Abstract
Much information is currently available for molecules in early odontogenesis, but there is limited knowledge regarding terminal cytodifferentiation of ameloblasts and odontoblasts for the determination of normal crown morphology. The present differential display PCR (DD-PCR) revealed that insulin-like growth factor-binding protein 5 (IGFBP5) was differentially expressed in molar tooth germs between the cap (before crown mineralization) and root formation (after crown mineralization) stages. Real-time PCR confirmed that the expression levels of IGFBP1–4 were not significantly changed but those of IGFBP5–7 were upregulated in a time-dependent manner. Immunoreactivities for IGFBP5–7 were hardly seen in molar germs at the cap/early bell stage and protective-stage ameloblasts at the root formation stage. However, the reactivity was strong in odontoblasts and maturation-stage ameloblasts, which are morphologically and functionally characterized by wide intercellular space and active enamel matrix mineralization. The localization of each IGFBP was temporospatial. IGFBP5 was localized in the nuclei of fully differentiated odontoblasts and ameloblasts, while IGFBP6 was localized in the apical cytoplasm of ameloblasts and odontoblasts with dentinal tubules, and IGFBP7 was mainly found in the whole cytoplasm of odontoblasts and the intercellular space of ameloblasts. IGFBP silencing using specific siRNAs upregulated representative genes for dentinogenesis and amelogenesis, such as DMP1 and amelogenin, respectively, and augmented the differentiation media-induced mineralization, which was confirmed by alizarin red s and alkaline phosphatase staining. These results suggest that IGFBP5–7 may play independent and redundant regulatory roles in late-stage odontogenesis by modulating the functional differentiation of ameloblasts and odontoblasts.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Odontogenesis, the progression of tooth germs through complicated and sequential molecular reactions, results in the proliferation, movement, differentiation, and apoptotic death of odontogenic cells. Currently, the bioengineering of tooth germs is suggested to be an ideal tooth rehabilitation approach. However, it faces hurdles, including the acquisition of normal tooth morphology (e.g., crown size and shape are faced with difficulty) and the establishment of odontogenic cells (Ono et al. 2017; Smith et al. 2018). Accordingly, the detection of molecules involved in the regulation of the enamel and dentin formation by odontogenic cells has been mainstream research in tooth developmental fields.
Odontogenesis begins from the oral epithelium and ectomesenchymal cells, which develop into the enamel (or dental) organ and dental papilla, respectively. Dental papilla cells rapidly increase in length and differentiate into dentin-forming odontoblasts; concurrent with this change, inner enamel epithelia differentiate into enamel-forming ameloblasts at the late bell stage. According to the morphology of the enamel organ, odontogenesis is subdivided into four stages: bud, cap, bell, and dental hard tissue formation (crown and root formation). Changes in its morphology are strictly regulated by genetic programs, and, therefore, the detection of regulatory genes responsible for these changes has continuously gained attention. Currently, more than 300 genes are reported to be involved in tooth morphogenesis, but most are associated with the patterning and morphogenesis of tooth germs, events of relatively early morphogenetic stages from initiation through cap/early bell stages (Thesleff 2006; Lan et al. 2014; Doshi et al. 2016; Sadier et al. 2019; Wang et al. 2019).
More importantly, further changes in odontogenic cells, such as differentiation and phenotypic protein expression, are needed for them to functionally mature to form and mineralize enamel and dentin matrices. In addition, tooth bioengineering using fate-determined odontogenic cells ultimately needs functional cell differentiation leading to amelogenesis and odontogenesis that may determine normal crown morphology (Oyanagi et al. 2019). Currently, however, there is a lack of information regarding molecules that regulate the terminal functional differentiation of odontogenic cells. These secrete growth factors, such as the insulin-like growth factor (IGF), which is a potent mitogen and transforming growth factor-β that regulate cell proliferation and differentiation. Additionally, receptors for growth factors and endothelin receptors were reported to be involved in odontogenesis (Klopcic et al. 2007; Neuhaus and Byers 2007), implying the importance of ligand-receptor axis in odontogenesis. This axis is further delicately regulated by the modulation of specific molecules or ligands (Chen et al. 2017; Juno et al. 2019).
This study addressed the hypothesis that developing tooth germs may express specific molecules that act on odontogenic cells to induce amelogenesis and dentinogenesis. To test this, molecules from tooth germs at two different developmental stages (cap and hard tissue formation) were compared. Our results revealed that IGF-binding proteins (IGFBPs) were differentially expressed between the two stages in molar germ development. These proteins regulate IGF signaling globally and locally under various conditions. IGFBP binding increases the half-life of IGF and blocks its potential binding to the insulin receptor (Allard and Duan 2018). IGFBPs have high affinity to IGFs that is equal to or greater than that of IGF receptors and play crucial pathophysiological roles in many tissues (Kang et al. 2015; Hoeflich et al. 2018; Mazerbourg and Monget 2018). IGFBPs are expressed in dental pulp stem cells (Magnucki et al. 2013; Al-Kharobi et al. 2018) and are dysregulated in human cleidocranial dysplasia (Greene et al. 2018). They regulate IGF-I-induced matrix mineralization in dental pulp cells (Al-Kharobi et al. 2016) and further enhance differentiation potentials of mesenchymal stem cells (Wang et al. 2017). Considering the functional significance of IGF-I in modulating bioengineered tooth morphogenesis (Oyanagi et al. 2019) and in mineralized tissues (Al-Kharobi et al. 2014), the determination of IGFBP roles in odontogenesis may provide a clue to develop strategies to control tooth morphology in tooth bioengineering.
Materials and methods
Histology and tooth germ sampling
All procedures were performed in accordance with the ethical standards formulated by the animal care and use committee in Chonnam National University. Newborn Sprague–Dawley rat pups were housed in approved facilities for laboratory animal care. Portions of the alveolar bone containing maxillary molar tooth germs were isolated at postnatal days 3, 6, and 9 (n = 5 for each day), and immediately fixed in 4% paraformaldehyde solution. They were then decalcified in ethylenediaminetetraacetic acid (pH 7.4) and routinely processed for embedding in paraffin. Five micrometer-thick sagittal sections were cut for hematoxylin and eosin, and immunofluorescence staining. For RNA and protein extraction, the maxillary 2nd and 3rd molar germs were taken out from their bony crypts and immediately frozen in liquid nitrogen. Extracted molar germs were also immediately used for dental papilla cell culture.
Differential display polymerase chain reaction (DD-PCR) and gene identification
The total RNA was extracted from maxillary 2nd and 3rd molar germs (n = 30 for each germ) using a TRIzol® Reagent (Invitrogen, Carlsbad, CA, USA) and treated with DNase I (Gibco BRL, Gaithersburg, MD, USA). DD-PCR was performed using RNAs extracted from the 2nd and 3rd molar germs at postnatal day 9 using the GeneFishing ™ DEG kit (Seegene, Del Mar, CA, USA) on a Palm-Cycler thermocycler (Corbett Life Science, Sydney, Australia). Products were resolved on 1.2% agarose gel and stained with SYBG® safe DNA gel stain (Invitrogen). Differentially expressed products were ligated with pGEM-T® Easy Vector using T4 DNA ligase (Promega, Madison, WI, USA) and transformed into DH5α cells. Isolated plasmid DNAs were sequenced using a T7 promoter primer.
Induction of differentiation and mineralization
1st and 2nd maxillary molar germ tissues at postnatal day 3 (n = 30 for each germ) were minced into small pieces and digested with 0.1% collagenase (Sigma-Aldrich, St. Louis, MO, USA) for 30 min at 37 °C, then filtered through a cell strainer (100 μm). Cells were incubated with Dulbecco’s Modified Eagle Medium (DMEM) containing 20% fetal bovine serum (FBS) and an antibiotic–antimycotic mixture (Invitrogen). Two to three passages of cells were differentiated into odontoblasts by culturing them in differentiation media, i.e., DMEM containing 10% FBS, supplemented with 170 μM ascorbic acid 2-phosphate, 5 mM β-glycerophosphate (Sigma-Aldrich), and 100 ng/ml BMP2 (R&D Systems, Minneapolis, MN, USA). The medium was changed every three days. For alkaline phosphatase (ALP) staining, cells at day 3 were fixed with 3.7% formaldehyde for 10 min, followed by the addition of the BCIP/NBT substrate (Sigma-Aldrich). The reaction was stopped with the addition of water. The relative staining density was measured using the Scion Image software (Scion, Frederick, MD, USA). For mineralization detection, cells on day 14 were fixed with 70% ethanol, and then stained with 40 mM alizarin red s (AR) solution (pH 4.2, Sigma-Aldrich). For quantitation, the stains were extracted using 10% (w/v) cetylpyridinium chloride in 10 mM sodium phosphate (pH 7.0) for 15 min and measured at 570 nm using a microplate reader (Bio-Tek Instruments, Winooski, VT, USA).
SF2 rat dental epithelial cells were kindly donated from Prof. Fukumoto at the Graduate School of Dentistry Tohoku University and grown in growth media: DMEM/F12 (1:1; Invitrogen) containing 10% FBS and antibiotic–antimycotic mixture. Cells were differentiated into ameloblasts by culturing them in differentiation media: the above growth media supplemented with 170 μM ascorbic acid 2-phosphate, 5 mM CaCl2, and 0.1 μM vitamin D3 (Arakaki et al. 2012). Differentiation media were changed every other day. ALP and AR staining were performed on day 4 and day 12, respectively.
Gene silencing
Cells at the second and third passage were transfected with specific siRNAs (30 nM) against IGBP5–7 (Bioneer, Dajeon, Korea) and scrambled RNA as control (Ambion, Carlsbad, CA, USA) using Lipofectamine RNAiMAX (Invitrogen), respectively. A day after the transfection, SF2 and dental papilla cells were treated with differentiation media for 6 and 9 days, respectively. Gene expression measurements were performed using quantitative real-time PCR.
RT-PCR
Target and reference genes from molar germs (n = 30 for each germ) were amplified in real-time on a Rotor-Gene RG-3000 (Corbett Research, Morklake, Australia) and detected using a SYBR Green PCR Master Mix Reagent kit (Qiagen, Valencia, CA, USA). Data were analyzed using the Corbett Robotics Rotor-Gene software (Rotor-Gene 6 version 6.1, build 90 software). Intensities ratios of the target genes and β-actin signals were used as a relative measurement of the target gene expression. The mean fold change of mRNA expression was calculated using the 2−ΔΔCt method. Base sequences of primers, amplicon size, and GenBank No. are summarized in Table 1.
Western blot
Protein lysates were extracted from molar germs (n = 50 for each germ) using the ReadyPrep protein extraction kit (Bio-RAD, Hercules, CA, USA) and transferred to a Protran nitrocellulose membrane (Whatman GmbH, Dassel, Germany). The membrane was then incubated with purified rabbit polyclonal primary antibodies raised against IGFBP5–7 amino acids of human origin (Santa Cruz Biotech, Delaware, CA, USA). A purified mouse monoclonal primary antibody against β-actin (Sigma-Aldrich) was used as the reference. The membrane was then incubated with horseradish peroxidase (HRP)-conjugated antibodies (Cell Signaling Technology, Beverly, MA, USA). Reactants were visualized with Immobilon Western Chemiluminescent HRP substrate (ECL; Millipore, Billerica, MA, USA).
Immunofluorescence stain
Immunofluorescence staining was performed using the TSATM Kit (Invitrogen). After endogenous peroxidase was blocked, tissue sections were stained with the same primary antibody used for western blot, followed by incubation in HRP-conjugated secondary antibody. Membranes were incubated in Alexa Fluor 488Ⓡ tyramide working solution, followed by propidium iodide stain for nuclei. Reactants were photographed using an LSM confocal microscope (Carl Zeiss, Standort Göttingen-Vertrieb, Deutschland). Immunological specificity was assessed by substituting the primary antibody with normal mouse or rabbit serum (Sigma-Aldrich).
Results
IGFBP5 is differentially expressed in developing tooth germs
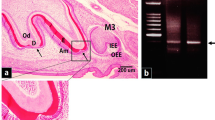
Microscopic observation revealed that the maxillary of 2nd and 3rd molar germs on postnatal day 9 were at the root formation (after the amelogenesis and dentinogenesis) and cap/early bell stages (before the amelogenesis and dentinogenesis) of development, respectively. The 2nd molar germs at the root stage were characterized by the presence of the epithelial diaphragm and had a well-defined crown outline. The crown enamel was covered with protective-stage ameloblasts in the cuspal tip and maturation-stage ameloblasts with wide intercellular space in most parts, which are functionally characterized by the mineralization role of the enamel matrix. In addition, active root dentin formation and the maturation of the crown dentin matrix by odontoblasts were noted (Fig. 1a). The maxillary 2nd molar germs were at the early bell and crown formation stages on postnatal days 3 and 6, respectively (data not shown). DD-PCR revealed that one gene was highly expressed in 2nd molar germs compared with the 3rd molar germs at postnatal day 9 (Fig. 1b). This gene was sequenced as a part (293 bp) of IGFBP5 (GenBank accession no. BC_087030). Other differentially expressed bands, which were detected using other primers, were meaningless or irrespective of histodifferentiation of odontogenic cells and accordingly excluded from this study.
Developing molars and differential expression of IGFBP5 mRNA. a Light micrographs of developing upper molar germs at postnatal day 9. Hematoxylin & Eosin (H&E) stain. (a) The 3rd molar germ (3rd) is at the cap stage in development, whereas the 2nd molar germ (2nd) is at the root formation stage. (b) Hertwig epithelial root sheath (HERS) is seen for the root formation. Ameloblasts (Am) and odontoblasts (Od) are actively engaged in the maturation of the enamel and the formation/maturation of dentin, respectively. (c) High magnification near the future cementoenamel junction is characterized by maturation-stage ameloblasts (Am[m]) with a flat apical region and wide intercellular space. (d) High magnification of the occlusal fissure region is also characterized by Am[m]. (e) High magnification of the cusp tip region is characterized by shortened protective-stage ameloblasts (Am[p]) and the papillary layer (PL) composed of the stratum intermedium and outer enamel epithelium. (f) High magnification of the 3rd molar at the cap stage. Either the enamel or dentin formation is not seen. AB alveolar bone, D dentin, DP dental papilla, E enamel, OEE outer enamel epithelium, SI stratum intermedium. b A gel image acquired from DD-PCR using a specific primer ACP73. A differentially expressed band (arrow), approximately 300 bp in size, between the 2nd molar (root formation stage) and 3rd molar (cap stage) germs is shown. A 100 bp molecular marker (M) ladder was used
The isoforms IGFBP5–7 are upregulated during tooth germ mineralization in vivo
Differential IGFBP5 regulation was confirmed by comparing its expression between the cap stage (2nd molar germs) and the root stage (3rd molar germs). Amplicons of the expected size (211 bp) were generated from both molar germs, but the mRNA level was much higher in the root stage germs. Further examination of IGFBP5 expression levels from the 2nd molar germs on postnatal days 3, 6, and 9 revealed that IGFBP5 was upregulated in a stage-dependent manner. In addition, the mRNA levels of IGFBP6–7 were much higher in the 2nd molar germs than in 3rd molar germs. In contrast, changes in the IGFBP1–4 mRNA levels were negligible. Expression levels of GFBP6–7 from the maxillary 2nd molar germs were also upregulated in a stage-dependent manner. However, mRNA levels of IGFBP1–4 were not significantly changed during the investigated stages (Fig. 2a). Protein levels of IGFBP5–7 in the 2nd molar germs were also examined by western blot. These were upregulated in a stage-dependent manner, consistent with RT-PCR results (Fig. 2b).
Expression of IGFBP isoforms. a mRNA levels determined by real-time PCR (RT-PCR). Generation and comparison of IGFBPs amplicons between the 2nd molar germs (root formation stage) and 3rd molar germs (cap stage) on postnatal day 9, and the generation and comparison of IGFBPs amplicons between 2nd molar germs on postnatal days 3 (early bell stage), 6 (crown stage), and 9 (root formation stage). Data (mean ± SD) were acquired from three independent experiments. A p < 0.05 was considered significant. A 100 bp molecular marker (M) ladder was used. b Protein levels of IGFBP5–7 at three different stages of 2nd molar germs were determined by Western blotting
IGFBPs are differentially localized in ameloblasts and odontoblasts
Differentially expressed IGFBP5–7 were localized in developing molar tooth germs. Immunoreactivity against IGFBP5 was hardly seen in 3rd molar germs at the cap/early bell stages. In contrast, a strong reactivity was sporadically found in fully differentiated odontoblasts and maturation staged-ameloblasts in 2nd molar germs at the root stage, specifically localized in the nuclei of both odontogenic cells (Fig. 3a). Immunoreactivity against IGFBP6 was also hardly seen in 3rd molar germs at the cap/early bell stages (data not shown). However, there was sporadic reactivity in differentiated odontoblasts and maturation-stage ameloblasts in 2nd molar germs at the root stage, specifically localized in the apical cytoplasm of ameloblasts and the apical cytoplasm and dentinal tubules of odontoblasts (Fig. 3b). Immunoreactivity against IGFBP7 was also hardly seen in 3rd molar germs at the cap/early bell stage (Fig. 3c). The reactivity against IGFBP7 seemed to be sporadically localized in the intercellular space of maturation-stage ameloblasts at the occlusal fissure (a) and cervical region (b) of 2nd molar germs at the root stage. The reactivity in odontoblasts was not localized but generalized and seen in the cytoplasm and dentinal tubules of fully differentiated odontoblasts in 2nd molar germs at the root stage (c, d). However, scarce reactivity was found in the inner enamel epithelium and dental papilla of the 3rd molar and protective-stage ameloblasts in the 2nd molar (d). The negative control did not show any reactivity (Fig. 3d).
Localization of IGFBP5–7 in molar germs on postnatal day 9. a Reactivity against IGFBP5 is seen in the 2 molar (2nd) and developing alveolar bone (AB). (a) High magnification. The reactivity is not seen in either dental papilla cells (*) or inner enamel epithelium (#) in 3rd molar germs (3rd) at the cap stage. Strong reactivity is seen in nuclei of fully differentiated odontoblasts (Od), but not in the protective-stage ameloblasts (Am[p]) at the cusp tip. (b) Higher magnification showing reactivity in the nuclei of maturation-stage ameloblasts (Am[m]) but rarely in the dental follicle (DF) and Hertwig epithelial root sheath (HERS). b Immunoreactivity against IGFBP6 is localized in ameloblasts and odontoblasts but not in the dental papilla and dental follicle in 2nd molar germ at the root stage. (a) Strong reactivity is seen in the cytoplasm, in the dentinal tubules (DT) processes of odontoblasts (Od), and (b) the distal cytoplasm of maturation-stage ameloblasts (Am[m]) at the occlusal fissure region. c Immunoreactivity against IGFBP7 in the root stage molar germ. The reactivity is found intercellularly in maturation-stage ameloblasts (Am[m]) at the occlusal fissure region (a) and cervical region (b). (c) The reactivity is also seen in the whole cytoplasm and processes in dentinal tubules of odontoblasts (Od). (d) The reactivity is not found in dental papilla cells (*) or the inner enamel epithelium (#) in the 3rd molar and protective-stage ameloblasts (Am[p]) in the 2nd molar. d Negative control did not show any reactivity. Am ameloblasts, D dentin, DF dental follicle, DP dental papilla, E enamel, Od odontoblasts, OF occlusal fissure
IGFBPs may negatively regulate the enamel and dentin formation
Dental papilla cells acquired from developing molars were primary cultured for analyzing the expression of IGFBP and dentinogenesis marker molecules during mineralization using RT-PCR. As shown in Fig. 4a, marker genes, such as osteocalcin (OCN), osteopontin (OPN), and dentin matrix acidic phosphoprotein 1 (DMP1) were upregulated on day 8 and further enhanced on day 11 by media differentiation. ALP and dentin sialophosphoprotein (DSPP) were significantly upregulated on day 11. The expression levels of IGFBP5–6 was unchanged on day 8 but significantly upregulated on day 11. In contrast, the mRNA levels of IGFBP7 remained unchanged during the investigated time points (Fig. 4b). To elucidate a functional relationship of IGFBPs with dentinogenesis marker molecules, we treated dental papilla cells with siRNA for IGFBP5–7 for 9 days, respectively. Notably, all treatments affected the investigated genes DMP1, DSPP, and OCN, which are late stage odontoblast differentiation markers that regulate matrix mineralization (Papagerakis et al. 2002) and were significantly upregulated (Fig. 4c). This result was further confirmed by ALP and AR staining. Differentiation media with control siRNA induced dentin matrix mineralization, and the induced mineralization was further augmented by the silencing of IGFBP5–7 with the specific siRNA, respectively (Fig. 4d).
Functional regulation of dentinogenesis by IGFBPs in dental papilla cells. mRNA levels were measured by real-time PCR (RT-PCR). a Cells were treated with differentiation media for 8 and 11 days. Each gene marker expression was compared with its expression on day 0. b Gene expression levels of IGFBPs were compared with their respective expression level on day 0. c Cells were transfected with IGFBPs or control siRNAs. The mRNA levels of each gene were compared. d Differentiation media (DM) treatments with control siRNA were compared with those with specific siRNA for each IGFBP by alkaline phosphatase (ALP) and alizarin red s (AR) stain. GM: growth media only. All data were acquired from three independent experiments. A p < 0.05 was considered significant
SF2 rat dental epithelial cells were differentiated into ameloblasts for analyzing the expression of IGFBPs and amelogenesis marker molecules during mineralization. As shown in Fig. 5a, amelogenin (Amelx), a representative gene for amelogenesis was upregulated in a time-dependent manner and OPN was highly upregulated in all investigated times using the treatment with differentiation media. ALP was upregulated at a later stage, day 6, but OCN expression was unchanged. Changes in IGFBP5 expression during amelogenesis were negligible, but expression levels of IGFBP6–7 were upregulated on day 5 and further enhanced on day 7 (Fig. 5b). To elucidate a functional relationship of IGFBPs with amelogenesis marker expression, we treated SF2 with siRNA for IGFBP5–7 for 6 days, respectively. Notably, all treatments upregulated Amelx, ameloblastin (Ambn), OPN, and ALP mRNA levels except OCN (Fig. 5c). This result was further confirmed by ALP and AR staining. Treatments using differentiation media with control siRNA induced enamel matrix mineralization and this induced mineralization was further augmented by the silencing of IGFBP5–7 with the specific siRNA, respectively (Fig. 5d).
Functional regulation of amelogenesis by IGFBPs in SF2 epithelial cells. mRNA levels were measured by real-time PCR (RT-PCR). a Cells were treated with differentiation media for 3, 4, and 6 days. Each gene marker expression was compared with its expression on day 0. b Gene expression levels of IGFBPs were compared with their respective expression level on day 0. c Cells were treated with IGFBPs or control siRNAs. The mRNA levels of each gene were compared. d Differentiation media (DM) treatments with control siRNA were compared with those with specific siRNA for each IGFBP by alkaline phosphatase (ALP) and alizarin red s (AR) stain. GM: growth media only. All data were acquired from three independent experiments. A p < 0.05 was considered significant
Discussion
The present study revealed the involvement and significance of IGFBP5–7 in late stage odontogenesis after initiation of the crown enamel and dentin mineralization, i.e., negative regulation of late stage amelogenesis and dentinogenesis. IGFs were shown to have a working mechanism in coordinating mineralized matrix formation in osteoblasts and dental pulpal cells (Al-Kharobi et al. 2014, 2016). Furthermore, IGF-I increases the size and cusp number of bioengineered teeth via the induction of enamel knot formation (Oyanagi et al. 2019). The strong binding affinity of IGFBPs to IGFs and the key regulation of IGFs actions by IGFBPs in various tissues (Hoeflich et al. 2018) imply that IGFBPs may provide a way to control tooth morphology in IGF-aided tooth bioengineering and regeneration.
The IGF family is a system composed of IGFs, IGF receptors, IGFBPs, and IGFBP proteases. IGF-I and IGF-II stimulate growth, differentiation, and survival of various cells, including ameloblasts and odontoblasts. IGFs were related to the expression of amelogenin, ameloblastin, and enamelin at the secretory stage (Catón et al. 2005). However, IGFs alone are limited to explain molecular mechanism which works for odontogenic cell differentiation during amelogenesis and dentinogenesis (Tompkins 2006; Lee et al. 2018; Xiong et al. 2019). IGFs are known to be modulated by a family of high-affinity IGFBPs, but few studies have addressed how IGFs are functionally modulated in amelogenesis and odontogenesis. The present study revealed that IGFBP5–7 were upregulated, whereas changes in levels of IGFBP1–4 were negligible during enamel and dentin mineralization. This result contrasts a report that IGFBP2 stimulates differentiation in osteoblasts (Xi et al. 2014). In particular, expression levels of IGFBP5–7 increased in a time-dependent manner from the cap through the root stage, implying that these molecules may have functional significance in the maturation of the enamel matrix via the regulation of ameloblasts differentiation or by maintaining their differentiation state, since ameloblasts at this stage have completed the formation of the enamel matrix. This suggestion is evidenced by the expression of IGFBP5–7 in maturation-stage ameloblasts (either ruffled ended or smooth ended), which are functionally characterized by their involvement in enamel matrix mineralization and morphologically characterized by their wide intercellular space and long columnar shape. In contrast, they were hardly expressed in either inner enamel epithelium and dental papilla cells or protective-stage ameloblasts, which are characterized by short cuboidal shape and no further involvement in matrix mineralization.
IGFBP5 possesses a nuclear localization sequence in its carboxy-terminal domain and interacts with LIM protein-2, which facilitates its transport into the nucleus where the complex then modulates the transcription of genes involved in osteoblast proliferation and/or differentiation (Schedlich et al. 2000; Amaar et al. 2002). The present study also demonstrated that IGFBP5 was localized in the nuclei of differentiated odontoblasts and ameloblasts, providing a possibility that IGFBP5 may act by the IGF-independent mechanism in these cells. This suggestion is also supported by the report that locally injected IGFBP5 had an effect on osteoblasts in IGF-I knockout mice (Miyakoshi et al. 2001).
Regarding the molecular function of IGFBP5, there have been more suggestions for their role than the simple sequestering of IGF. IGFBP5 was expressed in the differentiated spinous cells of the gingival epithelium and was able to enhance their migration, differentiation, and anti-apoptosis processes (Hung et al. 2008). Furthermore, a "loss of function” approach to the knockdown of endogenous IGFBP5 expression in osteosarcoma cells revealed that endogenous IGFBP5 was important in maintaining bone cell survival and differentiation but had little effect on cell proliferation (Yin et al. 2004). About 25% of ameloblasts in rats die during postsecretory transition, whereas another 25% do so during proper maturation (Smith and Warshawsky 1977; Smith 1979). Our results revealed the localization of IGFBP5 in ameloblasts and odontoblasts at the root formation stage and its absence in either proliferating cells, preodontoblasts, or preameloblasts, suggest that this factor may locally regulate the differentiation of matured odontogenic cells or maintain the differentiation state rather than proliferation. This suggestion is supported by reports showing that IGFBP5 expression is associated with non-proliferative states, such as cell differentiation and quiescence (Bach 2005), and its mRNA expression is enhanced in serum withdrawal-induced cell death (Mazerbourg and Monget 2018).
The expression levels of IGFBP6 were higher at the hard tissue forming stage than the cap stage and increased in a time-dependent manner. IGFBP6 was localized in the distal region in the cytoplasm of ameloblasts and odontoblasts, and it was involved in growth arrest with features of senescence and was associated with non-proliferative states, such as cell differentiation and quiescence (Han et al. 1999; Han and Carter 2000). In addition, IGFBP6 overexpression inhibited Caco-2 cell growth and mediated the growth-inhibitory effect of all-trans retinoic acid by suppressing the IGF-II autocrine loop (Kim et al. 2002). The present IGFBP6 localization suggests its secretion into matrices to regulate IGF action as an autocrine or paracrine factor.
IGFBP7, also known as IGFBP-related protein 1, can negatively modulate the stimulatory effect of VEGF on angiogenesis by interfering with its expression and signaling (Tamura et al. 2009; Chen et al. 2010). Recombinant IGFBP7 was found to inhibit the proliferation and enhance differentiation of human keratinocytes (Nousbeck et al. 2010). IGFBP7 has been shown to mediate senescence and apoptosis in melanocytes, suppress melanoma growth in vivo (Wajapeyee et al. 2008), and suppress cell survival in cholangiocarcinoma (Yue et al. 2018). The RT-PCR and western blot data revealed that IGFBP7 expression also corresponds to the hard tissue formation stage. IGFBP7 was localized in the whole cytoplasm of fully differentiated odontoblasts at the secretory stage, along with their processes and the intercellular space of maturation-stage ameloblasts. This molecule was not found either in the cap stage or protective-stage ameloblasts, which ended the enamel matrix mineralization.
In attempting to elucidate a potential functional relationship between IGFBP5–7 and odontoblastic differentiation of dental papilla cells, our results showed that odontogenic marker genes were upregulated from day 8 and further enhanced on day 11 as shown in Fig. 4. Consistent with these results, the expression levels of IGFBP5–6 were unchanged on day 8 but significantly upregulated on day 11. This suggested that IGFBPs may have been involved in odontoblast differentiation at later stages of mineralization. In contrast, mRNA levels of IGFBP7 remained unchanged, which might be due to high basal level of IGFBP7 expression (data not shown). The high basal level of IGFBP7 was also reflected from its unlocalized but whole area of immunoreactivity in odontoblasts as shown in Fig. 3c. Secondly, when we treated dental papilla cells with siRNA for IGFBPs for days 4 and 10, all treatments significantly upregulated marker genes, except for OPN. DMP1 is critical for proper dentin mineralization and is exported to the extracellular matrix where it orchestrates mineralized matrix formation. OCN is a non-collagenous, vitamin K-dependent protein that contains three gamma-carboxyglutamic acid (Gla) motif. OCN is a later stage bone formation and differentiation marker of osteoblasts (Papagerakis et al. 2002), regulates mineralization in the bone matrix (Zoch et al. 2016), and facilitates mineral deposition in the presence of calcium and bone remodeling (Razzaque 2011; Burr et al. 2015; Shan et al. 2019). Thirdly, the involvement of IGFBPs in mineralization was also confirmed by ALP and AR staining. Mineralization induced by the differentiation media was further augmented by IGFBP silencing. Altogether, these results suggest that IGFBP5–7 may play a key role in the regulation of in dentin matrix mineralization by downregulating genes, such as OC and DMP1, during later stage odontoblasts differentiation either directly or indirectly through IGF binding.
In evaluating the relationship between IGFBP5–7 and ameloblastic differentiation of SF2 rat dental epithelial cells, IGFBP5–7 expression levels were upregulated as shown in Fig. 5. During the induced mineralization, Amelx (a representative gene for amelogenesis), OPN (a highly expressed gene in ameloblast-like cells) (Hyun et al. 2019), and ALP levels were upregulated. In contrast, IGFBP5 was unexpectedly unchanged. This may due to different culture conditions or culture times. Regarding the functional regulation of amelogenesis by IGFBPs, all IGFBP siRNA treatments upregulated genes, such as Amelx and ALP, and augmented mineralization, which was further confirmed by ALP and AR staining. Altogether, these results suggest that IGFBP5–7 may play a role in the regulation of mineralization in enamel matrix by downregulating the expression of representative genes, such as Amelx and ALP, during later stage ameloblasts differentiation either directly or indirectly through IGF binding.
This study is the first to provide the differential expression of IGFBPs and their functional relationships in ameloblasts and odontoblasts in developing tooth germs. Further studies are needed to elucidate their precise mechanism and function, including their redundancy in tooth development using differentiation-inducing agents, such as retinoic acid (Esteban-Pretel et al. 2010) or knockout animal models. This study may provide practical knowledge when considering IGFBPs as mineralization-regulating agents in tooth bioengineering and regeneration.
References
Al-Kharobi H, El-Gendy R, Devine DA, Beattie J (2014) The role of the insulin-like growth factor (IGF) axis in osteogenic and odontogenic differentiation. Cell Mol Life Sci 71(8):1469–1476
Al-Kharobi H, Alhodhodi A, Hawsawi Y, Alkafaji H, Devine D, El-Gendy R, Beattie J (2016) IGFBP-2 and -3 co-ordinately regulate IGF1 induced matrix mineralisation of differentiating human dental pulp cells. Stem Cell Res 17(3):517–522
Al-kharobi HE, Al-Khafaji H, Beattie J, Devine DA, El-Gendy R (2018) Insulin-like growth factor axis expression in dental pulp cells derived from carious teeth. Front Bioeng Biotechnol. https://doi.org/10.3389/fbioe.2018.00036
Allard JB, Duan C (2018) IGF-binding proteins: why do they exist and why are there so many? Front Endocrinol. https://doi.org/10.3389/fendo.2018.00117
Amaar YG, Thompson GR, Linkhart TA, Chen ST, Baylink DJ, Mohan S (2002) Insulin-like growth actor-binding protein 5 (IGFBP-5) interacts with a four and a half LIM protein 2 (FHL2). J Biol Chem 277:12053–12060
Arakaki M, Ishikawa M, Nakamura T, Iwamoto T, Yamada A, Fukumoto E, Saito M, Otsu K, Harada H, Yamada Y, Fukumoto S (2012) Role of epithelial-stem cell interactions during dental cell differentiation. J Biol Chem 287(13):10590–10601
Bach LA (2005) IGFBP-6 five years on; not so “forgotten”? Growth Horm IGF Res 15:185–192
Burr DB, Bellido T, White KE (2015) 6—bone structure and function. In: Hochberg MC, Silman AJ, Smolen JS, Weinblatt ME, Weisman MH, (eds) Rheumatology, 6th edn, content repository only! Philadelphia, pp. 42–55. https://doi.org/10.1016/B978-0-323-09138-1.00006-1
Catón J, Bringas P Jr, Zeichner-David M (2005) IGFs increase enamel formation by inducing expression of enamel mineralizing specific genes. Arch Oral Biol 50(2):123–129
Chen RY, Chen HX, Lixu PJ, Li J, Fan YM, Tu YT (2010) Intratumoral injection of pEGFC1-IGFBP7 inhibits malignant melanoma growth in C57BL/6J mice by inducing apoptosis and down-regulating VEGF expression. Oncol Rep 23:981–988
Chen J, Almo SC, Wu Y (2017) General principles of binding between cell surface receptors and multi-specific ligands: a computational study. PLoS Comput Biol 13(10):e1005805. https://doi.org/10.1371/journal.pcbi.1005805
Doshi R, Kulkarni U, Shinde S, Sabane A, Patil A (2016) Role of genes in odontogenesis. Br J Med Med Res 14(6):1–9
Esteban-Pretel G, Marín MP, Renau-Piqueras J, Barber T, Timoneda J (2010) Vitamin A deficiency alters rat lung alveolar basement membrane: reversibility by retinoic acid. J Nutr Biochem 21(3):227–236
Greene SL, Mamaeva O, Crossman DK, Lu C, MacDougall M (2018) Gene-expression analysis identifies IGFBP2 dysregulation in dental pulp cells from human cleidocranial dysplasia. Front Genet. https://doi.org/10.3389/fgene.2018.00178
Han VK, Carter AM, Chandarana S, Tanswell B, Thompson K (1999) Ontogeny of expression of insulin-like growth factor (IGF) and IGF binding protein mRNAs in the guinea-pig placenta and uterus. Placenta 20(4):361–377
Han VK, Carter AM (2000) Spatial and temporal patterns of expression of messenger RNA for insulin-like growth factors and their binding proteins in the placenta of man and laboratory animals. Placenta 21:289–305
Hoeflich A, David R, Hjortebjerg R (2018) Current IGFBP-related biomarker research in cardiovascular disease—We need more structural and functional information in clinical studies. Front Endocrinol. https://doi.org/10.3389/fendo
Hung PS, Kao SY, Liu CJ, Tu HF, Wu CH, Lin SC (2008) Insulin-like growth factor binding protein-5 enhances the migration and differentiation of gingival epithelial cells. J Periodontal Res 43(6):673–680
Hyun SY, Mun S, Kang KJ, Lim JC, Kim SY, Han K, Jang YJ (2019) Amelogenic transcriptome profiling in ameloblast-like cells derived from adult gingival epithelial cells. Sci Rep. https://doi.org/10.1038/s41598-019-40091-x
Juno JA, Wragg KM, Kristensen AB, Lee WS, Selva KJ, van der Sluis RM, Kelleher AD, Bavinton BR, Grulich AE, Lewin SR, Kent SJ, Parsons MS (2019) Modulation of the CCR5 receptor/ligand axis by seminal plasma and the utility of in vitro versus in vivo models. J Virol 15:93. https://doi.org/10.1128/JVI.00242-19
Kang HS, Kim MY, Kim SJ, Lee JH, Kim YD, Seo YK, Bae JH, Oh GT, Song DK, Ah YH, Im SS (2015) Regulation of IGFBP-2 expression during fasting. Biochem J 467:453–460
Kim EJ, Kang YH, Schaffer BS, Bach LA, MacDonald RG, Park JH (2002) Inhibition of Caco-2 cell proliferation by all-trans retinoic acid: role of insulin-like growth factor binding protein-6. Cell Physiol 190:92–100
Klopcic B, Maass T, Meyer E, Lehr HA, Metzger D, Chambon P, Mann A, Blessing M (2007) TGF-beta superfamily signaling is essential for tooth and hair morphogenesis and differentiation. Eur J Cell Biol 86(11–12):781–799
Lan Y, Jia S, Jiang R (2014) Molecular patterning of the mammalian dentition. Semin Cell Dev Biol 25–26:61–70. https://doi.org/10.1016/j.semcdb.2013.12.003
Lee DS, Roh SY, Park JC (2018) The Nfic-osterix pathway regulates ameloblast differentiation and enamel formation. Cell Tissue Res 374(3):531–540
Magnucki G, Schenk U, Ahrens S, Santos AN, Gernhardt CR, Schaller HG, Hoang-Vu C (2013) Original expression of the IGF-1, IGFBP-3 and IGF-1 receptors in dental pulp stem cells and impacted third molars. J Oral Sci 55(4):319–327
Mazerbourg S, Monget P (2018) Insulin-like growth factor binding proteins and IGFBP proteases: a dynamic system regulating the ovarian folliculogenesis. Front Endocrinol (Lausanne). https://doi.org/10.3389/fendo.2018.00134
Miyakoshi N, Richman C, Linkhart TA, Baylink DJ, Mohan S (2001) Evidence that insulin-like growth factor binding protein-5 functions as a growth factor. J Clin Invest 107:73–81
Neuhaus SJ, Byers MR (2007) Endothelin receptors and endothelin-1 in developing rat teeth. Arch Oral Biol 52:655–662
Nousbeck J, Sarig O, Avidan N, Indelman M, Bergman R, Ramon M, Enk CD, Sprecher E (2010) Insulin-like growth factor-binding protein 7 regulates keratinocyte proliferation, differentiation and apoptosis. J Invest Dermatol 130(2):378–387
Ono M, Oshima M, Ogawa M, Sonoyama W, Hara ES, Oida Y, Shinkawa S, Nakajima R, Mine A, Hayano S, Fukumoto S, Kasugai S, Yamaguchi A, Tsuji T, Kuboki T (2017) Practical whole-tooth restoration utilizing autologous bioengineered tooth germ transplantation in a postnatal canine model. Sci Rep. https://doi.org/10.1038/srep44522
Oyanagi T, Takeshita N, Hara M, Ikeda E, Chida T, Seki D, Yoshida M, Seiryu M, Takano I, Kimura S, Oshima M, Tsuji T, Takano-Yamamoto T (2019) Insulin-like growth factor 1 modulates bioengineered tooth morphogenesis. Sci Rep. https://doi.org/10.1038/s41598-018-36863-6
Papagerakis P, Berdal A, Mesbah M, Peuchmaur M, Malaval L, Nydegger J, Simmer J, Macdougall M (2002) Investigation of osteocalcin, osteonectin, and dentin sialophosphoprotein in developing human teeth. Bone 30(2):377–385
Razzaque MS (2011) Osteocalcin: a pivotal mediator or an innocent bystander in energy metabolism? Nephrol Dial Transplant. https://doi.org/10.1093/ndt/gfq721
Sadier A, Twarogowska M, Steklikova K, Hayden L, Lambert A, Schneider P, Laudet V, Hovorakova M, Calvez V, Pantalacci S (2019) Modeling Edar expression reveals the hidden dynamics of tooth signaling center patterning. PLoS Biol 17(2):e3000064. https://doi.org/10.1371/journal.pbio.3000064
Schedlich LJ, Le Page SL, Firth SM, Briggs LJ, Jans DA, Baxter RC (2000) Nuclear import of insulin-like growth factor-binding protein-3 and -5 is mediated by the importin beta subunit. J Biol Chem 275(31):23462–23470
Shan C, Ghosh A, Guo XZ, Wang SM, Hou YF, Li ST, Liu JM (2019) Roles for osteocalcin in brain signalling: implications in cognition- and motor-related disorders. Mol Brain 12:23. https://doi.org/10.1186/s13041-019-0444-5
Smith CE (1979) Ameloblasts: secretory and resorptive functions. J Dent Res 25:695–707
Smith CE, Warshawsky H (1977) Quantitative analysis of cell turnover in the enamel organ of the rat incisor. Evidence for ameloblast death immediately after enamel matrix secretion. Anat Rec 187:63–98
Smith EE, Angstadt S, Monteiro N, Zhang W, Khademhosseini A, Yelick PC (2018) Bioengineered tooth buds exhibit features of natural tooth buds. Practical whole-tooth restoration utilizing autologous bioengineered tooth germ transplantation in a postnatal canine model. J Dent Res 97(10):1144–1151
Tamura K, Hashimoto K, Suzuki K, Yoshie M, Kutsukake M, Sakurai T (2009) Insulin-like growth factor binding protein-7 (IGFBP7) blocks vascular endothelial cell growth factor (VEGF)-induced angiogenesis in human vascular endothelial cells. Eur J Pharmacol 610(1–3):61–67
Thesleff I (2006) The genetic basis of tooth development and dental defects. Am J Med Gene Part A 140A:2530–2535
Tompkins K (2006) Molecular mechanism of cytodifferentiation in mammalian tooth development. Connect Tissue Res 47:111–118
Wajapeyee N, Serra RW, Zhu X, Mahalingam M, Green MR (2008) Oncogenic BRAF induces senescence and apoptosis through pathways mediated by the secreted protein GFBP7. Cell 132(3):363–374
Wang Y, Liu Y, Fan Z, Liu D, Wang F, Zhou Y (2017) IGFBP2 enhances adipogenic differentiation potentials of mesenchymal stem cells from Wharton’s jelly of the umbilical cord via JNK and Akt signaling pathways. PLoS ONE 12(8):e0184182. https://doi.org/10.1371/journal.pone.0184182
Wang F, Li G, Wu Z, Fan Z, Yang M, Wu T, Wang J, Zhang C, Wang S (2019) Tracking diphyodont development in miniature pigs in vitro and in vivo. Biol Open 8(2):236. https://doi.org/10.1242/bio.037036
Xi G, Wai C, DeMambro V, Rosen CJ, Clemmons DR (2014) IGFBP-2 directly stimulates osteoblast differentiation. J Bone Miner Res 29(11):2427–2438
Xiong Y, Fang Y, Qian Y, Liu Y, Yang X, Huang H, Huang H, Li Y, Zhang X, Zhang Z, Dong M, Qiu M, Zhu XJ, Zhang Z (2019) Wnt production in dental epithelium is crucial for tooth differentiation. J Dent Res 98(5):580–588
Yin P, Xu Q, Duan C (2004) Paradoxical actions of endogenous and exogenous insulin-like growth factor-binding protein-5 revealed by RNA interference analysis. J Biol Chem 279:32660–32666
Yue C, Yang M, Tian Q, Mo F, Peng J, Ma Y, Huang Y, Wang D, Wang Y, Hu Z (2018) IGFBP7 is associated to prognosis and could suppress cell survival in cholangiocarcinoma. Artif Cells Nanomed Biotechnol 46:817–825
Zoch ML, Clemens TL, Riddle RC (2016) New insights into the biology of osteocalcin. Bone 82:42–49
Funding
This work was supported by the National Research Foundation of Korea (NRF) Grant funded by the Korea government (MSIT) (No. 2019R1A5A2027521) and (No. 2019R1A2C1003520). The authors report no conflicts of interest related to this study.
Author information
Authors and Affiliations
Contributions
JSM and YSN contributed to conception, data acquisition, analysis and interpretation; JHK, SYL, and DYK drafted and revised the manuscript; DWY, HMK, and MSK contributed to conception and design; SHK contributed to design and data interpretation and critically revised the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Moon, JS., Nam, YS., Kang, JH. et al. Regulatory role of insulin-like growth factor-binding proteins in odontogenic mineralization in rats. J Mol Histol 52, 63–75 (2021). https://doi.org/10.1007/s10735-020-09923-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10735-020-09923-3