Abstract
Previous studies have revealed that a basic helix–loop–helix domain transcription factor AtTT8 is not only involved in biosynthesis of anthocyanins and proanthocyanidins, but also regulates seed development and oil biosynthesis in Arabidopsis. However, the functions of BnTT8 in accumulation of seed storage reserves and seedling establishment remain largely unclear. Here, we obtained the BnTT8 coding domain sequence from the Brassica napus cultivar QINYOU Seven, and analyzed its conserved protein domains. A subcellular localization experiment in tobacco leaves indicated that BnTT8 functioned as a transcription factor. Expression analysis indicated that TT8 was widely expressed in various tissues, and was also predominantly present in developing seeds of QINYOU Seven. Our results strongly demonstrated that introduction of BnTT8 into the tt8-4 mutant could fully complement many phenotypes of tt8-4 seeds, such as yellow seed coat, higher fatty acid content, and lower storage protein content. We also found that ectopic expression of BnTT8 successfully corrected the sensitivity of the tt8-4 mutant to salinity and glucose stresses during seed germination and seedling establishment. Moreover, altered expression levels of many stress-responsive genes in tt8-4 seedlings were rescued to wild-type levels by ectopic expression of BnTT8. These results should improve understanding of the entire function of BnTT8 and provide promising targets for genetic manipulations of B. napus to improve oil quantity in seeds.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Canola (Brassica napus L., AACC, 2n = 38), a major oil-producing crop, is an important source of vegetative oil. Canola oil modified through breeding practices has high nutritional value with high levels of unsaturated C18 fatty acids (FAs; >60%), traces of erucic acid (C22:1; <1%), and low concentrations of glucosinolates (Dupont et al. 1989; Prakash et al. 2011). Moreover, it is a suitable source for biofuels and raw industrial materials (Thelen and Ohlrogge 2002). In addition, canola meal as the byproduct after oil refining of canola seeds contains abundant protein and fiber, and is suitable for use as animal feed. In canola seeds, the storage reserves are mainly oils stored as triacylglycerols, proteins, and carbohydrates, which not only play essential roles in maintaining this species by joining two distinct sporophytic generations, but also provide energy for seed germination and seedling establishment (Sullivan and Deng 2003). Canola seeds include embryo, endosperm, and seed coat, with most of the seed reserves stored in the embryo. Demand for vegetable oil and proteins has greatly increased in recent years, and the increase of oil and protein contents is thus a major challenge for genetic improvement of oilseed crops. Therefore, understanding the functions of key canola genes on seed development and accumulation of seed storage reserves is important in terms of social and economic significance.
For angiosperms, including Brassicaceae species like Arabidopsis thaliana and B. napus, seed development starts with a double fertilization event that produces a diploid zygote and triploid endosperm. Seed development consists of two major stages: embryonic morphogenesis and maturation (Baud and Lepiniec 2009). After double fertilization, the embryo enters the embryonic morphogenesis stage, during which the endosperm generally occupies most of the total seed volume (Baud et al. 2002). During development, the endosperm supplies nutrition for the growing embryo and is almost degraded and reduced to a single cell layer surrounding the embryo, which acquires the basic architecture of the plant (Jenik et al. 2007; Millar et al. 2015). The seed maturation stage is characterized by the accumulation of seed storage reserves and acquisition of desiccation tolerance (Baud et al. 2002; Fait et al. 2006). The seed coat plays important roles in releasing nutrients and transmitting signals from the maternal tissues to the embryo and the endosperm during seed development and in protecting the embryo and the endosperm against detrimental events outside, and is necessary for proper seed development (Weber et al. 1996; Debeaujon and Koornneef 2000; Millar et al. 2015).
Seed coat color is determined by the type of pigment accumulated in seed coat cells. It is related to important agronomic traits of seeds such as seed dormancy, longevity, and seed oil and protein contents. In canola, yellow seed is a desirable quality trait. The genes controlling seed coat color mainly correspond to enzymes and regulatory factors involved in flavonoid biosynthesis (Li et al. 2012). Previous studies demonstrated that some of these are also involved in accumulation of seed storage reserves in canola. TRANSPARENT TESTA 2 (TT2) was significantly associated with seed coat color and linoleic acid and total FA contents in canola (Zhou et al. 2016). BnTT16 plays multiple roles in regulating the development of inflorescences, flowers, siliques, seed embryo development, and FA biosynthesis in canola (Deng et al. 2012). Ectopic expression analysis of BnaC.GLABRA2.b from the C genome of B. napus in Arabidopsis indicated that it can negatively regulate oil accumulation in seeds (Chai et al. 2010). The functions and mechanisms of genes controlling seed coat color in canola and underlying seed storage reserves remain largely unknown and require more investigation.
Previous studies revealed that AtTT8, which encodes a basic helix–loop–helix domain transcription factor, is involved in the biosynthesis of anthocyanins and proanthocyanidins (Nesi et al. 2000; Baudry et al. 2006). More recently, we found that it could target LEC1, LEC2, and FUS3 to regulate seed development and/or oil biosynthesis in Arabidopsis (Chen et al. 2014b). TT8 from B. rapa could rescue the yellow seed phenotype of the Arabidopsis tt8 mutant by regulating flavonoid accumulation in the seed coat (Li et al. 2012). In the present study, we showed that ectopic expression of BnTT8 in the Arabidopsis tt8-4 mutant restored the yellow seed coat, and altered seed oil and storage protein contents during seed development. In addition, we provided evidence that BnTT8 exhibited similar functions to AtTT8 on seed germination and seedling establishment under abiotic stresses.
Materials and methods
Plant material and growth condition
Arabidopsis wild-type (WT) Columbia (Col-0) and the AtTT8-deficient Arabidopsis mutant tt8-4 (Chen et al. 2014b) were used in this study. Arabidopsis plants were grown in a growth chamber under long-day (16/8 h of light/dark) conditions at 22 °C. The light intensity was 160 µmol m−2 s−1, as detected at the middle of plants.
The canola cultivar QINYOU Seven was grown in the Agricultural Farm of North Campus, Northwest A&F University, China. All canola plant materials (roots, stems, leaves, flowers, and developing seeds) were collected from the farm-grown plants, immediately frozen in liquid nitrogen, and then stored at −80 °C until use.
Gene cloning and plasmid construction
According to the full-length coding domain sequence (CDS) of TT8 from B. napus, which are BnaA.TT8 (Accession, ADV03941) and BnaC.TT8 (Accession, ADV03942), primers BnTT8_P1 (5′-ATGGATGAATTAAGTAT-3′) and BnTT8_P2 (5′-GAGTTTATTATTATATATGATTTGATGG-3′) were used to amplify the BnTT8 gene from total RNA isolated from QINYOU Seven developing seeds at 15 days after pollination (DAP). The PCR products were cloned into the pMD19-T vector (TaKaRa Bio, Dalian, China), and then eight randomly selected single colonies were sequenced by Sangon Biotechnology (Shanghai, China).
To construct 35S:BnTT8, the BnTT8 cDNA was amplified with primers BnTT8_P3_XhoI (5′-ATTCTCGAGATGGATGAATTAAGTAT-3′) and BnTT8_P4_ HindIII (5′-ATTAAGCTTGAGTTTATTATTATATATGATTTGATGG-3′). The PCR products were digested by XhoI and HindIII and then cloned into pGreen-35S to obtain an in-frame fusion of BnTT8 under control of the 35S promoter. Similarly, BnTT8 cDNA was also amplified with primers BnTT8_P5_XmaI (5′-TATTCCCGGGATGGATGAATTAAGTAT-3′) and BnTT8- _P6_SpeI (5′-GGACTAGTGAGTTTATTATTATATATGATTTGATGG-3′) and then cloned into pGreen-35S–eGFP (where GFP is green fluorescent protein) to obtain an in-frame fusion of BnTT8–eGFP under control of the 35S promoter (35S:BnTT8–eGFP).
Subcellular localization of BnTT8–eGFP fusion protein
The 35S:BnTT8–eGFP construct was transiently expressed in Nicotiana benthamiana leaves as previously reported (Yang et al. 2000). Images were collected on a confocal laser scanning microscope (Zeiss LSM 700, Germany) within 72 h following agroinfiltration.
Generation of Arabidopsis transgenic plants
The construct 35S:BnTT8 was transformed into Agrobacterium tumefaciens strain GV3101, which was then used for the transformation of Arabidopsis tt8-4 plants via floral dip (Clough and Bent 1998). The transgenic plants were selected on soil using Basta® until T3 transgenic plants were generated.
Morphological observations of mature seeds
The seeds were harvested from siliques at the basal part of a major inflorescence. The mature seeds from different lines were randomly selected and photographed using an Olympus SZ 61 stereomicroscope.
Determination of seed FAs and storage proteins
Seed FAs were detected as reported by Chen et al. (2012a). In brief, total FAs were converted to FA methyl esters in methanol solution containing 1 M hydrochloric acid for 2 h at 80 °C. FAs in seeds were subsequently determined using a gas chromatograph (GC-2014; Shimadzu). Storage proteins in seeds were measured as described by Chen et al. (2012b). Briefly, in the presence of Polyclar, seeds were homogenized in the extraction buffer: 50 mM HEPES, 5 mM MgCl2, 5 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, 1 mM EDTA, and 10% (v/v) ethylene glycol (pH 7.5). After centrifugation (18,000×g for 10 min at 4 °C), the supernatant was utilized for storage protein measurements. Gammaglobulin (Bio-Rad) was used for calibration.
Analysis of gene expression by reverse transcription PCR and real-time quantitative PCR
The RNA extraction was conducted using a TaKaRa MiniBest Plant RNA Extraction Kit (TaKaRa Bio) following the manufacturer’s instructions. The RNA samples were treated with RNase-free DNAse I supplied by the kit to remove any trace of genomic DNA. The first-strand cDNA was synthesized in a 20-µL reaction solution containing approximately 2.0 µg of total RNA, using PrimerScript™ reverse transcriptase (TaKaRa Bio) and oligo (dT) 12–18 as a primer. AtACTIN7 (At5g09810) and BnACTIN7 were regarded as the internal control. The real-time quantitative PCR (qPCR) and the calculation of relative gene expression levels were performed as described in detail by Chen et al. (2014a). Primers used for reverse transcription PCR (RT-PCR) and qPCR are listed in Supplementary Table S1.
Analysis of seed germination rate and seedling establishment under stressed conditions
The Arabidopsis seeds used for the germination assays were collected at the same time from plants grown under the same growth conditions as previously described, allowed to mature for 6 weeks at room temperature, and stored at −20 °C. The seeds were pre-chilled at 4 °C for 5 d and then surface sterilized twice with 75% (v/v) ethanol for 30 s. Then, seeds were washed five times with ddH2O and subsequently sown onto solid Murashige and Skoog (MS) stock medium containing 3% (w/v) glucose (Glc) and 100 mM sodium chloride (NaCl). The seed germination rate (defined as radicle emergence) was scored daily (Cernac et al. 2006). Arabidopsis seedlings at 17 days after sowing (DAS) were photographed and collected for RNA analysis.
Statistical analysis
Completely randomized block designs with at least three biological replicates were used in the study. Data were classified with Win-Excel and analyzed via ANOVA using the SPSS statistical package (version 8.0). Seed size, seed weight, and seed oil and protein contents for the WT and tt8-4 mutant were compared using Student’s t-test. Tukey’s highly significant difference test and Student’s t-test were used to analyze gene expression and seedling fresh and dry weights under abiotic stresses. P-values ≤ 0.05 were considered statistically significant.
Results
Sequence analysis of the BnTT8 protein
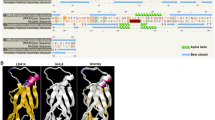
The BnTT8 CDS isolated from developing seeds of B. napus cultivar QINYOU Seven is designated BnTT8 in this study. We used the BnTT8 CDS to BLAST with the B. napus nucleotide database in NCBI (National Center for Biotechnology Information), and found that BnTT8-like located on the A8 genome shared the highest identity in nucleotide sequence with BnTT8 CDS, except for BnaA.TT8 and BnaC.TT8. The TT8 from canola and Arabidopsis shared high nucleotide and protein sequence similarities (Fig. 1, Supplementary Figure S1). All four canola TT8 proteins had 521 amino acids, and BnTT8 shared highest identity in amino acid sequence with BnaC.TT8 (99.62%), compared with BnaA.TT8 (98.85%) and BnTT8-like (98.85%) (Fig. 1). Consistently, BnTT8 shared highest nucleotide sequence identity with BnaC.TT8 (99.23%), compared with BnaA.TT8 (99.04%) and BnTT8-like (99.04%) (Supplementary Figure S1). These indicated that BnTT8 should be located on the C genome of QINYOU Seven. Moreover, they were highly conserved in the N-terminal region of the family of MYB and MYC transcription factors and the helix–loop–helix DNA-binding region (Fig. 1).
Alignment of protein sequences of TT8 from canola and Arabidopsis. The alignments of AtTT8, BnaA.TT8 (ADV03941), BnaC.TT8 (ADV03942), BnTT8-like (NM_001315974.1), and BnTT8 were analyzed using the MUSCLE program (http://www.ebi.ac.uk/Tools/msa/muscle/). Their conserved regulatory domains were predicted by Conserved Domain Search program (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi). Asterisks represent different amino acids. The two domains 20–201 and 366–417 indicated by black lines are the N-terminal regions of a family of MYB and MYC transcription factors and the helix–loop–helix DNA-binding regions, respectively
Subcellular localization of BnTT8 protein
Using laser scanning confocal microscopy, the fluorescence signal was specifically localized in the nucleus of N. benthamiana leaves transformed using the fusion construct, 35S:BnTT8–eGFP (Fig. 2a). This result supported the suggestion that BnTT8 functions as a transcription factor.
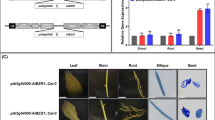
Expression analysis of canola TT8. a Subcellular localization of BnTT8 protein fused with GFP (35S:BnTT8–eGFP) in N. benthamiana leaves. DAPI, fluorescence of 4′,6-diamino-2-phenylindole; Merge, merging of GFP, DAPI, and bright field images. qPCR analysis of canola TT8 expression in various tissues (b) and developing seeds at different developmental stages (c) in cultivar QINYOU Seven. The qPCR result was normalized against BnACTIN7 expression as an internal control. Values are means of three replicates carried out on cDNA dilutions obtained from three independent RNA extractions. Error bars denote standard deviations. DAP, days after pollination
Expression pattern of TT8 in the canola genome
At least two copies of TT8 were present in the A and C genomes of QINYOU Seven, and may have had different expression patterns. However, the expression pattern of all TT8 copies in QINYOU Seven would still help in better understanding the biological function of TT8. Therefore, the temporal and spatial mRNA distributions of all TT8 copies were analyzed using qPCR in QINYOU Seven. TT8 was widely expressed in various tissues (Fig. 2b), and was predominantly present in developing seeds (Fig. 2c). The TT8 transcript level was higher in stems and flowers than in leaves and roots (Fig. 2b). During seed development, TT8 expression progressively increased at 15 DAP to a maximum at 20 DAP, then remained stable, and slightly decreased at 35 DAP (Fig. 2c).
BnTT8 affects the accumulation of seed storage reserves in Arabidopsis
To explore the BnTT8 function, we transformed the construct 35S:BnTT8 into the Arabidopsis mutant tt8-4. At least 20 T1 plants were obtained by Basta® selection. Four selected independent tt8-4 35S:BnTT8 T3 homozygous transgenic lines were identified by PCR amplification of BnTT8 with the specific primers 35S_P/BnTT8_R (Supplementary Figure S2; Table S1). Furthermore, heterologous expression of BnTT8 was also detected in these lines by RT-PCR (Supplementary Figure S2).
Previous studies indicated that loss of function of AtTT8 resulted in a yellow seed coat (Nesi et al. 2000) and higher oil and lower storage protein contents in Arabidopsis seeds (Chen et al. 2014b). Ectopic expression of BnTT8 corrected the yellow seed coat phenotype of tt8-4 (Fig. 3a). However, seed length and width (Fig. 3b) and seed weight (Fig. 3c) did not significantly differ among the WT, tt8-4, and tt8-4 35S:BnTT8 plants. In addition, the higher FA and lower storage protein contents in tt8-4 seeds could be fully rescued to WT levels by ectopic expression of BnTT8 (Fig. 4). These results together indicated that BnTT8 had similar functions to AtTT8 in the accumulation of seed storage reserves in Arabidopsis.
Analysis of seed coat color, seed size, and seed weight among WT (Col-0), tt8-4, and tt8-4 35S:BnTT8 plants. a Microscopic observations of randomly selected mature seeds. Comparison of seed size (b) and seed weight (c) of mature seeds. Values of seed size and weight are means of three biological replicates, and each of the three assays for each biological replicate contained 200 seeds from 12 individual plants grown in different pots arranged randomly within one of three blocks. There were no significant differences in seed size and weight between WT and the tt8-4 mutant (Student’s t-test, P ≤ 0.05). Error bars denote standard deviations
Comparison of seed storage reserves including seed storage proteins (a) and seed fatty acids (b) among the WT (Col-0), tt8-4, and tt8-4 35S:BnTT8 plants. Data presented are means of five biological replicates. Error bars denote standard deviations. Asterisks denote significant differences between the WT and tt8-4 mutant (Student’s t-test, P ≤ 0.05)
BnTT8 affects tolerance to abiotic stresses during seedling establishment in Arabidopsis
The effects of BnTT8 on stress responses, seed germination, and seedling establishment of WT, tt8-4, and tt8-4 35S:BnTT8 plants were examined in MS medium containing 100 mM NaCl or 3% (w/v) Glc. These lines exhibited similar seed germination rates and seedling establishment (defined as the formation of true leaves and root elongation) on the medium without any stress treatment (Fig. 5). Under the stress of 100 mM NaCl, seed germination vigor was lower in the tt8-4 mutant than the WT (Fig. 5a). The true leaves and roots developed poorly in the tt8-4 mutant, suggesting that the loss of AtTT8 function induced sensitivity to salinity stress (Fig. 5b). In the medium containing 3% (w/v) Glc, the rate and vigor of seed germination were significantly lower for the tt8-4 mutant line than the WT. Almost all WT seeds germinated at 7 DAS; in contrast, only about 75% of tt8-4 mutant seeds germinated (Fig. 5a). All WT plants successfully finished seedling establishment at 17 DAS, whereas most tt8-4 plants failed to develop true leaves in the medium (Fig. 5b). Both fresh and dry weights of tt8-4 seedlings were significantly lower compared with WT seedlings for each abiotic stress (Supplementary Figure S3). The phenotypes of seed germination and seedling establishment in the tt8-4 mutant under the abiotic stresses could be fully rescued by introduction of BnTT8 into the tt8-4 mutant (Fig. 5; Supplementary Figure S3). This indicated that both AtTT8 and BnTT8 had similar functions in responses to abiotic stresses of high concentrations of NaCl and Glc.
Analysis of seed germination and seedling establishment of the WT (Col-0), tt8-4, and tt8-4 35S:BnTT8 plants on MS agar medium containing 100 mM NaCl (left) or 3% (w/v) glucose (right). a Comparison of seed germination. Seed germination was scored daily after the radical tip had fully emerged from the seed coat. Data are the means from three independent experiments evaluating 200 seeds. Error bars denote standard deviations. b Comparison of seedling establishment. The pictures were taken at 17 DAS, showing the situations on the control medium (without stress, CK), MS agar medium containing 100 mM NaCl, and MS agar medium containing 3% (w/v) glucose
To better understand how BnTT8 affects seedling establishment, we analyzed the transcript levels of some stress-responsive genes in WT, tt8-4, and tt8-4 35S:BnTT8 seedlings at 17 DAS. Under control conditions, expressions of all detected stress-responsive genes were not significant, except for AtABA1, AtNCED3, and AtRAB18 (Fig. 6). Under 100 mM NaCl and 3% Glc stresses, AtABA1 expression was significantly reduced both in tt8-4 mutant and WT seedlings, with its relative level uniformly higher in WT than in the tt8-4 mutant. Expressions of AtABA2 and AtNCED3 in the tt8-4 mutant were significantly decreased under high concentrations of Glc and NaCl, respectively. AtABI2 was greatly induced in the WT and slightly induced in the mutant under both stresses. For 100 mM NaCl and 3% Glc stresses, AtADH was significantly induced both in the tt8-4 mutant and WT plants, and its relative expression level was uniformly lower in the WT than the tt8-4 mutant. In WT seedlings, the transcript level of AtRD29A was much higher under Glc stress than control conditions. The expression of AtRAB18 was significantly elevated in both the tt8-4 mutant and WT, and its relative level was uniformly much higher in the WT than the tt8-4 mutant. AtKIN1 expression was significantly increased in the WT under both stresses. In addition, expressions of AtERD15, AtRD22, and AtCBL1 in WT seedlings was significantly increased under Glc stress, whereas their expressions in the tt8-4 mutant and WT seedlings did not significantly change under both stresses. AtCBL9 expression was greatly reduced under the two stresses. Under 100 mM NaCl and 3% Glc stresses, all the measured stress-responsive genes exhibited similar expression patterns in the tt8-4 35S:BnTT8 compared with WT seedlings (Fig. 6), indicating that ectopic expression of BnTT8 could control expression of the genes involved in abiotic stresses, thus regulating the responses to abiotic stresses in Arabidopsis.
Comparison of relative transcript levels of stress-responsive genes among the WT (Col-0), tt8-4, and tt8-4 35S:BnTT8 plants under 100 mM NaCl or 3% (w/v) glucose stresses. Total RNA was isolated from whole seedlings at 17 DAS. The qPCR result was normalized against AtACTIN7 expression as an internal control, and the transcript levels are relative to the WT on the MS medium, which was set to 1. Values are the means of two replicates carried out on cDNA dilutions obtained from two independent RNA extractions. Different letters within each treatment indicate significant differences at P ≤ 0.05 (Tukey’s honestly significant difference test); each kind of letters should only be compared with like letters. Asterisks denote significant differences between indicated samples (Student’s t-test, P ≤ 0.05). Error bars denote standard deviations
Discussion
Studies have shown that AtTT8, as a basic helix–loop–helix domain transcription factor, not only regulates the biosynthesis of anthocyanins and proanthocyanidins, but also controls seed development and accumulation of seed storage reserves in Arabidopsis (Nesi et al. 2000; Baudry et al. 2006; Chen et al. 2014b). The TT8 from B. rapa is involved in the accumulation of proanthocyanidins in the seed coat of Arabidopsis (Li et al. 2012). However, limited information is available about the role of BnTT8 protein in the accumulation of seed storage reserves and seedling establishment. In this study, BnTT8 obtained from the cultivar QINYOU Seven was present in the C genome (Fig. 1; Supplementary Figure S1). TT8 proteins from Arabidopsis and B. napus shared highly conserved MYB and MYC transcription factor and helix–loop–helix DNA-binding regions (Fig. 1). The subcellular localization experiment indicated that TT8 functioned as a transcription factor (Fig. 2a). Transcripts of all TT8 copies were broadly present in different vegetative tissues (Fig. 2b), and highest in developing seeds (Fig. 2c) of cultivar QINYOU Seven, indicating that it might regulate many aspects of plant growth and development, especially seed development.
We compared the phenotypes of seed coat color and seed storage reserves including FAs and storage proteins among the WT, tt8-4, and tt8-4 35S:BnTT8 plants. Ectopic expression of BnTT8 fully rescued many phenotypes of tt8-4 seeds (Nesi et al. 2000; Chen et al. 2014b), such as yellow seed coat (Fig. 3a), higher seed FAs, and lower storage protein contents (Fig. 4). In canola developing seeds at 15 DAP, sucrose and other nutrients are delivered to the seed through the vascular tissue and then accumulate in the seed coat or endosperm. Large amounts of soluble sugar and nitrogen are present in the endosperm, accounting for about 80% of the total weight of seeds at 20 DAP (King et al. 1997; Lohaus and Moellers 2000). The seed embryo is fully developed and almost occupies the whole seed at 25 DAP, and then the seed storage reserves enter the rapid accumulation period (Dong et al. 2004; Xu et al. 2015). These, together with the observation of the highest and stable expression of all TT8 copies in canola developing seeds during 15–33 DAP (Fig. 2c), suggested that BnTT8 is important in the regulatory network that suppresses the accumulation of seed storage reserves.
This study showed that the tt8-4 mutant line was very sensitive to abiotic stresses caused by high concentrations of Glc and NaCl in the growth medium (Fig. 5; Supplementary Figure S3). The hormone abscisic acid (ABA) plays a major role in optimizing vegetative growth under stress conditions and, at the cellular level, ABA can promote tolerance to some abiotic stresses, including salinity, drought, and cold (Finkelstein et al. 2002). High concentrations of Glc inhibit seedling growth in an ABA-dependent manner, mediated by the osmotic effects of sugar and some aspects of sugar signaling (Gibson 2001; Finkelstein et al. 2002). The transcription levels of many stress-responsive genes differed between the WT and tt8 seedlings under stressful environments (Fig. 6). Under Glc and NaCl stresses, expressions of all these genes were higher in the WT and tt8-4 35S:BnTT8 seedlings than in the tt8-4 mutant except for AtADH for both stresses, and AtABA2 and AtERD15 for NaCl stress (Fig. 6). Moreover, the introduction of BnTT8 driven by the 35S promoter in the tt8-4 mutant not only fully rescued impaired seed germination and seedling establishment of the tt8-4 mutant under both salinity and Glc stresses (Fig. 5; Supplementary Figure S3), but also restored expression patterns of these stress-responsive genes in the tt8-4 mutant to WT levels (Fig. 6). FAs or FA derivatives serve as essential dietary nutrients for human beings and animals, and also facilitate successful seed germination and seedling establishment (Sullivan and Deng 2003; Li et al. 2006; Mu et al. 2008). In addition, they act as essential components of cellular membranes and cellular signal or hormone molecules protecting the plant from environmental stresses (Ohlrogge and Jaworski 1997; Hong et al. 2008; Chen et al. 2012a, b). Flavonoids play a major role in plant responses to abiotic stresses (Winkel-Shirley 2002; Petrussa et al. 2013). It is possible that deficiency of flavonoids alters the expression patterns of stress-responsive genes during seedling establishment. However, the stress-sensitive phenotypes of the tt8-4 mutant may also be caused by increased diffusion of stressful compounds through the tt8-4 seed coat. Another possible explanation is that AtTT8 and BnTT8 function as important mediators in the regulatory network by directly regulating stress-responsive genes. The FA composition linolenic acid is the primary substrate for jasmonic acid biosynthesis in plants, and its content in tt8-4 seeds was significantly increased (Supplementary Table S2; Chen et al. 2014b). A complex of bHLH factors (AtTT8, AtGL3, and AtEGL3) and MYB factors (AtMYB75 and AtGL1) was found to be required for jasmonic acid-dependent anthocyanin accumulation (Qi et al. 2011). Therefore, AtTT8 and BnTT8 probably combine these possibilities together to affect the tolerance to abiotic stresses during Arabidopsis seedling establishment.
In B. napus, yellow seeds have a significantly thinner seed coat than black seeds, and so have a lower hull proportion and higher proportions of oil and protein (Mahmood et al. 2006). The investigation and manipulation of transcription factors controlling seed coat color are of great significance for agricultural production. In this regard, BnTT8 is a promising target to genetically manipulate B. napus to increase seed oil content. However, breeders should fully consider that the loss of BnTT8 might lead to a decreased ability of B. napus against abiotic stresses during seed germination and seedling establishment.
References
Baud S, Lepiniec L (2009) Regulation of de novo fatty acid synthesis in maturing oilseeds of Arabidopsis. Plant Physiol Biochem 47(6):448–455
Baud S, Boutin JP, Miquel M, Lepiniec L, Rochat C (2002) An integrated overview of seed development in Arabidopsis thaliana ecotype WS. Plant Physiol Biochem 40(2):151–160
Baudry A, Caboche M, Lepiniec L (2006) TT8 controls its own expression in a feedback regulation involving TTG1 and homologous MYB and bHLH factors, allowing a strong and cell-specific accumulation of flavonoids in Arabidopsis thaliana. Plant J 46(5):768–779
Cernac A, Andre C, Hoffmann-Benning S, Benning C (2006) WRI1 is required for seed germination and seedling establishment. Plant Physiol 141(2):745–757
Chai GH, Bai ZT, Wei F, King GJ, Wang CG, Shi L, Dong CH, Chen H, Liu SY (2010) Brassica GLABRA2 genes: analysis of function related to seed oil content and development of functional markers. Theor Appl Genet 120(8):1597–1610
Chen MX, Du X, Zhu Y, Wang Z, Hua SJ, Li ZL, Guo WL, Zhang GP, Peng JR, Jiang LX (2012a) Seed Fatty Acid Reducer acts downstream of gibberellin signalling pathway to lower seed fatty acid storage in Arabidopsis. Plant Cell Environ 35(12):2155–2169
Chen MX, Wang Z, Zhu YN, Li ZL, Hussain N, Xuan LJ, Guo WL, Zhang GP, Jiang LX (2012b) The effect of TRANSPARENT TESTA2 on seed fatty acid biosynthesis and tolerance to environmental stresses during young seedling establishment in Arabidopsis. Plant Physiol 160(2):1023–1036
Chen MX, Maodzeka A, Zhou LH, Ali E, Wang Z, Jiang LX (2014a) Removal of DELLA repression promotes leaf senescence in Arabidopsis. Plant Sci 219:26–34
Chen MX, Xuan L, Wang Z, Zhou L, Li Z, Du X, Ali E, Zhang G, Jiang L (2014b) TRANSPARENT TESTA 8 inhibits seed fatty acid accumulation by targeting several seed development regulators in Arabidopsis. Plant Physiol 165:905–916
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16(6):735–743
Debeaujon I, Koornneef M (2000) Gibberellin requirement for Arabidopsis seed germination is determined both by testa characteristics and embryonic abscisic acid. Plant Physiol 122(2):415–424
Deng W, Chen GQ, Peng F, Truksa M, Snyder CL, Weselake RJ (2012) Transparent Testa16 plays multiple roles in plant development and is involved in lipid synthesis and embryo development in canola. Plant Physiol 160(2):978–989
Dong JZ, Keller WA, Yan W, Georges F (2004) Gene expression at early stages of Brassica napus seed development as revealed by transcript profiling of seed-abundant cDNAs. Planta 218(3):483–491
Dupont J, White PJ, Johnston KM, Heggtveit HA, McDonald BE, Grundy SM, Bonanome A (1989) Food safety and health effects of canola oil. J Am Coll Nutr 8(5):360–375
Fait A, Angelovici R, Less H, Ohad I, Urbanczyk-Wochniak E, Fernie AR, Galili G (2006) Arabidopsis seed development and germination is associated with temporally distinct metabolic switches. Plant Physiol 142(3):839–854
Finkelstein RR, Gampala SSL, Rock CD (2002) Abscisic acid signaling in seeds and seedlings. Plant Cell 14:S15–S45
Gibson SI (2001) Plant sugar-response pathways. Part of a complex regulatory web. Plant Physiol 125(4):2203–2203
Hong JK, Choi HW, Hwang IS, Kim DS, Kim NH, Choi du S, Kim YJ, Hwang BK (2008) Function of a novel GDSL-type pepper lipase gene, CaGLIP1, in disease susceptibility and abiotic stress tolerance. Planta 227(3):539–558
Jenik PD, Gillmor CS, Lukowitz W (2007) Embryonic patterning in Arabidopsis thaliana. Annu Rev Cell Dev Biol 23:207–236.
King SP, Lunn JE, Furbank RT (1997) Carbohydrate content and enzyme metabolism in developing canola siliques. Plant Physiol 114(1):153–160
Li YH, Beisson F, Pollard M, Ohlrogge J (2006) Oil content of Arabidopsis seeds: The influence of seed anatomy, light and plant-to-plant variation. Phytochemistry 67(9):904–915
Li X, Chen L, Hong M, Zhang Y, Zu F, Wen J, Yi B, Ma C, Shen J, Tu J, Fu T (2012) A large insertion in bHLH transcription factor BrTT8 resulting in yellow seed coat in Brassica rapa. PLoS ONE 7(9):e44145.
Lohaus G, Moellers C (2000) Phloem transport of amino acids in two Brassica napus L. genotypes and one B. carinata genotype in relation to their seed protein content. Planta 211(6):833–840
Mahmood T, Rahman MH, Stringam GR, Yeh F, Good AG (2006) Identification of quantitative trait loci (QTL) for oil and protein contents and their relationships with other seed quality traits in Brassica juncea. Theor Appl Genet 113(7):1211–1220
Millar JL, Khan D, Becker MG, Chan A, Dufresne A, Sumner M, Belmonte MF (2015) Chalazal seed coat development in Brassica napus. Plant Sci 241:45–54
Mu J, Tan H, Zheng Q, Fu F, Liang Y, Zhang J, Yang X, Wang T, Chong K, Wang XJ (2008) LEAFY COTYLEDON1 is a key regulator of fatty acid biosynthesis in Arabidopsis. Plant Physiol 148:1042–1054
Nesi N, Debeaujon I, Jond C, Pelletier G, Caboche M, Lepiniec L (2000) The TT8 gene encodes a basic helix-loop-helix domain protein required for expression of DFR and BAN genes in Arabidopsis siliques. Plant Cell 12(10):1863–1878
Ohlrogge JB, Jaworski JG (1997) Regulation of fatty acid synthesis. Annu Rev Plant Physiol Plant Mol Biol 48:109–136
Petrussa E, Braidot E, Zancani M, Peresson C, Bertolini A, Patui S, Vianello A (2013) Plant flavonoids-biosynthesis, transport and involvement in stress responses. Int J Mol Sci 14(7):14950–14973
Prakash S, Wu X, Bhat SR (2011) History, evolution, and domestication of Brassica crops. Plant Breed Rev 35:19–70.
Qi T, Song S, Ren Q, Wu D, Huang H, Chen Y, Fan M, Peng W, Ren C, Xie D (2011) The Jasmonate-ZIM-domain proteins interact with the WD-Repeat/bHLH/MYB complexes to regulate Jasmonate-mediated anthocyanin accumulation and trichome initiation in Arabidopsis thaliana. Plant Cell 23(5):1795–1814
Sullivan JA, Deng XW (2003) From seed to seed: the role of photoreceptors in Arabidopsis development. Dev Biol 260(2):289–297
Thelen JJ, Ohlrogge JB (2002) Metabolic engineering of fatty acid biosynthesis in plants. Metab Eng 4(1):12–21
Weber H, Borisjuk L, Wobus U (1996) Controlling seed development and seed size in Vicia faba: a role for seed coat-associated invertases and carbohydrate state. Plant J 10(5):823–834
Winkel-Shirley B (2002) Biosynthesis of flavonoids and effects of stress. Curr Opin Plant Biol 5(3):218–223
Xu HM, Kong XD, Chen F, Huang JX, Lou XY, Zhao JY (2015) Transcriptome analysis of Brassica napus pod using RNA-Seq and identification of lipid-related candidate genes. BMC Genomics 16(1):858
Yang YN, Li RG, Qi M (2000) In vivo analysis of plant promoters and transcription factors by agroinfiltration of tobacco leaves. Plant J 22(6):543–551
Zhou LH, Li YL, Hussain N, Li ZL, Wu DZ, Jiang LX (2016) Allelic variation of BnaC.TT2.a and its association with seed coat color and fatty acids in rapeseed (Brassica napus L.). PLoS ONE 11 (1): 351–363.
Acknowledgements
The work of our lab was supported by the Natural Science Foundation of China (Grant no. 31501336), Chinese Universities Scientific Fund, Science Fund for The Cultivation of The Excellent Youth Scholars (Z109021517) and Startup Fund for Talents (Z111021402) of Northwest A&F University.
Author information
Authors and Affiliations
Corresponding author
Additional information
Shuanghui Qi and Kaige Liu they have contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Qi, S., Liu, K., Gao, C. et al. The effect of BnTT8 on accumulation of seed storage reserves and tolerance to abiotic stresses during Arabidopsis seedling establishment. Plant Growth Regul 82, 271–280 (2017). https://doi.org/10.1007/s10725-017-0257-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10725-017-0257-4