancy in mice.
Notch 3 was very strong positive in stromal cells, and it was highest on the 4th day of pregnancy, according to our immunohistochemistry and QRT-PCR results. Zhou et al., showed that interaction of Notch 3 and Jagged 1 significantly reduced the expression of HEY 1 and classical receptivity markers- Forkhead box protein O1 (Foxo1), Lifr, and Stat3 when JAG 1 was knocked down in human endometrial epithelial cells (Zhou et al. 2021). Also, Wang et al. performed single-cell sequencing analyses of human endometrium across the menstrual cycle. They showed the increased expression of Notch 3 in the epithelial cells, specifically in the receptive window (Wang et al. 2020). Mikhailik et al. showed expression of Notch 3 in isolated human endometrial epithelial and stromal cells (Mikhailik et al. 2009). Shawber et al. investigated the expression of some Notch signaling pathway members during the peri-implantation period in mouse uterus, and they showed Notch3 expression in the pericytes before implantation (Shawber et al. 2015).
According to our results, Notch4 was highly expressed on days 4, 5, 6, and 8 of pregnancy when P4 levels were high. In Murta et al.‘s study, Notch4 transcription in the uterus was shown to be correlated with plasma P4 concentrations (Murta et al. 2015). Shawber et al. denoted the expression of Notch 4 in endothelial cells of newly formed decidual capillaries. Notch4 has unique roles during decidual angiogenesis and early placentation (Shawber et al. 2015). Since we also showed high expression of Notch4 during decidualization, our results are compatible.
According to a study by Su et al., Notch 4 was decreased in the eutopic endometrium of women and baboons with endometriosis (Su et al. 2015). In contrast to mice, Notch 4 decreases from the proliferative to the secretory phase in human endometrium. Also, in menopause, the expression level of Notch4 was reduced. In the pathological human endometrium, the researchers observed a decrease in Notch-4 expression from polyps to carcinoma (Cobellis et al. 2008). Also, Su et al. showed a decline of Notch4 in the eutopic endometrium of women and baboons with endometriosis (Su et al. 2015). So Notch-4 may be involved in controlling proliferation in human endometrium.
Zhang et al. concluded that uterine Rbpj is required for embryonic-uterine orientation before embryo attachment and decidual remodeling at post-implantation stages. They showed that Rbpj directly regulates the expression of uterine matrix metalloproteinase at post-implantation stages, which are required for normal post-implantation decidual remodeling (Zhang et al. 2014). Our results showed that Rbpj mRNA and protein levels were highest on day 6 of pregnancy. Our results also confirm that Rbpj is involved in decidualization during mouse pregnancy. In another study by Strug et al., Rbpj was shown to mediate uterine repair in the mouse and be reduced in women with recurrent pregnancy loss (Strug et al. 2018).
Our Hes 1 results showed that mRNA and protein levels were at the lowest on the 4th day of pregnancy. After the 4th day, it increased gradually and reached the highest level on the 8th day of pregnancy. Murta et al. investigated the dynamics of Notch signaling in the mouse uterus during the estrous cycle. They found that the transcription levels of Hes1 peak during oestrus, and they concluded that Hes1 in the uterus follows the trend of plasma E2 concentrations (Murta et al. 2015). Our study showed high expression of Hes1 on day 1 uteri and day 5 implantation site where preovulatory and preimplantation estrogen is high, respectively. Murta et al. observed that Hes1 expression increased significantly in stromal cells during estrus. The authors concluded that these proteins regulate stroma proliferation (Murta et al. 2015). Hes1 expression pattern at implantation sites on the 5th day of pregnancy was interesting. It was only localized to the antimesometrial side of the luminal epithelium, close to the embryo. Since implantation always occurs on the antimesometrial side, unique Hes1 expression in the luminal epithelium should be paramount.
Similarly, Van Sinderen et al. evaluated the localization of Hes1 cycling endometrium. They found that Hes1 was moderately expressed in the glandular and luminal epithelium throughout the menstrual cycle, with significantly elevated levels in the late secretory phase when implantation occurs (Van Sinderen et al. 2014). There was strong Hes 1 expression in the stroma of implantation sites of our experimental groups, notably on days 6 and 8 of pregnancy. Since Hes1 was strongly expressed in decidua cells on days 6 and 8 of pregnancy, we think Hes 1 may have a role in stromal cell proliferation. On the other hand, Mikhailik et al. reported scarce Hes1 expression in isolated stromal and endometrial epithelial cells, respectively (Mikhailik et al. 2009). This might be a difference between the human and mouse uterus.
Hes 7 protein expression gradually increased from D1 to D8 in our experimental groups’ uteri and implantation sites. On the 8th day of pregnancy, it was at the highest level. For this reason, Hes7 may be involved in decidualization and placentation. Our results about Hes7 are the first since there is no study about the role of Hes7 during pregnancy in the literature.
Hey 2 expression was at the highest level on the 1st and the lowest on the 4th day of pregnancy. On the 4th day, Hey 2 expression was positive in the luminal epithelium; it was much weaker than on other days of pregnancy. The preovulatory expression of E2 is high on the 1st, and preimplantation E2 is high on the 4th day of the pregnancy (Fukui et al. 2019). Since Hey2 expression was variable on days of pregnancy, we suggest no correlation between Hey2 expression and E2 levels. On the contrary, Nakamura et al. showed estrogen-dependent down-regulation of Hey2 in the uterus (Nakamura et al. 2012). Hey2 was also high on days 6 and 8 of pregnancy, during which primary and secondary decidual zones were formed.
On the other hand, Shawber et al. reported that Hey2 was not expressed by CD31+ endothelial cells in the peri-implantation period mouse uterus. Hey2 was strongly expressed in smooth muscle cells in the myometrium and vascular smooth muscle cells surrounding large arteries during both pre-and post-implantation periods (Shawber et al. 2015). Fischer et al. showed strong Hey1/2 expression in fetal endothelial cells in the chorioallantoic layer, which gives rise to the placental labyrinth. They also demonstrated the failure of double-knockout Hes/Hey mutant mice to undergo chorioallantoic branching (Fischer et al. 2004).
In a study by Nakamura et al., they showed estrogen-dependent up-regulation of HeyL in the uterus (Nakamura et al. 2012). According to our results, Hey L expression showed a similar pattern between experimental groups. This protein was in both nuclear and cytoplasmic localization. Hey L expression at the apical of the glands on the 4th day of pregnancy was remarkable. We think this expression pattern of Hey L might be involved in the excretion of secretions deposited in the glands.
Our Fbxw7 results showed that expression was high on the 1st and 4th days of pregnancy, then decreased on the 5th, 6th, and 8th days. Fbxw 7 was in both cytoplasmic and nuclear localization. Interestingly, Fbxw 7 expression reduced in the periphery of the luminal epithelium but increased in the neighborhood of myometrium. Since Fbxw 7 takes a role in ubiquitin-mediated degradation, it should be substantial. In their study, Cuevas et al. concluded that Fbxw7 is a driver of uterine carcinosarcoma by promoting epithelial-mesenchymal transition (Cuevas et al. 2019). It is known that mesenchymal-epithelial transition exists during decidualization (Zhang et al. 2013). Our study showed low expression of Fbxw7 on days 6 and 8 of pregnancy when decidual cells are formed.
In the literature, there are studies about the expression of Notch signaling members in the endometrium (Cobellis et al. 2008; Mitsuhashi et al. 2012; Mazella et al. 2008; Mikhailik et al. 2009), during the preimplantation period of embryo development (Cormier et al. 2004; Cuman et al. 2014; Adjaye et al. 2005; Aghajanova et al. 2012; Wang et al. 2004). There are also studies on the role of Notch1 in mouse placental development (Gasperowicz et al. 2013) and the decision-making process (Hess et al. 2007; Wu et al. 2021; Afshar et al. 2012a, b). In addition, our study reveals the localization and expression of some Notch signaling members during implantation and decidualization processes in the mouse. Our results suggest that Notch signaling is crucial for regulating endometrial receptivity. We believe these results lay the groundwork for loss- and gain-of-function studies that will determine the cell-type-specific requirements for Notch proteins during implantation and decidualization. Our results will help to reveal the underlying causes of implantation failure and pregnancy losses.
Data availability
All the data supporting the findings of this study are available within the article and its supplementary materials.
Abbreviations
- Dll:
-
Delta-like ligand
- Fbxw 7:
-
F-Box And WD Repeat Domain Containing 7
- Hes 1:
-
Hes Family BHLH Transcription Factor 1
- Hes 7:
-
Hes Family BHLH Transcription Factor 7
- Hey 2:
-
Hes Related Family BHLH Transcription Factor With YRPW Motif 2
- Hey L:
-
Hes Related Family BHLH Transcription Factor With YRPW Motif 3
- NECD:
-
Notch extracellular domain
- NICD:
-
Notch intracellular domain
- RBPJ:
-
Recombination signal binding protein for immunoglobulin kappa J region
References
Adjaye J, Huntriss J, Herwig R, BenKahla A, Brink TC, Wierling C, Hultschig C, Groth D, Yaspo ML, Picton HM, Gosden RG, Lehrach H (2005) Primary differentiation in the human blastocyst: comparative molecular portraits of inner cell mass and trophectoderm cells. Stem Cells 23(10):1514–1525. https://doi.org/10.1634/stemcells.2005-0113
Afshar Y, Jeong JW, Roqueiro D, DeMayo F, Lydon J, Radtke F, Radnor R, Miele L, Fazleabas A (2012) Notch1 mediates uterine stromal differentiation and is critical for complete decidualization in the mouse. FASEB J 26(1):282–294. https://doi.org/10.1096/fj.11-184663
Afshar Y, Miele L, Fazleabas AT (2012b) Notch1 is regulated by chorionic gonadotropin and progesterone in endometrial stromal cells and modulates decidualization in primates. Endocrinology 153(6):2884–2896. https://doi.org/10.1210/en.2011-2122
Aghajanova L, Shen S, Rojas AM, Fisher SJ, Irwin JC, Giudice LC (2012) Comparative transcriptome analysis of human trophectoderm and embryonic stem cell-derived trophoblasts reveal key participants in early implantation. Biol Reprod 86(1):1–21. https://doi.org/10.1095/biolreprod.111.092775
Cha J, Sun X, Dey SK (2012) Mechanisms of implantation: strategies for successful pregnancy. Nat Med 18(12):1754–1767. https://doi.org/10.1038/nm.3012
Chu PW, Wang YP, Chen IC, Pan HM, Wu GJ (2011) Notch 1 signaling pathway effect on implantation competency. Fertil Steril 96(5):1225–1229. https://doi.org/10.1016/j.fertnstert.2011.08.032
Cobellis L, Caprio F, Trabucco E, Mastrogiacomo A, Coppola G, Manente L, Colacurci N, De Falco M, De Luca A (2008) The pattern of expression of notch protein members in normal and pathological endometrium. J Anat 213(4):464–472. https://doi.org/10.1111/j.1469-7580.2008.00963.x
Cormier S, Vandormael-Pournin S, Babinet C, Cohen-Tannoudji M (2004) Developmental expression of the notch signaling pathway genes during mouse preimplantation development. Gene Expr Patterns 4(6):713–717. https://doi.org/10.1016/j.modgep.2004.04.003
Cuevas IC, Sahoo SS, Kumar A, Zhang H, Westcott J, Aguilar M, Cortez JD, Sullivan SA, Xing C, Hayes DN, Brekken RA, Bae-Jump VL, Castrillon DH (2019) Fbxw7 is a driver of uterine carcinosarcoma by promoting epithelial-mesenchymal transition. Proc Natl Acad Sci USA 116(51):25880–25890. https://doi.org/10.1073/pnas.1911310116
Cuman C, Menkhorst E, Winship A, Van Sinderen M, Osianlis T, Rombauts LJ, Dimitriadis E (2014) Fetal-maternal communication: the role of notch signalling in embryo implantation. Reproduction 147(3):R75–86. https://doi.org/10.1530/REP-13-0474
De Falco M, Cobellis L, Giraldi D, Mastrogiacomo A, Perna A, Colacurci N, Miele L, De Luca A (2007) Expression and distribution of notch protein members in human placenta throughout pregnancy. Placenta 28(2–3):118–126. https://doi.org/10.1016/j.placenta.2006.03.010
Dey SK (2010) How we are born. J Clin Investig 120(4):952–955. https://doi.org/10.1172/JCI42380
Enders AC, Schlafke S (1969) Cytological aspects of trophoblast-uterine interaction in early implantation. Am J Anat 125(1):1–29. https://doi.org/10.1002/aja.1001250102
Fischer A, Schumacher N, Maier M, Sendtner M, Gessler M (2004) The Notch target genes Hey1 and Hey2 are required for embryonic vascular development. Genes Dev 18(8):901–911. https://doi.org/10.1101/gad.291004
Fukui Y, Hirota Y, Matsuo M, Gebril M, Akaeda S, Hiraoka T, Osuga Y (2019) Uterine receptivity, embryo attachment, and embryo invasion: multistep processes in embryo implantation. Reprod Med Biol 18(3):234–240. https://doi.org/10.1002/rmb2.12280
Gasperowicz M, Otto F (2008) The notch signalling pathway in the development of the mouse placenta. Placenta 29(8):651–659. https://doi.org/10.1016/j.placenta.2008.06.004
Gasperowicz M, Rai A, Cross JC (2013) Spatiotemporal expression of notch receptors and ligands in developing mouse placenta. Gene Expr Patterns 13(7):249–254. https://doi.org/10.1016/j.gep.2013.04.006
Herr F, Schreiner I, Baal N, Pfarrer C, Zygmunt M (2011) Expression patterns of notch receptors and their ligands jagged and delta in human placenta. Placenta 32(8):554–563. https://doi.org/10.1016/j.placenta.2011.04.018
Hess AP, Hamilton AE, Talbi S, Dosiou C, Nyegaard M, Nayak N, Genbecev-Krtolica O, Mavrogianis P, Ferrer K, Kruessel J, Fazleabas AT, Fisher SJ, Giudice LC (2007) Decidual stromal cell response to paracrine signals from the trophoblast: amplification of immune and angiogenic modulators. Biol Reprod 76(1):102–117. https://doi.org/10.1095/biolreprod.106.054791
Koot YE, Boomsma CM, Eijkemans MJ, Lentjes EG, Macklon NS (2011) Recurrent pre-clinical pregnancy loss is unlikely to be a ‘cause’ of unexplained infertility. Hum Reprod 26(10):2636–2641. https://doi.org/10.1093/humrep/der217
Massimiani M, Lacconi V, La Civita F, Ticconi C, Rago R, Campagnolo L (2019) Molecular signaling regulating endometrium-blastocyst crosstalk. Int J Mol Sci. https://doi.org/10.3390/ijms21010023
Mazella J, Liang S, Tseng L (2008) Expression of delta-like protein 4 in the human endometrium. Endocrinology 149(1):15–19. https://doi.org/10.1210/en.2007-0477
Mikhailik A, Mazella J, Liang S, Tseng L (2009) Notch ligand-dependent gene expression in human endometrial stromal cells. Biochem Biophys Res Commun 388(3):479–482. https://doi.org/10.1016/j.bbrc.2009.07.037
Mitsuhashi Y, Horiuchi A, Miyamoto T, Kashima H, Suzuki A, Shiozawa T (2012) Prognostic significance of notch signalling molecules and their involvement in the invasiveness of endometrial carcinoma cells. Histopathology 60(5):826–837. https://doi.org/10.1111/j.1365-2559.2011.04158.x
Murta D, Batista M, Trindade A, Silva E, Mateus L, Duarte A, Lopes-da-Costa L (2015) Dynamics of Notch signalling in the mouse oviduct and uterus during the oestrous cycle. Reprod Fertil Dev. https://doi.org/10.1071/RD15029
Nakamura T, Miyagawa S, Katsu Y, Sato T, Iguchi T, Ohta Y (2012) Sequential changes in the expression of wnt- and notch-related genes in the vagina and uterus of ovariectomized mice after estrogen exposure. In vivo 26(6):899–906
Rozenberg JM, Taylor JM, Mack CP (2018) RBPJ binds to consensus and methylated cis elements within phased nucleosomes and controls gene expression in human aortic smooth muscle cells in cooperation with SRF. Nucleic Acids Res 46(16):8232–8244. https://doi.org/10.1093/nar/gky562
Sahin Z, Acar N, Ozbey O, Ustunel I, Demir R (2011) Distribution of Notch family proteins in intrauterine growth restriction and hypertension complicated human term placentas. Acta Histochem 113(3):270–276. https://doi.org/10.1016/j.acthis.2009.10.006
Shawber CJ, Lin L, Gnarra M, Sauer MV, Papaioannou VE, Kitajewski JK, Douglas NC (2015) Vascular notch proteins and notch signaling in the peri-implantation mouse uterus. Vascular Cell 7:9. https://doi.org/10.1186/s13221-015-0034-y
Strug MR, Su RW, Kim TH, Mauriello A, Ticconi C, Lessey BA, Young SL, Lim JM, Jeong JW, Fazleabas AT (2018) RBPJ mediates uterine repair in the mouse and is reduced in women with recurrent pregnancy loss. FASEB J 32(5):2452–2466. https://doi.org/10.1096/fj.201701032R
Su RW, Strug MR, Joshi NR, Jeong JW, Miele L, Lessey BA, Young SL, Fazleabas AT (2015) Decreased notch pathway signaling in the endometrium of women with endometriosis impairs decidualization. J Clin Endocrinol Metab 100(3):E433–442. https://doi.org/10.1210/jc.2014-3720
Tan J, Paria BC, Dey SK, Das SK (1999) Differential uterine expression of estrogen and progesterone receptors correlates with uterine preparation for implantation and decidualization in the mouse. Endocrinology 140(11):5310–5321. https://doi.org/10.1210/endo.140.11.7148
Tetzlaff MT, Yu W, Li M, Zhang P, Finegold M, Mahon K, Harper JW, Schwartz RJ, Elledge SJ (2004) Defective cardiovascular development and elevated cyclin E and notch proteins in mice lacking the Fbw7 F-box protein. Proc Natl Acad Sci USA 101(10):3338–3345. https://doi.org/10.1073/pnas.0307875101
Tu Z, Ran H, Zhang S, Xia G, Wang B, Wang H (2014) Molecular determinants of uterine receptivity. Int J Dev Biol 58(2–4):147–154. https://doi.org/10.1387/ijdb.130345wh
Van Sinderen M, Cuman C, Gamage T, Rainczuk K, Osianlis T, Rombauts L, Dimitriadis E (2014) Localisation of the notch family in the human endometrium of fertile and infertile women. J Mol Histol 45(6):697–706. https://doi.org/10.1007/s10735-014-9587-y
Wang H, Dey SK (2006) Roadmap to embryo implantation: clues from mouse models. Nat Rev Genet 7(3):185–199. https://doi.org/10.1038/nrg1808
Wang QT, Piotrowska K, Ciemerych MA, Milenkovic L, Scott MP, Davis RW, Zernicka-Goetz M (2004) A genome-wide study of gene activity reveals developmental signaling pathways in the preimplantation mouse embryo. Dev Cell 6(1):133–144. https://doi.org/10.1016/s1534-5807(03)00404-0
Wang W, Vilella F, Alama P, Moreno I, Mignardi M, Isakova A, Pan W, Simon C, Quake SR (2020) Single-cell transcriptomic atlas of the human endometrium during the menstrual cycle. Nat Med 26(10):1644–1653. https://doi.org/10.1038/s41591-020-1040-z
Wu Y, He JP, Xie J, Wang KZ, Kang JW, Fazleabas AT, Su RW (2021) Notch1 is crucial for decidualization and maintaining the first pregnancy in the mousedagger. Biol Reprod 104(3):539–547. https://doi.org/10.1093/biolre/ioaa222
Ye X (2020) Uterine luminal epithelium as the transient gateway for embryo implantation. Trends Endocrinol Metab 31(2):165–180. https://doi.org/10.1016/j.tem.2019.11.008
Zhang XH, Liang X, Liang XH, Wang TS, Qi QR, Deng WB, Sha AG, Yang ZM (2013) The mesenchymal-epithelial transition during in vitro decidualization. Reprod Sci 20(4):354–360. https://doi.org/10.1177/1933719112472738
Zhang S, Kong S, Wang B, Cheng X, Chen Y, Wu W, Wang Q, Shi J, Zhang Y, Wang S, Lu J, Lydon JP, DeMayo F, Pear WS, Han H, Lin H, Li L, Wang H, Wang YL, Li B, Chen Q, Duan E, Wang H (2014) Uterine rbpj is required for embryonic-uterine orientation and decidual remodeling via notch pathway-independent and -dependent mechanisms. Cell Res 24(8):925–942. https://doi.org/10.1038/cr.2014.82
Zhou W, Menkhorst E, Dimitriadis E (2021) Jagged1 regulates endometrial receptivity in both humans and mice. FASEB J 35(8):e21784. https://doi.org/10.1096/fj.202100590R
Acknowledgements
We thank TUBITAK most sincerely. The authors also thank Dr. Derya Mutlu for his excellent knowledge and help with quantitative real-time PCR experiments.
Funding
This work was supported by The Scientific and Technological Research Council of Turkey (TUBITAK) (No: 213S032).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Acar, N., Soylu, H., Avci, S. et al. Expressions of Notch signalling pathway members during early pregnancy in mice. J Mol Histol 54, 297–312 (2023). https://doi.org/10.1007/s10735-023-10132-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10735-023-10132-x