Abstract
Grapevine buds exhibit endodormancy (ED), triggered by short days (SD) and terminated through prolonged exposure to low temperature (LT). This study aims to unravel the genetic underpinnings the initiation and cessation of ED. Through previously reported SD-related gene expression changes and comparison with RNA-seq analysis of grapevine buds subjected to LT, we elucidated distinct gene clusters with contrasting regulation patterns. One cluster termed “Thermoregulatory Clock Cluster” encompassed 46 differential expressed genes (DEGs) upregulated by SD but downregulated under LT condition. Functional analysis revealed enrichment in circadian clock and temperature responsiveness functions. In contrast, the “Growth Control Cluster” comprising 292 DEGs downregulated under SD but upregulated under LT, is enriched in functions associated with cell division and transcription regulation. This investigation underscores the pivotal roles of genes associated with circadian clock, temperature responsiveness, cell division and transcriptional regulation in orchestrating the entry and exit of grapevine buds from ED. Furthermore, we propose a mechanistic model wherein LT-induced repression of circadian clock genes provokes the upregulation of SD-suppressed genes that promote ED. Thus, the role of LT in releasing buds from ED can be understood at the molecular level. This study not only advances our understanding of the genetic basis of bud ED, but also holds implications for viticulture and broader insights into plant dormancy regulation.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Dormancy is a state of temporarily halted growth in meristem containing structures such as seeds and buds (Rohde and Bhalerao 2007; Singh et al. 2017). In buds, it is classified into three types: para-, endo- and ecodormancy based on the factors responsible for the arrest of growth (Lang et al. 1987; Yamame et al. 2021). Paradormancy (PD) is caused by external factors affecting the buds, such as the phenomenon of apical dominance. Endodormancy (ED), results from internal inhibition factors within the bud, while ecodormancy (ECD) is brought on by environmental factors, primarily low temperature (LT). Grapevine (Vitis vinifera L.) buds enter winter recess or ED at the end of summer to reduce winter injury. Grapevine drop their shoot tips and develop ED in the compound axillary buds that have grown continuously during the growing season (Mathiason et al. 2009; Fennell et al. 2015). The transition to ED is a critical event in bud tissue, marked by decreased respiration (Parada et al. 2016), cessation of cell division (Vergara et al. 2017), increased cell wall thickness of bud meristematic cells (Dantas et al. 2020), and increased levels of sucrose and the sucrose–glucose ratio (Noriega et al. 2022). Grapevine’s compound buds are photoperiod sensitive and the transition from PD to ED is triggered by decreasing photoperiod (Kühn et al. 2009; Grant et al. 2013; Cragin et al. 2017). Therefore, genes that are regulated by the SD photoperiod could play a key role in the induction of ED. To break ED, buds must be exposed to LT (2–9 °C) for an extended period, and this phenomenon is known as chilling requirement (CR) (Campoy et al. 2011; Singh et al. 2017). Exposure to LT shifts grapevine buds from ED to ECD, and ecodormant buds can break when exposed to favorable growth conditions (Mathiason et al. 2009; Kovaleski et al. 2018). Therefore, genes that are regulated by LT could play a key role in the release of buds from ED. Lack of winter cold in subtropical regions limits grapevine cultivation due to prolonged ED, leading to uneven budbreak (Erez 1987). The dormancy of perennial plants is mainly regulated by photoperiod and temperature (Petterle et al. 2013; Ding and Nilsson. 2016). Despite the key role of these two environmental cues in regulating bud ED in grapevines, the underlying mechanisms and genes that control this process remain largely unknown. Studies using microarray and RNA-seq technologies have characterized the expression of differential expressed genes (DEGs) during various grape bud dormancy stages (Díaz-Riquelme et al. 2012; Khalil-Ur-Rehman et al. 2017). The impact of LT (Mathiason et al. 2009) and SD (Fennell et al. 2015; Smita et al. 2021) on grape bud gene expression has also been investigated. Bud ED is a complex physiological process that is essential for the survival, growth and development of deciduous perennial plants. The timely release of ED is important for flowering and fruit production of deciduous fruit trees. A better understanding of the mechanism of induction and release of bud from ED will help to cope with climate change (Yang et al. 2021).
Considering the divergent impacts of SD and LT conditions on bud ED in grapevines, we hypothesize that genes displaying contrasting responses to SD and LT within grapevine buds are pivotal in coordinating the onset and cessation of bud ED. This research aims to identify these genes, and ascertain their functionality aligns with the entry into and exit from ED of grapevine buds.
Materials and methods
Plant material and experimental conditions
For the SD experiment, 6-year-old Vitis riparia Manitoba 37 Michx grapevines were grown for 30 days post budbreak reaching a shoot length of 10–15 nodes. Three replicates of ten-vine experimental unit were randomly assigned to each photoperiod treatment of continued (LD, 15 h; paradormancy) or short photoperiod (SD, 13 h; endodormancy) for 28 days, as previously described (Smita et al. 2021). For LT experiment, 10-year-old Thompson Seedless grapevines from the Chilean Institute of Agriculture Research (INIA) were used. Canes were collected at ED stage (Rubio et al. 2016), excised into single-bud cuttings, and subjected to a 1-week treatment of either LT (4 °C in the dark) or control (14 °C in the dark).
Preparation of RNA-seq libraries
Total RNA was extracted from three biological replicates of LD and SD V. riparia Manitoba 37 Michx and three biological replicates of LT and control Thompson Seedless buds using a modified Chang et al. (1993) method. RNA-seq libraries for LD and SD transcriptomes were prepared and sequenced as described in Smita et al. (2021). For LT experiment, DNA was removed by RNase-free DNase treatment (Thermo Scientific, USA) and RNA quality was checked through agarose gel electrophoresis. The libraries were sequenced using Illumina HiSeq (PE 2×150) with a 200 bp insert following the manufacturer’s protocol.
Mapping sequence reads to the reference genome
For the SD experiment, full raw sequencing data were submitted to the GEO database with the accession number GSE95429 and mapped reads in each library were assembled into transcripts using cufflinks (V.2:2.1) (Smita et al. 2021). For the LT experiment, sequence reads were processed with Trimmomatic v0.36 to remove low-quality nucleotides and adapter sequences. They were then aligned to the V. vinifera reference genome GCF_000003745.3 on NCBI database using STAR aligner v2.5.2b, a splice aligner that includes splice junctions. The resulting BAM files were used to calculate unique gene hit counts using feature Counts from the Subread package v1.5.2, considering only reads that lay within exon regions (Table S1).
DEGs and cluster analysis
After gene hit count extraction, a DEG analysis was performed using the gene hit count table and DESeq2. The Wald test was used to determine p-values and log2(fold changes) for comparisons of gene expression between SD-treated and LD-treated, and LT-treated and control grapevine buds. DEGs were identified as genes with an adjusted p-value < 0.05 and absolute log2(fold change) > 1. Cluster analysis of DEGs was performed using the heatmap R package, with the “complete” method and five main clusters were extracted using the cutree function and height 6, as described by Wang et al. (2010).
GO analysis of DEGs
GO enrichment analysis was conducted using g:Profiler (Raudvere et al. 2019) using the hypergeometric distribution adjusted for multiple hypothesis correction by set count sizes (SCSs). SCS threshold was applied to eliminate false-positive GO terms and prioritizes significant results. Each gene was assigned a GO term if it crossed the adjusted p-value threshold (SCS) ≤ 0.05. Difference between treatments were analysed by ANOVA, and multiple comparison analysis was carried out using Dunnett’s test.
Real-time quantitative PCR (RT-qPCR) analysis of DEGs
For the SD experiment, DEGs from a microarray study with similar photoperiod treatments (Fennell et al. 2015) was utilized to calculate correlation with current RNA-seq data (Smita et al. 2021). For the LT experiment, six genes were validated by RT-qPCR using an Eco Real-Time PCR system (Illumina, San Diego, USA) and KAPA SYBR FAST (KK 4602) qPCR Master Mix (2×). The cDNA was amplified using the following conditions: 2 min denaturation at 94 °C, followed by 40 cycles of 94 °C for 30 s, 55 °C for 30 s, and 72 °C for 45 s. Gene expression was quantified using the 2−ΔΔCT method (Livak and Schmittgen 2001). Each reaction was conducted with three biological replicates, each with three technical replicates. VvUBIQUITIN and VvACTIN were used as reference genes for normalization.
Results
DEGs in grapevine buds under exposure to short-day and low temperature conditions
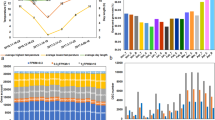
A group of 1336 DEGs were detected in grapevine buds subjected to SD condition in contrast to LD condition, wherein 400 DEGs were upregulated and 936 were downregulated. While a group of 6121 DEGs was identified in grapevine buds exposed to LT in comparison to control buds. Among these, 4098 DEGs were upregulated while 2093 were downregulated (Fig. 1a). To identify shared DEGs between the LT and SD experiments, a Venn diagram was generated using the link: http://bioinfogp.cnb.csic.es/tools/venny/index.html. The outcomes of this analysis revealed a prominent group of overlapping DEGs (292) between LT and SD. Remarkably, this group exhibited a consistent pattern of being downregulated under SD condition and upregulated under LT condition and was named “Growth Control Cluster” (Fig. 1b). Another subset of overlapping DEGs (46) displayed an opposite regulatory pattern, being upregulated under SD condition but downregulated under LT condition and was named “Thermoregulatory Clock Cluster”. Both set of DEGs, exhibiting contrasting regulation response to LT and SD signals, are likely pivotal in the intricate processes governing the entry and exit of grapevine buds from the ED state. In contrast, the shared sets of DEGs that display either up- or downregulation response to both signals are unrelated to ED and constitute the smallest groups of overlapping DEGs (Fig. 1b). Employing log2(ratio) values of transcript expression, cluster analysis was performed on the subset of DEGs responding to LT and SD signals in grapevine buds. The resulting heatmap (Fig. 1c) illustrated that most of the SD DEGs were downregulated, while most of LT DEGs were upregulated compared to the controls. This analysis revealed the presence of five distinct clusters characterized by distinct expression pattern. Cluster 1 (Thermoregulatory Clock Cluster) encompassed genes upregulated by SD and downregulated by LT (Supplemental Table S1). Cluster 2 contained genes that exhibited upregulation in response to both SD and LT (Supplemental Table S2). Cluster 3 and 4 (Growth Control Cluster) comprised genes that were downregulated by SD and upregulated by LT, respectively (Supplemental Tables S3 and S4) and Cluster 5 comprised genes downregulated by both SD and LT (Supplemental Table S5). Thermoregulatory Clock Cluster and Growth Control Cluster exhibited DEGs with contrasting regulation by LT and SD implying their involvement in the processes of governing the entry and exit of buds from the ED state.
Analysis of differentially expressed genes (DEGs) in grapevine buds subjected to low temperature (LT) vs. control, and short days (SD) vs. long days (LD). a The upregulated and downregulated DEGs were examined in both cases, b Venn diagram showing the unique and overlapping gene expression pattern observed in grapevine buds in response to different expression patterns in grapevine buds in response to LT and SD treatments. c Cluster analysis of differentially expressed genes (DEGs) based on log2(ratio) in grapevine buds. Cluster1 (Thermoregulatory Clock Cluster) and Cluster 3, 4 (Growth Control Cluster) show DEGs contrastingly regulated by SD and LT conditions
Enrichment analysis of DEGs of thermoregulatory clock cluster and growth control cluster
To explore the potential biological significance of DEGs contrastingly regulated in grapevine buds by SD and LT conditions, we performed GO enrichment analysis of cluster 1 and cluster 3, 4 DEGs. Cluster 1 (Thermoregulatory Clock Cluster) encompasses a set of genes that exhibited upregulation in response to SD conditions and downregulation when subjected to LT conditions. This intriguing behavior implies their potential significance in orchestrating the intricate process of entry and exit of grapevine buds from the ED state. Notably, only 46 DEGs meet both criteria. Upon subjecting this selected group of DEGS to Gene Ontology (GO) analysis, the molecular function (MF) and cellular component (CC) categories did not exhibit any significant enrichment. However, an intriguing outcome emerged in terms of biological process (BP) categories. Specifically, seven BP categories exhibited substantial enrichment, all centered around rhythmic processes (GO:0048511), circadian rhythm (GO:0007623), and responsive reactions to temperature stimuli (G:0009266) (Fig. 2). These results prompt us to name Cluster 1 as “Thermoregulatory Clock Cluster” to underscore the pivotal role of circadian rhythm function in the onset and cessation of ED. Clusters 3 and 4 (Growth Control Cluster) encompasses a discrete set of genes with a distinct behavior, downregulation in response to SD condition and upregulation when exposed to LT condition. This intriguing pattern also implicated their potential contribution to the intricate control of grapevine bud entry into and exit from the ED state. A more extensive set of 292 DEGs fulfill both discerning criteria. Upon Gene Ontology (GO) analysis, these DEGs revealed enrichment across molecular function (MF), biological process (BP), and cellular component (CC) categories. Notably, the (GO:0008017) microtubule binding category displayed the highest level of enrichment within MF, the (GO:0000278) cellular mitotic cycle category had the highest enrichment within BP, and the (GO:0000793) condensed chromosome category had the highest enrichment within CC (Fig. 3). These compelling findings indicate that the regulation of cell division and transcriptional regulation plays a pivotal role steering grapevine bud entry into and exit from the ED state. Therefore, we named cluster 3 and 4 as “Growth Control Cluster”.
Gene Ontology (GO) analysis of Growth Control Cluster (Cluster 3, 4). Differentially expressed genes (DEGs) in this cluster are downregulated by short day (SD) but upregulated by low temperature (LT). GO categories corresponding to a molecular function, b biological process, and c cellular component were enriched in this cluster
Validation of RNA-seq by RT-qPCR
Six DEGs were chosen for RT-qPCR analysis to test the reproducibility of the RNA-seq analysis results. In each case, the results from the RT-qPCR and the RNA-seq assays exhibited the same trends (Fig. 4).
Validation of RNA-seq by RT-qPCR. Expression patterns of DEGs a 2C protein phosphatase 2 LOC100263754 (VvPP2C2); b dehydration-responsive element-binding protein LOC100253072 (VvCBF6); c dehydration-responsive element-binding protein LOC100248181 (VvCBF3); d gibberellin 2-beta-dioxigenase1 LOC100245010 (VvGA2ox1); e superoxide dismutase, chloroplastic LOC100258067 (VvSOD2); f CBF4 transcription factor LOC100245286 (VvCBF4)
Discussion
Unveiling the regulation of grapevine bud endodormancy
The progression of grapevine buds through endodormancy (ED), from initiation to cessation, involves a finely tuned interplay between environmental cues, particularly photoperiod and temperature, along with intricate physiological and molecular processes. SD photoperiod initiates ED in grapevine buds (Kühn et al. 2009; Grant et al. 2013; Cragin et al. 2017), resulting in an overall reduction in metabolic and cellular activities (Parada et al. 2016; Vergara et al. 2017; Dantas et al. 2020). Conversely, low temperature (LT), besides inducing cold acclimation, acts as a catalyst, prompting the release of buds from ED and their transition from endo- to ecodormancy (ECD) (Kovaleski et al. 2018; Carbonneau et al. 2015; Pérez and Rubio. 2022). However, the molecular mechanisms and genes involved in the SD induction of bud ED and their release by LT remain unknown. In this study, we conducted a comparative analysis of the DEGs under SD and LT conditions, leading to the identification of five distinct clusters exhibiting different expression patterns. Notably, Clusters 2 and 5 exhibited synchronous responses to both SD and LT stimuli, in contrast to the divergent responses observed in Clusters 1, 3, 4. Upon closer examination of the DEGs within the clusters, we designated Cluster 1 as the “Thermoregulatory Clock Cluster”, while Clusters 3 and 4 were designated as the “Growth Control Cluster”.
Thermoregulatory clock cluster
Our comprehensive transcriptomic analysis reveals a small cluster of DEGs named the Thermoregulatory Clock Cluster. The DEGs of this cluster show an intriguing expression pattern, being upregulated under SD conditions while exhibiting downregulation in response to LT conditions. These small subset of genes holds promise in deciphering the initiation and cessation of ED. Gene Ontology (GO) analysis of the 46 DEGs within this cluster unveils their enrichment in biological process (BP) associated with circadian rhythmic and cold response (Fig. 2). Of particular interest are key genes of the circadian clock network such as ADAGIO PROTEIN 1, PRR5, COR27 as well as of temperature-responsive pathway genes such as, HSFC1 STH2 (Supplemental Table S1). COR27 and COR28 are known for their role in regulating period length of the circadian clock and for associating with chromatin regions surrounding clock genes such as PSEUDO-RESPONSE REGULATOR (PRR5) and TIMING OF CAB2 EXPRESSION (TOC1) to modulate transcription (Li et al. 2016). Recent studies have identified COR27 and COR28 as novel regulators of the COP1-HY5 regulatory hub and photomorphogenesis, affecting the expression of numerous genes in Arabidopsis (Li et al. 2020). The established correlation between circadian regulation and cell division across diverse organisms has been extensively documented (Farshadi et al. 2020). Nonetheless, the mechanisms underlying the upregulation of circadian clock genes during SD conditions, leading to the suppression of cell division genes and the induction of ED remain elusive. Previous reports have suggested that LT can disrupt the functioning of the circadian clock, affecting both its components and output genes under normal light–dark conditions (Bieniawska et al. 2008). This phenomenon have been observed in chestnut trees, where cold temperatures disrupt the behavior of circadian clock genes, resulting in sustained expression of core clock genes such as LHY, CCA1, TOC1 and PSEUDO-RESPONSE REGULATOR (PRR), which are essential components of the circadian oscillator network (Ibañez et al. 2008). The potential disruption of circadian oscillation by LT could potentially fine-tune the expression levels of circadian-regulated genes, which in turn may impact overall plant fitness (Bieniawska et al. 2008). Our previous study also demonstrated a cessation in the oscillation of phytochrome A and B gene expression during the day in grapevine buds, with static expression emerging around mid-April, coinciding with the onset of declining temperature (Kühn et al. 2009). Considering the circadian clock profound influence on numerous genes, alongside its heightened activity under SD conditions and diminished activity during LT exposure, it is plausible that both environmental cues, SD and LT regulate ED by modulating the expression of circadian clock genes.
HEAT SHOCK TRANSCRIPTION FACTOR C1 (HSFC1) is associated with dormancy in plants (Nishitani et al. 2012; Tarancón et al. 2017). The upregulation of cold response genes by SD could potentially amplify the sensitivity of bud to LT. This set the stage for the subsequent influence of LT on activating cell cycle genes, thereby promoting the release of buds from the ED (Fig. 5).
Growth control cluster
This cluster, notably larger, encompasses 292 DEGs that are downregulated by SD and upregulated by LT. Predominantly, the DEGs within this cluster show downregulation under SD conditions, aligning with the reduced cellular and metabolic activities characteristic of the ED state. This trend underscores their likely involvement in these biological functions. Particularly intriguing is their contrasting response under LT exposure, wherein their expression is upregulated. This highlight the critical role of LT in upregulating genes typically downregulated by the SD photoperiod to facilitate the transition of buds from ED to ECD. A Gene Ontology (GO) analysis of the 292 DEGs from the Growth Control Cluster reveals enrichment in molecular function (MF) categories linked to microtubule binding, tubulin binding, biological process (BP) concerning the mitotic cell cycle and cellular components (CC) categories tied to condensed chromosome and microtubule cytoskeleton (Fig. 3). Overall, the primary enriched functions of the Growth Control Cluster are linked to cell division and transcription regulatory activity, firmly underscoring the central role these genes play in governing ED. While dormancy and quiescence naturally involve cell cycle arrest (Velappan et al. 2017), the activation of cell cycle by LT is surprising, given that LT is conventionally associated with growth arrest. Our results shed light on the molecular mechanism underlying the well-established phenomenon of LT triggering the release of grapevine buds from ED (Londo and Johnson 2014; Cragin et al. 2017; Pérez and Rubio 2022). By counteracting the SD-mediated repression of gene expression in the Growth Control Cluster, LT triggers the transition of grapevine buds from ED to ECD through a circadian clock mechanism. The scheme in Fig. 5 visually portrays how the regulatory networks governed by SD and LT act upon distinct gene clusters to either induce the initiation of ED or facilitate the subsequent release from this dormancy state in grapevine buds. This dynamic interplay underscores the intricate molecular mechanisms underlying the bud’s responsiveness to changing environmental cues.
Conclusions
Two distinct clusters of differentially expressed genes (DEGs) in grapevine buds, coined as the “Thermoregulatory Clock Cluster” and the “Growth Control Cluster”, have been discerned, each responding differently to SD and LT stimuli. The SD photoperiod activates the Thermoregulatory Clock Cluster, upregulating the expression of circadian clock genes and cold-responsive genes. This activation suppresses the Growth Control Cluster and promotes the development of ED through the inhibition of cell cycle and transcription regulatory genes. Conversely, LT stimuli suppress the Thermoregulatory Clock Cluster, causing a downregulation of circadian and cold responsive genes. This suppression, in turn, activates the Growth Control Cluster, resulting in the upregulation of cell cycle genes and transcription regulatory genes. This upregulation promotes the release of buds from ED. These findings highlight the pivotal roles of circadian clock and cell cycle genes in the regulation of ED in grapevine buds. Additionally, they provide a molecular explanation for the involvement of LT in the release of buds from ED.
References
Bieniawska Z, Espinoza C, Schlereth A, Sulspice R, Hincha DK, Hannah M (2008) Disruption of the Arabidopsis circadian clock responsible for the extensive variation in the cold-responsive transcriptome. Plant Physiol 147:263–279
Campoy JA, Ruiz D, Egea J (2011) Dormancy in temperate fruit trees in a global warming context: a review. Sci Hortic 130:357–372
Carbonneau A, Deloire A, Torregrosa L, Jaillard B, Pellegrino A, Métay A, Ojeda H, Lebon E, Abbal P (2015) Traité de la Vigne, Physiologie, Terroir, Culture, 2nd edn. Editions Dunod, p 573
Chang S, Puryear J, Cairney J (1993) A simple and efficient method for isolating RNA from pine trees. Plant Mol Biol Rep 11:113–116
Cragin J, Serpe M, Keller M, Shellie K (2017) Dormancy and cold hardiness transition in wine grape cultivars Chardonnay and Cabernet Sauvignon. Am J Enol Viti 68:195–202
Dantas D, Bressan-Smith R, Noriega X, Pérez FJ (2020) Italia melhorada grapevines grown under tropical conditions develop a quiescent state. Environ Exp Bot 171:103951
Díaz-Riquelme J, Grimplet J, Martínez-Zapater JM, Carmona MJ (2012) Transcriptome variation along bud development in grapevine (Vitis vinifera L.). BMC Plant Biol 12:181
Ding J, Nilsson O (2016) Molecular regulation of phenology in trees—because the seasons they are a-changing. Curr Opin Plant Biol 29:73–79
Erez A (1987) Chemical control of budbreak. HortScience 22:1240–1243
Farshadi E, van der Horst GTJ, Chaves I (2020) Molecular links between the circadian clock and the cell cycle. J Mol Biol 432:3515–3524
Fennell A, Schlauch KA, Gouthu S, Khadka S, Sreekantan L, Grimplet J, Cramer, Mathiason KL (2015) Short day transriptomic programming during induction of dormancy in grapevine. Front Plant Sci 6:834
Grant TNL, Gargrave J, Dami IE (2013) Morphological physiological, and biochemical changes in Vitis genotype in responses to photo-period regimes. Am J Enol Viti 64:466–475
Ibañez C, Ramos A, Acebo P, Contreras A, Casado R, Allona I, Argoncillo C (2008) Overall alteration of circadian clock gene expression in the Chesnut cold response. PLoS ONE 3:e3567
Khalil-Ur-Rehman M, Sung L, Li CX, Faheem M, Wang W, Tao JM (2017) Comparative RNA-seq based transcriptome analysis of bud dormancy in grape. BMC Plant Biol 17:18
Kovaleski AP, Reisch BI, Londo JP (2018) Deacclimation kinetics as a quantitative phenotype for delineating the dormancy transition and thermal efficiency for budbreak in Vitis species. AoB Plants 10:1–12
Kühn N, Ormeño-Nuñez J, Jaque-Zamora G, Pérez FJ (2009) Photoperiod modifies the diurnal expression profile of VvPHYA and VvPHYB transcripts in field grown grapevine leaves. J Plant Physiol 166:1172–1180
Lang GA, Early JD, Martin GC, Darnell RL (1987) Endo-, para-, and ecodormancy: physiological terminology and classification for dormancy research. HortScience 22:371–377
Li X, Ma D, Lu SX, Hu X, Huang R, Liang T, Xu T, Tobin EM, Liu H (2016) Blue-light and low temperature-regulated COR27 and COR28 play roles in the circadian clock. Plant Cell 28:2755–2769
Li X, Zhiwei Z, Ma D, Zhang J, Yang Y, Liu Y, Liu H (2020) COR27 and COR28 are novel regulators of the COP1-HY5 hub and photomorphogenesis in Arabidopsis. Plant Cell 32:3139–3154
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real time quantitative PCR and the 2-ΔΔCT methods. Methods 25:402–408
Londo JP, Johnson LM (2014) Variation in the chilling requirement and budburst rate of wild Vitis species. Environ Exp Bot 106:138–147
Mathiason K, He D, Grimplet J, Venkateswari D, Galbraith DW, Or E (2009) Transcript profiling in Vitis riparia during chilling requirement fulfillment reveals coordination of gene expression pattern with optimized budbreak. Funct Integr Genomics 9:81–96
Nishitani C, Siro T, Ubi BE, Shimazu T, Itai A, Saito T, Yamamoto T, Moriguchi T (2012) Transcriptome analysis of Pyrus pyrifolia leaf during transition from endodormancy to ecodormancy. Sci Hort 147:49–55
Noriega X, Rubio S, Pérez FJ (2022) Sucrose accumulation and endodormancy are synchronized events induced by short-day photoperiod in grapevine buds. Plant Physiol Biochem 190:101–108
Parada F, Noriega X, Dantas D, Bressam-Smith, Pérez FJ (2016) Differences in respiration between dormant and non-dormant buds suggest the involvement of ABA in the development of endodormancy in grapevines. J Plant Physiol 201:71–78
Pérez FJ, Rubio S (2022) Relationship between bud cold hardiness and budbreak in two Vitis vinifera cultivars, Chardonnay, and Thompson seedless. J Plant Growth Regul 41:840–847
Petterle A, Karlberg A, Bhalerao RP (2013) Daylength mediated control of seasonal growth patterns in perennials trees. Curr Opin Plant Biol 16:301–306
Raudvere U, Kolsberg L, Kuzmin I, Arak T, Adler P, Peterson H, Vilo J (2019) g:Profiler: a web server for functional enrichment analysis and conversions of gene list. Nucleic Acids Res. https://doi.org/10.1093/nar/gkw199
Rohde A, Bhalerao RP (2007) Plant dormancy in the perennial context. Trends Plant Sci 12:217
Rubio S, Dantas D, Bressan-Smith R, Pérez FJ (2016) Relationship between endodormancy and cold hardiness in grapevine buds. J Plant Growth Regul 35:266–275
Singh RK, Svystun T, AlDahmash B, Jonsson AM, Bhalerao RP (2017) Photoperiod and temperature-mediated control of phenology in trees—a molecular perspective. New Phytol 213:511–524
Smita S, Robben M, Deuja A, Accerbi M, Green PJ, Subramanian S, Fennell A (2021) Integrative analysis of gene expression and miRNA reveal biological pathways associated with bud paradormancy and endodormancy in grapevine. Plants 10:669
Tarancón C, Gonzáles-Grandío E, Oliveros JC, Nicolas M, Cubas P (2017) A conserved carbon starvation response underlies bud dormancy in woody and herbaceous species. Front Plant Sci 8:788
Velappan Y, Signorelli S, Considine MJ (2017) Cell cycle arrest in plants: what distinguishes quiescence, dormancy and differentiated G1? Ann Bot 120:495–509
Vergara R, Noriega X, Aravena KS, Prieto H, Pérez FJ (2017) ABA represses the expression of cell cycle genes and may modulate the development of endodormancy in grapevine buds. Front Plant Sci 8:812
Wang L, Feng Z, Wang X, Zhang X (2010) DEGseq: an Rpackage for identifying differentially expressed genes from RNA-seq data. Bioinformatics 261(1):136–138
Yamame H, Singh AK, Cooke JEK (2021) Plant dormancy research: from environmental control to molecular regulatory networks. Tree Physiol 41:523–528
Yang Q, Gao Y, Wu X, Moriguchi T, Bai S, Teng Y (2021) Bud endodormancy in deciduous fruit trees: advances and prospects. Hortic Res 8:139
Acknowledgements
The financial support of Fondo Nacional de Ciencia y Tecnología (FONDECYT), is gratefully acknowledged.
Author information
Authors and Affiliations
Contributions
XN perform RT-qPCR experiments. SR perform analysis of RNA-seq results and FJP plan the experiments and wrote the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
There is no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Rubio, S., Noriega, X. & Pérez, F.J. Genes contrastingly regulated by short days and low temperature are key players in the onset and cessation of endodormancy in grapevine buds. Plant Growth Regul (2024). https://doi.org/10.1007/s10725-024-01156-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10725-024-01156-8