Abstract
MicroRNAs (miRNAs) are small non-coding RNAs that play essential functions in abiotic stress responses. High temperature is a serious stress that affects crop yield and quality; however, the regulatory mechanisms of heat-responsive miRNAs remain poorly understood in maize (Zea mays L.). Here, 340 miRNAs (215 known and 125 novel) were identified from maize seedlings under heat stress and control conditions using high-throughput sequencing in elite inbred line CM1. The 215 known miRNAs were divided into 40 families, and 21-nt miRNAs were the most abundant. In total, 35 miRNAs (26 known and 9 novel) exhibited significantly different expression levels between the heat stress and control samples. Furthermore, 174 targets were predicted to be cleaved by 115 miRNAs using degradome sequencing. The biological roles and pathways of these targets were investigated using Gene Ontology and Kyoto Encyclopedia of Genes and Genomes analyses. On the basis of relationships among miRNAs and target genes, as well as the enrichment results, a regulatory network was constructed, and 16 significantly differentially expressed miRNAs were involved in the network. Our study shows a comprehensive analysis of heat-responsive miRNAs and their targets in the maize inbred line CM1. The results are helpful for the functional research of the heat stress responsive miRNAs, and provide novel insights into the miRNA-mediated regulatory network and the underlying mechanisms in maize during heat stress response.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Maize (Zea mays L.), an important cereal crops, has become a model plant in functional genomics and genetic research (Schnable et al. 2009). However, its yield and quality are frequently affected by various abiotic stresses, such as drought and high temperature. During evolution, plants have developed complex and elaborate regulatory mechanisms to cope with various stresses at the morphological, physiological and molecular levels (Tuteja 2007; Yamaguchi-Shinozaki and Shinozaki 2006). Multiple genes may be regulated under stress conditions (Shinozaki and Yamaguchi-Shinozaki 2007), for example, many transcription factors (TFs) play significant roles in stress responses by regulating the expression of a series of downstream genes (Casaretto et al. 2016; Chen and Zhu 2004; Hoang et al. 2017; Lee et al. 2016; Nakashima et al. 2007; Sun et al. 2013). However, the regulatory mechanisms and networks involved in stress responses need to be further explored in plants.
MicroRNAs (miRNAs) are a group of approximately 18–24 nucleotide (nt) non-coding endogenous small RNAs (sRNAs) that regulate the expression of target genes at the post-transcriptional level (Bartel 2004; Covarrubias and Reyes 2010). In plants, primary miRNAs are initially generated through transcription by RNA polymerase II and then further processed to generate miRNA/miRNA* duplexes after being processed in the cell nucleus (Bartel 2004; Krutzfeldt et al. 2006; Kurihara and Watanabe 2004). Finally, the duplexes are transported into the cytoplasm, and mature miRNAs are loaded onto Argonaute protein to form an RNA-induced silencing complex that guides target mRNA regulation (Bartel 2004; Baumberger and Baulcombe 2005; Mallory et al. 2008). The first known miRNA, lin4, was discovered in Caenorhabditis elegans (Lee et al. 1993), and improved sequencing technology has led to the identification of many more miRNAs in eukaryotes. Furthermore, degradome sequencing technology has been extensively used to identify the target mRNAs of miRNAs, and the results indicated that single miRNA can regulate one or multiple targets. Thus, predicting target genes can contribute to the understanding of the biological functions of miRNAs (Li et al. 2017; Wang et al. 2019).
miRNAs have important roles in diverse plant developmental stages and stress responses. For example, Arabidopsis miR169 targets NF-YA2 and NF-YA10 genes to regulate primary root growth (Sorin et al. 2014). In tomato, overexpressing miR167 down-regulates the expression levels of auxin response factors (ARFs) 6 and 8, which are involved in flower organs development (Liu et al. 2014a, b). In particular, numerous miRNAs that respond to abiotic stresses have been identified in plants using sRNA sequencing. For example, multiple drought-responsive miRNAs have been identified based on their expression patterns in different rice cultivars (Balyan et al. 2017). In tomato, Cao et al. (2014) reported hundreds of chilling-responsive miRNAs, and 49 miRNAs are significantly differentially expressed (Cao et al. 2014). In wheat, 12 miRNAs that respond to heat stress have been reported (Xin et al. 2010). Importantly, functional analyses of some stress-responsive miRNAs have also been performed in plants (Denver and Ullah 2019; Dong and Pei 2014; Fang et al. 2014; Wang et al. 2012; Yang et al. 2013; Yue et al. 2017; Zhang et al. 2011).
Recently, high temperature has become a serious stress that negatively influences maize yield and quality in Huang-Huai-Hai area of China. Our previous study indicated that the hybrid An’nong 591 exhibits a high tolerance to high temperature in this area, and the heat tolerance was mainly contributed by its paternal line CM1 based on their phenotypic characters and RNA-seq analysis (Zhao et al. 2019). Although the gene expression profiles of protein-coding genes were investigated in CM1, heat-responsive miRNAs and their targets, especially the miRNA-medicated regulatory network remain poorly understood. In this study, expression patterns of miRNAs were investigated under control and heat stress conditions using sRNA sequencing, and the target genes were predicted using degradome sequencing. Importantly, a miRNA-medicated regulatory network of maize heat response was constructed. The results provide valuable information in the exploration of heat-response mechanisms in maize inbred line CM1, which will laid an important foundation for creating heat-tolerant germplasm through molecular breeding.
Materials and methods
Plant materials, heat treatments and RNA isolation
The elite maize inbred line CM1 was obtained from the Maize Engineering Technology Research Center of Anhui Province, Anhui Agricultural University. CM1 is a paternal line of hybrid An’nong 591, which shows strong tolerance to heat stress in production (Zhao et al. 2019). Seedlings of the inbred line CM1 were grown in a plant growth chamber under the culture conditions of 28 °C/23 °C (day/night temperature) and 16-h light/8-h dark photoperiod. To identify heat-responsive miRNAs, seedlings were subjected to a 42 °C treatment for 24 h at the three-leaf stage, and the third leaves were collected for RNA isolation. Total RNA was extracted using a total RNA purification kit (LC Sciences, USA) according to the manufacturer’s protocols. The quantity and purity of the total RNAs were checked using an Agilent Bioanalyzer 2100 (Agilent, USA) and RNA 6000 Nano LabChip Kit (Agilent). Three independent biological replicates were taken for both the heat stress (MH) and control (MC) groups.
Library construction, sequencing and bioinformatics analysis
Six sRNA libraries were constructed using a TruSeq Small RNA Sample Prep kit (Illumina, USA) in accordance with the manufacturer’s protocol. Briefly, 1 µg of total RNA per sample was used for library construction and ligated to 3′ adaptors and 5′ adaptors (Illumina). Subsequently, the first-strand cDNA was synthesized and used for PCR amplification. After the PCR products were gel purified, the sRNA library was sequenced on an Illumina HiSeq 2500 (Illumina) platform, and 1 × 50 bp single-end reads were generated. Raw reads were first processed using ACGT101-miR (v4.2) software (LC Sciences) to remove low-quality reads, adapter sequences, junk reads and reads having lengths < 18 nt and > 25 nt. The remaining reads were aligned to mRNA, RFam and Repbase databases, and then, common RNA families (rRNA, tRNA, snRNA, and snoRNA), repeats and sequences matching maize mRNAs were filtered. The remaining valid reads were mapped to maize precursors in miRBase 22.0 (Griffiths-Jones et al. 2008) to identify known miRNAs and novel 3p- and 5p-derived miRNA candidates. Finally, the unmapped sequences were aligned to the maize genome (AGPv4) to identify potential novel miRNAs, and RNAfold software was used to predict miRNA secondary structures (http://rna.tbi.univie.ac.at/cgi-bin/RNAWebSuite/RNAfold.cgi).
Identification of heat-responsive differentially expressed miRNAs (DEMs)
To identify the DEMs between control and heat stress conditions, the expression levels of miRNAs were normalized as described in a previous study (Li et al. 2016). Differentially expressed sequences were analyzed using a t-test. DEMs were identified using following criteria: |log2(Fold change)|> 1 and p-value ≤ 0.05. Normalized miRNA counts were calculated using log2 (norm + 1) to construct a heatmap.
Quantitative real-time PCR (qRT-PCR) analysis of miRNAs
The qRT-PCR analysis was conducted to validate the abundance changes in detected miRNAs. Total RNA was reverse transcribed with a Mir-X™ miRNA First-Strand Synthesis kit (Clontech, USA) using the poly (A)-tailing method. All the primers used for qRT-PCR are listed in Table S1. qRT-PCR reactions were performed using methods published in our previous study (Zhao et al. 2019). The 5S rRNA was used as internal control for the expression analysis (Nie et al. 2016), and the 2−ΔΔCT method was used to analyze the relative change in each miRNA’s expression level (Livak and Schmittgen 2001).
Identification of miRNA targets using degradome sequencing
Two degradome libraries (MC and MH) were constructed from the mixed total RNAs of the three biological samples, which had been used for sRNA library construction. The degradome library was constructed as described previously (Ma et al. 2010) with some modifications, and library sequencing was conducted on an Illumina HiSeq 2500 (Illumina) platform. After sequencing, raw data were filtered to remove adaptor sequences and low-quality reads. CleaveLand v3.0 (Addo-Quaye et al. 2009) and TargetFinder (http://targetfinder.org/) were used to detect potentially sliced targets of miRNAs. And t-plots were generated to analyze the degradation patterns based on the signature distributions and abundance levels along the transcripts. The targets were divided into five pattern-based categories (0–4) as described in previous study (Cao et al. 2014).
Gene ontology (GO) and Kyoto encyclopedia of genes and genomes (KEGG) enrichment analyses and regulatory network construction
GO and KEGG analyses were conducted to determine the biological roles of miRNA targets using the GOSeq R package (v1.34.1) (Young et al. 2010) and KEGG database (http://www.genome.ad.jp/kegg/) (Kanehisa et al. 2008), respectively. To explore the regulatory mechanisms of miRNAs in response to heat stress, relationships of miRNAs, targets and enriched GO terms and KEGG pathways were used to construct a network using Cytoscape (v3.7.2) software as described previously (Xu et al. 2018).
Results
sRNA sequencing and data analysis
In our previously study (Zhao et al. 2019), we reported that the maize inbred line CM1 exhibits a high tolerance to heat stress. To identify heat-responsive miRNAs, six sRNA libraries were constructed (MH1, MH2, MH3, MC1, MC2 and MC3) and sequenced on an Illumina HiSeq 2500 platform. In total, averages of 17,568,632 and 21,192,599 raw reads from the three biological replicates were obtained from MC and MH libraries, respectively (Table 1). Finally, 5,522,709–19,120,848 valid reads of 18–25 nt were obtained from each sample, and they were subsequently used to identify known and novel miRNAs. The length distributions of the sRNAs were similar between the MC and MH libraries (Fig. 1A). The majority of the sRNAs were 21–22 nt in length. Furthermore, the 24-nt sRNAs accounted for a large percentage. The percentage of 18–21 nt sRNAs in the MH library was greater than in the MC library, while the opposite was found for 22–25 nt sRNAs.
Identification of known and novel miRNAs in maize CM1
In total, 215 known miRNAs were identified from the MC and MH libraries (Table S2). The known miRNAs were 18–25 nt in length, with the 21-nt miRNAs being the most numerous, accounting for 55.81%, followed by the 22-nt miRNAs accounting for 18.60% (Fig. 1B). These known miRNAs were divided into 40 families, with a large range of members in each miRNA family. The miR166 and miR169 families had the most members (22 each), followed by the miR156 and miR171 families, which had 21 and 20 members, respectively. Among the 40 miRNA families, 14 families contained only one member, such as miR11969, miR395 and miR6199 (Fig. 2). After the identification of known miRNAs, the remaining sequences were aligned to the maize genome to screen for potential novel miRNAs. In total, 125 sequences were identified as the novel miRNAs. By comparison of the size distribution, significant difference was found between the known and novel miRNAs. The length distributions of the novel miRNAs ranged from 18 to 24 nt, with 24-nt being the most abundant length representing 64.00%, followed by 21 nt representing 20.80%. In addition, only one member of 18-nt length was identified among the novel miRNAs (Table S3).
Identification of heat-responsive miRNAs and qRT-PCR validation
Expression patterns of the identified miRNAs were analyzed using the normalized method reported previously (Li et al. 2016). A Pearson’s correlation analysis was used to analyze the correlations among the biological replicates of MC and MH samples. As shown in Fig. 3A, the correlation coefficients (R values) of the three biological replicates were greater than 0.913 and 0.945 for the MC and MH libraries, respectively, suggesting the high reproducibility of the high-throughput sequencing. A total of 35 miRNAs (26 known and 9 novel) were significantly differentially expressed between MC and MH samples. Compared with the expression in the MC library, 21 miRNAs were up-regulated and 14 were down-regulated (Fig. 3B, C. The 35 DEMs belonged to different miRNA families, and some DEMs showed large fold change, such as zma-miR160f-5p_R-1, mtr-miR171c and zma-miR168b-3p_R + 1 (Table S4). Additionally, miRNAs in the same family showed similar expression patterns. For example, the expression levels of the five miR164 family members were up-regulated, while those of the three miR168 family members were down-regulated. To validate the DEM expression patterns, 10 DEMs were selected for a qRT-PCR analysis. Our results indicated that most of the 10 selected miRNAs shared similar expression trends compared to those indicated by the sequencing data (Fig. 4), suggesting the reliability of the latter.
Identification of miRNA target genes
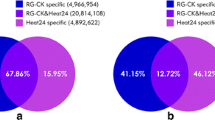
Degradome sequencing was used to predict the target genes of detected miRNAs. The CleaveLand and TargetFinder packages were used to identify sliced targets of miRNAs in the MC and MH libraries. In total, 21,136,209 and 11,630,262 raw reads were generated from the MC and MH libraries, respectively. Additionally, 9,395,171 (44.45%) and 4,861,092 (41.80%) reads were mapped to the maize transcript in the MC and MH libraries, respectively (Table S5). On the basis of the relative abundance of the signatures at the cleavage sites, cleavage products were divided into five categories (0–4) (Fig. 5). In the MC library, 1,563 cleavage events were identified, with 683 (43.70%), 72 (4.61%), 437 (27.96%), 29 (1.86%) and 342 (21.88%) being classified into categories 0–4, respectively. In the MH library, 1,386 cleavage events were identified, with 596 (43.00%), 97 (7.00%), 459 (33.12%), 9 (0.65%) and 225 (16.23%) being classified into categories 0–4, respectively (Fig. 6; Table S6).
Among these cleavage events, 174 (527 transcripts) targets were predicted to be cleaved by 115 miRNAs (104 known and 11 novel). Many of the targets encoded different TF families, such as auxin response factor (ARF), squamosa promoter binding protein-like (SPL) transcription factor, and NAM, ATAF and CUC (NAC) domain-containing protein (Table S6). We noted that the same miRNA family tend to share the same type of target genes. For example, the targets of the zma-miR160 family encode ARF TFs, and the targets of the zma-miR171 family encode GRAS TFs. These findings may suggest a conserved regulatory mechanism of miRNAs. Notably, many miRNAs have multiple target genes. For example, there are 15 genes predicted to be the targets of zma-miR156k-5p and 8 targets were predicted for zma-miR166a-3p. Of the 35 DEMs, degradome sequencing showed that only 17 members (16 known and 1 novel miRNAs) had target genes (Table S4).
GO and KEGG enrichment analyses of miRNA targets
GO and KEGG analyses were performed for the 174 targets. GO enrichment analysis indicated that the targets were significantly enriched in 31 terms (FDR < 0.05), and the 30 most significant GO terms are shown in Fig. 7. In the molecular function category, the targets were mainly enriched in the GO terms “DNA binding” (GO:0003677), “sequence-specific DNA binding transcription factor activity” (GO:0003700), and “protein dimerization activity” (GO:0046983). In the cellular component category, the targets were mainly enriched in “nucleus” (GO:0005634) and “nucleolus” (GO:0005730). In the biological process category, the targets were mainly enriched in “regulation of transcription, DNA-templated” (GO:0006355) and “auxin-activated signaling pathway” (GO:0009734). Besides, some genes were enriched in “response to hormone” (GO:0009725), “heat acclimation” (GO:0010286), and “response to hydrogen peroxide” (GO:0042542), suggesting that these genes may have potential roles in heat stress responses.
The KEGG analysis indicated that 42 pathways were enriched for the 174 target genes, and 30 pathways were shown in Fig. 8. Among these 30 pathways, four pathways, including “ubiquinone and other terpenoid-quinone biosynthesis” (ko00130), “selenocompound metabolism” (ko00450), “monobactam biosynthesis” (ko00261), and “sulfur metabolism” (ko00920), were significantly enriched (q-value < 0.05). Among the enriched pathways, the “metabolic pathways” (ko01100) contained 18 genes, while “biosynthesis of secondary metabolites” (ko01110) and “ribosome” (ko03010) pathways contained 10 and 7 genes, respectively. Additionally, four and three genes were enriched in “plant hormone signal transduction” (ko04075) and “spliceosome” (ko03040) pathways, respectively, and one gene was enriched in the “MAPK signaling pathway-plant” (ko04016) pathway.
Regulatory network of heat-responsive miRNAs in maize
A single miRNA may affect several different targets, and one target may also be cleaved by one or more miRNAs. On the basis of the relationships among miRNAs and targets, as well as enriched GO terms and pathways, a miRNA-mediated regulatory network of heat response was constructed in maize CM1. As shown in Fig. 9, 11 up-regulated DEMs (aof-miR160c_R-1_1ss20CT, aof-miR166b_L-2, mtr-miR171c, osa-miR164e_R + 1_2ss20AC21GA, zma-miR160a-5p, zma-miR160f-5p_R-1, zma-miR164a-5p, zma-miR164e-5p, zma-miR164f-5p_R-1, zma-miR164h-5p_R-2_1ss19GT, and zma-miR171l-3p_L-3_1ss21CT) and 5 down-regulated DEMs (PC-3p-89210_61, tae-MIR9774-p5_2ss16CT18AC, zma-miR166c-5p, zma-miR169i-3p_R + 1_1, and zma-miR169i-3p_R + 1_2) were involved in the network. The targets of the miRNAs were mainly assigned in several GO terms, such as “regulation of transcription, DNA-templated” (GO:0006355), “nucleus”(GO:0005634), and “DNA binding”(GO:0003677), and the most highly enriched pathways were “metabolic pathways” (ko01100), “biosynthesis of secondary metabolites” (ko01110), and “ribosome” (ko03010). The relationships among the miRNAs, targets, and enriched GO terms and pathways provides an important foundation for exploring the functional roles and regulatory mechanisms of heat-responsive miRNAs in maize CM1.
Regulatory network of heat-responsive miRNAs in maize inbred line CM1. Red and blue triangles represent the significantly up- and down-regulated miRNAs, respectively, while yellow represents no significant change. Black dots represent the targets of miRNAs. GO terms and KEGG pathways are represented by gray ellipses, and the enriched terms and pathways for the target genes of DEMs are represented by blue ellipses
Discussion
Recently, high temperature has become a frequently occurring environmental stress that affects maize growth and development. For example, high temperature occurred during the flowering period lead to serious yield reductions in the production. Thus, improving adaptability to high temperature has become a new focus of crop molecular breeding. Plants have developed elaborate regulatory mechanisms to cope with heat stress (Ravichandran et al. 2019; Zandalinas et al. 2018). Increasing evidences indicated that miRNAs have essential functions in plant growth, development and stress responses (Balyan et al. 2017; Cao et al. 2014; Ding et al. 2009; Liu et al. 2014a, b; Sorin et al. 2014; Sunkar et al. 2007). Previously, we showed that maize inbred line CM1 has a high tolerance to heat stress and the transcriptomic analysis was also performed to investigate the expression differences under heat stress condition compared with the maternal line and F1 plants (Zhao et al. 2019). However, the heat-responsive miRNAs and their regulatory mechanisms remain poorly understood in CM1.
In total, 340 miRNAs (215 known and 125 novel) were identified in the MC and MH libraries. The known miRNAs were divided into 40 families, and the roles of many miRNA families, such as miR156, miR167, miR169, and miR396, in stress responses have been demonstrated in previous studies (Covarrubias and Reyes 2010). Our results indicated that the 21-nt sequences of the known miRNAs were the most abundant, and 22-nt ranked in the second. Among the novel miRNAs, the 24-nt sequences were the most frequent (64.00%). Previous studies reported that there are greater numbers of 24-nt sRNAs than 21-nt sRNAs in plants. Distinct lengths of sRNAs are produced by different members of the Dicer-like (DCL) family. For example, DCL1 is mainly responsible for the biosynthesis of 21-nt miRNAs, while DCL3 is responsible for 24-nt miRNAs. The functional diversity among different DCL members may be responsible for this difference (Huang et al. 2019; Rogers and Chen 2013). Compared with the expression levels of the known miRNAs, none of the 125 novel miRNAs showed a high expression level (Tables S3). Among the 35 DEMs, 8 miRNAs, zma-miR164f-5p_R-1, zma-miR394a-5p, zma-miR168a-5p, zma-miR168b-3p_R + 1, zma-miR160f-5p_R-1, zma-miR164a-5p, tae-MIR9774-p3_1ss4AG, and zma-miR160a-5p, exhibited high expression levels in the MC or MH library. Like some previous studies, this study indicated that the expression levels of novel miRNAs were lower than those of known miRNAs (Li et al. 2017; Liu et al. 2014a, b). The expression patterns of the DEMs were also compared to those found in previous studies. Five conserved families (miR156, miR159, miR167, miR168 and miR169), exhibited down-regulated expression levels, while four families (miR156, miR159, miR167 and miR169) showed similar expression patterns to rice heat-responsive miRNAs. However, different expression patterns were also found. For example, the expression levels of miR164 and miR399 were up-regulated in our study, but down-regulated in rice (Li et al. 2015). Furthermore, miR160 exhibited up-regulated expression in our study and rice, but down-regulated expression in other species (Li et al. 2015; Pan et al. 2017; Wang et al. 2015). Different plant species, genetic background, developmental stages and heat treatment methods may be responsible for the differences among these studies. However, these observations also suggested the complex regulatory mechanisms of plant heat responses mediated by miRNAs, even though many miRNA families are evolutionarily conserved in different species.
miRNAs act as negative regulators of the expression levels of target mRNAs (Bartel 2004; Covarrubias and Reyes 2010). A total of 174 (527 transcripts) target genes were predicted to be cleaved by 115 miRNAs, while no target genes were detected for the other 225 miRNAs using degradome sequencing. We noted that many of the target genes encode TF families. For example, SPL TFs were targeted by the miR156 family, and GRAS TFs were targeted by the miR171 family, which might suggest the conserved regulatory mechanism for the miRNAs. Studies have increasingly indicated that TFs play crucial roles in stress responses (Chen and Zhu 2004; Huang et al. 2012). In Arabidopsis, the NTL4 TF participates in a reactive oxygen species mediated pathway that regulates programmed cell death in response to heat stress (Lee et al. 2014). Chao et al. (2017) reported that the overexpression of Arabidopsis SPL TFs SPL1 or SPL12 improves the thermotolerance of transgenic plants during the reproductive stage (Chao et al. 2017). We noted that two heat shock proteins were targeted by zma-miR396e-5p and PC-3p-184258_13, respectively, and were only detected in the MH library. The expression levels of heat shock proteins are rapidly induced by various environmental stresses and involving in protecting proteins and cells from damage under stressed conditions (Campbell et al. 2001; Kadota and Shirasu 2012; Wang et al. 2004). The two heat shock proteins were involved in posttranslational modification, protein turnover and chaperones as determined by a KOG annotation, suggesting the potential roles of zma-miR396e-5p and PC-3p-184258_13 in the regulation of heat responses.
GO and KEGG analyses were performed for the 174 targets, and we noted that some targets were involved in “response to hormone”, “response to hydrogen peroxide”, “heat acclimation” and “protein stabilization” terms in the GO biological process category. The pathway analysis indicated that targets were mainly enriched in “metabolic pathways”, “biosynthesis of secondary metabolites”, “ribosome”, “ubiquinone and other terpenoid-quinone biosynthesis” and “plant hormone signal transduction pathways” on the basis of the enriched gene numbers. Some target genes were involved in important pathways. For example, three genes were enriched in the “spliceosome” pathway. Heat stress may lead to an increase in alternative splicing events (Zhao et al. 2019), which are regulated by different plant developmental stages and environmental stresses (Filichkin et al. 2015; Laloum et al. 2018). Additionally, one gene targeted by a DEM (zma-miR169i-3p_R + 1_1) with down-regulated expression, was involved in the “MAPK signaling pathway-plant” pathway, which has important roles in abiotic stress response (Danquah et al. 2014). These findings suggest the potential functions of these targets in heat stress responses. To further explore the regulatory mechanisms of heat-responsive miRNAs, a regulatory network was constructed based on the direct relationships among miRNAs, targets, and enriched GO terms and KEGG pathways. A total of 16 DEMs, and their enriched terms and pathways, were highlighted in the regulatory network. The results provide an important foundation for the identification of heat-responsive miRNAs and their targets and for the exploration of the regulatory mechanisms of heat response in maize.
Conclusions
In this study, a total of 340 miRNAs, including 215 known and 125 novel members, were identified from the seedlings of maize inbred line CM1 using high-throughput sequencing. Expression profiles analysis indicated that expression levels of most known miRNAs were higher than those of the novel miRNAs, and 35 members (26 known and 9 novel) were found to be responsive to heat stress. Targets of the detected miRNAs were further predicted by degradome sequencing, and 174 genes (527 transcripts) were found to be targeted by 115 miRNAs. Among the 35 DEMs, 17 miRNAs (16 known and 1 novel miRNAs) had target genes in the MC or MH library. GO and KEGG analyses were further performed for the targets to explore their biological roles, and some important GO terms and pathways were enriched for these genes. Finally, a miRNA-mediated regulatory network was constructed based on the relationships among miRNAs, targets and the enriched GO terms and pathways. Our results provide valuable information for the identification of heat-responsive miRNAs and the related targets, which can be used for creating heat-tolerant germplasm through molecular breeding in maize.
Data availability
The generated raw reads were submitted to the Sequence Read Archive (SRA) of NCBI under Accession Number PRJNA643647.
References
Addo-Quaye C, Miller W, Axtell MJ (2009) CleaveLand: a pipeline for using degradome data to find cleaved small RNA targets. Bioinformatics 25(1):130–131. https://doi.org/10.1093/bioinformatics/btn604
Balyan S, Kumar M, Mutum RD, Raghuvanshi U, Agarwal P, Mathur S, Raghuvanshi S (2017) Identification of miRNA-mediated drought responsive multi-tiered regulatory network in drought tolerant rice, Nagina 22. Sci Rep 7(1):15446. https://doi.org/10.1038/s41598-017-15450-1
Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116(2):281–297. https://doi.org/10.1016/s0092-8674(04)00045-5
Baumberger N, Baulcombe DC (2005) Arabidopsis ARGONAUTE1 is an RNA Slicer that selectively recruits microRNAs and short interfering RNAs. Proc Natl Acad Sci USA 102(33):11928–11933. https://doi.org/10.1073/pnas.0505461102
Campbell JL, Klueva NY, Zheng HG, Nieto-Sotelo J, Ho TD, Nguyen HT (2001) Cloning of new members of heat shock protein HSP101 gene family in wheat (Triticum aestivum (L.) Moench) inducible by heat, dehydration, and ABA(1). Biochim Biophys Acta 1517(2):270–277. https://doi.org/10.1016/s0167-4781(00)00292-x
Cao X, Wu Z, Jiang FL, Zhou R, Yang ZE (2014) Identification of chilling stress-responsive tomato microRNAs and their target genes by high-throughput sequencing and degradome analysis. BMC Genomics 15(1):1130. https://doi.org/10.1186/1471-2164-15-1130
Casaretto JA, El-Kereamy A, Zeng B, Stiegelmeyer SM, Chen X, Bi YM, Rothstein SJ (2016) Expression of OsMYB55 in maize activates stress-responsive genes and enhances heat and drought tolerance. BMC Genomics 17:312. https://doi.org/10.1186/s12864-016-2659-5
Chao LM, Liu YQ, Chen DY, Xue XY, Mao YB, Chen XY (2017) Arabidopsis transcription factors SPL1 and SPL12 confer plant thermotolerance at reproductive stage. Mol Plant 10(5):735–748. https://doi.org/10.1016/j.molp.2017.03.010
Chen WJ, Zhu T (2004) Networks of transcription factors with roles in environmental stress response. Trends Plant Sci 9(12):591–596. https://doi.org/10.1016/j.tplants.2004.10.007
Covarrubias AA, Reyes JL (2010) Post-transcriptional gene regulation of salinity and drought responses by plant microRNAs. Plant Cell Environ 33(4):481–489. https://doi.org/10.1111/j.1365-3040.2009.02048.x
Danquah A, de Zelicourt A, Colcombet J, Hirt H (2014) The role of ABA and MAPK signaling pathways in plant abiotic stress responses. Biotechnol Adv 32(1):40–52. https://doi.org/10.1016/j.biotechadv.2013.09.006
Denver JB, Ullah H (2019) miR393s regulate salt stress response pathway in Arabidopsis thaliana through scaffold protein RACK1A mediated ABA signaling pathways. Plant Signal Behav 14(6):1600394. https://doi.org/10.1080/15592324.2019.1600394
Ding D, Zhang LF, Wang H, Liu ZJ, Zhang ZX, Zheng YL (2009) Differential expression of miRNAs in response to salt stress in maize roots. Ann Bot 103(1):29–38. https://doi.org/10.1093/aob/mcn205
Dong CH, Pei HX (2014) Over-expression of miR397 improves plant tolerance to cold stress in Arabidopsis thaliana. J Plant Biol 57:209–217. https://doi.org/10.1007/s12374-013-0490-y
Fang Y, Xie K, Xiong L (2014) Conserved miR164-targeted NAC genes negatively regulate drought resistance in rice. J Exp Bot 65(8):2119–2135. https://doi.org/10.1093/jxb/eru072
Filichkin S, Priest HD, Megraw M, Mockler TC (2015) Alternative splicing in plants: directing traffic at the crossroads of adaptation and environmental stress. Curr Opin Plant Biol 24:125–135. https://doi.org/10.1016/j.pbi.2015.02.008
Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ (2008) miRBase: tools for microRNA genomics. Nucl Acids Res 36:D154-158. https://doi.org/10.1093/nar/gkm952
Hoang XLT, Nhi DNH, Thu NBA, Thao NP, Tran LSP (2017) Transcription factors and their roles in signal transduction in plants under abiotic stresses. Curr Genomics 18(6):483–497. https://doi.org/10.2174/1389202918666170227150057
Huang GT, Ma SL, Bai LP, Zhang L, Ma H, Jia P, Liu J, Zhong M, Guo ZF (2012) Signal transduction during cold, salt, and drought stresses in plants. Mol Biol Rep 39(2):969–987. https://doi.org/10.1007/s11033-011-0823-1
Huang X, Feng JJ, Wang R, Zhang HS, Huang J (2019) Comparative analysis of microRNAs and their targets in the roots of two cultivars with contrasting salt tolerance in rice (Oryza sativa L.). Plant Growth Regul 87:139–148. https://doi.org/10.1007/s10725-018-0459-4
Kadota Y, Shirasu K (2012) The HSP90 complex of plants. Biochim Biophys Acta 1823(3):689–697. https://doi.org/10.1016/j.bbamcr.2011.09.016
Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T, Yamanishi Y (2008) KEGG for linking genomes to life and the environment. Nucl Acids Res 36:D480-484. https://doi.org/10.1093/nar/gkm882
Krutzfeldt J, Poy MN, Stoffel M (2006) Strategies to determine the biological function of microRNAs. Nat Genet 38:S14-19. https://doi.org/10.1038/ng1799
Kurihara Y, Watanabe Y (2004) Arabidopsis micro-RNA biogenesis through Dicer-like 1 protein functions. Proc Natl Acad Sci USA 101(34):12753–12758. https://doi.org/10.1073/pnas.0403115101
Laloum T, Martin G, Duque P (2018) Alternative splicing control of abiotic stress responses. Trends Plant Sci 23(2):140–150. https://doi.org/10.1016/j.tplants.2017.09.019
Lee RC, Feinbaum RL, Ambros V (1993) The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75(5):843–854. https://doi.org/10.1016/0092-8674(93)90529-y
Lee S, Lee HJ, Huh SU, Paek KH, Ha JH, Park CM (2014) The Arabidopsis NAC transcription factor NTL4 participates in a positive feedback loop that induces programmed cell death under heat stress conditions. Plant Sci 227:76–83. https://doi.org/10.1016/j.plantsci.2014.07.003
Lee DK, Jung H, Jang G, Jeong JS, Kim YS, Ha SH, Do Choi Y, Kim JK (2016) Overexpression of the OsERF71 transcription factor alters rice root structure and drought resistance. Plant Physiol 172(1):575–588. https://doi.org/10.1104/pp.16.00379
Li J, Wu LQ, Zheng WY, Wang RF, Yang LX (2015) Genome-wide identification of microRNAs responsive to high temperature in rice (Oryza sativa) by high-throughput deep sequencing. J Agron Crop Sci 201:379–388. https://doi.org/10.1111/jac.12114
Li X, Shahid MQ, Wu J, Wang L, Liu X, Lu Y (2016) Comparative small RNA analysis of pollen development in autotetraploid and diploid rice. Int J Mol Sci 17(4):499. https://doi.org/10.3390/ijms17040499
Li H, Peng T, Wang Q, Wu Y, Chang J, Zhang M, Tang G, Li C (2017) Development of incompletely fused carpels in maize ovary revealed by miRNA, target gene and phytohormone analysis. Front Plant Sci 8:463. https://doi.org/10.3389/fpls.2017.00463
Liu HJ, Qin C, Chen Z, Zuo T, Yang XR, Zhou HK, Xu M, Cao SL, Shen YO, Lin HJ, He XJ, Zhang YC, Li LJ, Ding HP, Lubberstedt T, Zhang ZM, Pan GT (2014a) Identification of miRNAs and their target genes in developing maize ears by combined small RNA and degradome sequencing. BMC Genomics 15:25. https://doi.org/10.1186/1471-2164-15-25
Liu N, Wu S, Van Houten J, Wang Y, Ding B, Fei Z, Clarke TH, Reed JW, van der Knaap E (2014b) Down-regulation of AUXIN RESPONSE FACTORS 6 and 8 by microRNA 167 leads to floral development defects and female sterility in tomato. J Exp Bot 65(9):2507–2520. https://doi.org/10.1093/jxb/eru141
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2 (-Delta Delta C(T)) method. Methods 25(4):402–408. https://doi.org/10.1006/meth.2001.1262
Ma ZR, Coruh C, Axtell MJ (2010) Arabidopsis lyrata small RNAs: transient MIRNA and small interfering RNA loci within the Arabidopsis genus. Plant Cell 22(4):1090–1103. https://doi.org/10.1105/tpc.110.073882
Mallory AC, Elmayan T, Vaucheret H (2008) MicroRNA maturation and action–the expanding roles of ARGONAUTEs. Curr Opin Plant Biol 11(5):560–566. https://doi.org/10.1016/j.pbi.2008.06.008
Nakashima K, Tran LS, Van Nguyen D, Fujita M, Maruyama K, Todaka D, Ito Y, Hayashi N, Shinozaki K, Yamaguchi-Shinozaki K (2007) Functional analysis of a NAC-type transcription factor OsNAC6 involved in abiotic and biotic stress-responsive gene expression in rice. Plant J 51(4):617–630. https://doi.org/10.1111/j.1365-313x.2007.03168.x
Nie Z, Ren Z, Wang L, Su S, Wei X, Zhang X, Wu L, Liu D, Tang H, Liu H, Zhang S, Gao S (2016) Genome-wide identification of microRNAs responding to early stages of phosphate deficiency in maize. Physiol Plant 157(2):161–174. https://doi.org/10.1111/ppl.12409
Pan Y, Niu MY, Liang JS, Lin EP, Tong ZK, Zhang JH (2017) Identification of heat-responsive miRNAs to reveal the miRNA-mediated regulatory network of heat stress response in Betula luminifera. Trees 31:1635–1652. https://doi.org/10.1007/s00468-017-1575-x
Ravichandran S, Ragupathy R, Edwards T, Domaratzki M, Cloutier S (2019) MicroRNA-guided regulation of heat stress response in wheat. BMC Genomics 20(1):488. https://doi.org/10.1186/s12864-019-5799-6
Rogers K, Chen X (2013) Biogenesis, turnover, and mode of action of plant microRNAs. Plant Cell 25(7):2383–2399. https://doi.org/10.1105/tpc.113.113159
Schnable PS, Ware D, Fulton RS, Stein JC, Wei F, Pasternak S, Liang C, Zhang J, Fulton L, Graves TA et al (2009) The B73 maize genome: complexity, diversity, and dynamics. Science 326(5956):1112–1115. https://doi.org/10.1126/science.1178534
Shinozaki K, Yamaguchi-Shinozaki K (2007) Gene networks involved in drought stress response and tolerance. J Exp Bot 58(2):221–227. https://doi.org/10.1093/jxb/erl164
Sorin C, Declerck M, Christ A, Blein T, Ma L, Lelandais-Briere C, Njo MF, Beeckman T, Crespi M, Hartmann C (2014) A miR169 isoform regulates specific NF-YA targets and root architecture in Arabidopsis. New Phytol 202(4):1197–1211. https://doi.org/10.1111/nph.12735
Sun L, Luo H, Bu D, Zhao G, Yu K, Zhang C, Liu Y, Chen R, Zhao Y (2013) Utilizing sequence intrinsic composition to classify protein-coding and long non-coding transcripts. Nucl Acids Res 41(17):e166. https://doi.org/10.1093/nar/gkt646
Sunkar R, Chinnusamy V, Zhu JH, Zhu JK (2007) Small RNAs as big players in plant abiotic stress responses and nutrient deprivation. Trends Plant Sci 12(7):301–309. https://doi.org/10.1016/j.tplants.2007.05.001
Tuteja N (2007) Abscisic acid and abiotic stress signaling. Plant Signal Behav 2(3):135–138. https://doi.org/10.4161/psb.2.3.4156
Wang W, Vinocur B, Shoseyov O, Altman A (2004) Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends Plant Sci 9(5):244–252. https://doi.org/10.1016/j.tplants.2004.03.006
Wang Y, Sun F, Cao H, Peng H, Ni Z, Sun Q, Yao Y (2012) TamiR159 directed wheat TaGAMYB cleavage and its involvement in anther development and heat response. PLoS ONE 7(11):e48445. https://doi.org/10.1371/journal.pone.0048445
Wang RH, Xu L, Zhu XW, Zhai LL, Wang Y, Yu RG, Gong YQ, Limera C, Liu LW (2015) Transcriptome-wide characterization of novel and heat-stress-responsive microRNAs in radish (Raphanus Sativus L.) using next-generation sequencing. Plant Mol Biol Rep 33:867–880. https://doi.org/10.1007/s11105-014-0786-1
Wang R, Yang Z, Fei Y, Feng J, Zhu H, Huang F, Zhang H, Huang J (2019) Construction and analysis of degradome-dependent microRNA regulatory networks in soybean. BMC Genomics 20(1):534. https://doi.org/10.1186/s12864-019-5879-7
Xin M, Wang Y, Yao Y, Xie C, Peng H, Ni Z, Sun Q (2010) Diverse set of microRNAs are responsive to powdery mildew infection and heat stress in wheat (Triticum aestivum L.). BMC Plant Biol 10:123. https://doi.org/10.1186/1471-2229-10-123
Xu YJ, Zhu SW, Liu F, Wang W, Wang XW, Han GM, Cheng BJ (2018) Identification of arbuscular mycorrhiza fungi responsive microRNAs and their regulatory network in maize. Int J Mol Sci 19(10):3201. https://doi.org/10.3390/ijms19103201
Yamaguchi-Shinozaki K, Shinozaki K (2006) Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu Rev Plant Biol 57:781–803. https://doi.org/10.1146/annurev.arplant.57.032905.105444
Yang CH, Li DY, Mao DH, Liu X, Ji CJ, Li XB, Zhao XF, Cheng ZK, Chen CY, Zhu LH (2013) Overexpression of microRNA319 impacts leaf morphogenesis and leads to enhanced cold tolerance in rice (Oryza sativa L.). Plant Cell Environ 36(12):2207–2218. https://doi.org/10.1111/pce.12130
Young MD, Wakefield MJ, Smyth GK, Oshlack A (2010) Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol 11:R14. https://doi.org/10.1186/gb-2010-11-2-r14
Yue E, Liu Z, Li C, Li Y, Liu Q, Xu JH (2017) Overexpression of miR529a confers enhanced resistance to oxidative stress in rice (Oryza sativa L.). Plant Cell Rep 36(7):1171–1182. https://doi.org/10.1007/s00299-017-2146-8
Zandalinas SI, Mittler R, Balfagon D, Arbona V, Gomez-Cadenas A (2018) Plant adaptations to the combination of drought and high temperatures. Physiol Plant 162(1):2–12. https://doi.org/10.1111/ppl.12540
Zhang X, Zou Z, Gong P, Zhang J, Ziaf K, Li H, Xiao F, Ye Z (2011) Over-expression of microRNA169 confers enhanced drought tolerance to tomato. Biotechnol Lett 33(2):403–409. https://doi.org/10.1007/s10529-010-0436-0
Zhao Y, Hu F, Zhang X, Wei Q, Dong J, Bo C, Cheng B, Ma Q (2019) Comparative transcriptome analysis reveals important roles of nonadditive genes in maize hybrid An’nong 591 under heat stress. BMC Plant Biol 19(1):273. https://doi.org/10.1186/s12870-019-1878-8
Acknowledgements
We would like to thank Shanshan Xie for her help in this study.
Funding
This research was supported by the National Natural Science Foundation of China (Nos. 31871627 and 31971895).
Author information
Authors and Affiliations
Contributions
YZ and QM conceived and designed the research. QYW, TCC, LJX, and JL performed the data analysis. QYW and XGZ conducted the gene expression experiments; YZ and GMH performed the construction of regulatory network. YZ drafted the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Additional information
Communicated by Zhong-Hua Chen.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhao, Y., Wei, Q., Chen, T. et al. Identification and characterization of heat-responsive miRNAs and their regulatory network in maize. Plant Growth Regul 96, 195–208 (2022). https://doi.org/10.1007/s10725-021-00769-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10725-021-00769-7