Abstract
Shock-like water stress using hydroponics and gradual water deficit in soil are the two widely used treatments to analyze transcriptional response of many crops to drought. In this study, we investigated the effects of shock drought (ShD) (0, 0.5, 1, 4, and 8 h) and slow drought (SDD) [soil water content (SWC) of 35 and 50 %] on the expression of well-known drought-responsive genes supplemented with physiological changes in barley. Two barley cultivars with contrasting leaf relative water content (RWC) and water loss rate (WLR) values were selected as Martı (MR; 60 % RWC and 0.046 gh−1 g−1 DW) and Erginel90 (ER; 38.3 % RWC and 0.350 gh−1 g−1 DW) under 38 % of SWC condition. According to the results, 0.5 h ShD was the critical time point for stress perception in leaves defined by the increase in WLR, ion leakage and H2O2 concentration. Expressions of antioxidant-related genes (Cu–Zn/SOD, HvCAT2, HvGST6, HvAPX) were rapidly induced in MR at 8 h shock, while only slightly upregulated in ER. We have also observed higher induction of expressions of HvBAS1, HvMT-2, HvABA7 and a photosynthesis-related gene HvLHCB during ShD compared to SDD. Contrarily, transcription factors (TFs), HvWRKY12 and HvDRF1 were expressed with lower values during shock-dehydration. Slow-drought treatments in both cultivars were characterized with high leaf RWCs and osmotic adjustment with low cell membrane damage, suggesting that barley maintains a basal tolerance to long-term water deficit. Our results confirmed that type of water stress treatment is crucial to measure gene expression, and a shock-like dehydration method should be the treatment of choice in evaluating barley plants with different physiological characteristics for water tolerance.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
To a great extent, grain yield in barley depends on annual precipitation and soil water availability, as it is mainly cultivated in marginal drylands. Climate changes around the world with decreasing regional annual rainfall and increasing temperatures emphasize the importance of the development or selection of drought tolerant cultivars from germplasm collections, especially for agriculturally important crops (Cattivelli et al. 2008; Mir et al. 2012).
Plants mainly respond to water deficit by reducing water loss in leaves through stomatal closure causing reduction in photosynthesis and by metabolic protection via accumulation of compatible solutes or production of antioxidant enzymes against the adverse effects of oxidative stress (Cushman and Bohnert 2000; Ramanjulu and Bartels 2002; Bandurska et al. 2013; Grzesiak et al. 2013). A rapid accumulation of reactive oxygen species (ROS), mainly hydrogen peroxide (H2O2), was observed during drought stress adaptation in many plants (Apel and Hirt 2004; Miller et al. 2010). H2O2 may also act as a signaling molecule for stress acclimation, programmed cell death (Gechev and Hille 2005; Balazadeh et al. 2011) or other stress responses including stomatal regulation (Xu et al. 2010) and signaling between root and shoots (Yang et al. 2006). A diverse group of enzymes including ascorbate peroxidases (APXs), superoxide dismutases (SODs), and catalases (CATs) are synthesized in the cell to prevent ROS accumulation. Glutathione-S-transferases (GSTs) are involved in the protection of tissues against oxidative damage and their expression is induced by pathogens, cellular stress and/or growth factors (Chen et al. 1996). Some GSTs have also shown to be highly transcribed in drought tolerant barley cultivars in transcriptomic studies (Guo et al. 2009; Rezaei et al. 2013).
Water stress in plants can be developed slowly (within days, weeks, and months) or rapidly (within hours to days) (Chaves et al. 2003). Rapid or slow-developing water stress may have different results in a particular species, even in cultivars of the same species, and mostly depends on the genotype and environment interactions (McDonald and Davies 1996; de Mezer et al. 2014). Slow-developing stress can easily be mimicked in the laboratory by withholding the water from pots, and rapid-developing water stress treatment can be performed by taking plants out of hydroponics, or even pots, and leaving them out for a certain period of time; a shock-like water deficit treatment. Both slow-developing and shock-like dehydration stress treatments have been previously used for barley in evaluation of expressional changes of water stress responsive genes through cDNA microarrays or RNA-sequencing (Table 1). To our best of knowledge, there is only one study in literature comparing the transcriptome of barley in response to two different water stress conditions through a microarray platform (Talame et al. 2007).
The role of water stress regime on the gene expression and physiological response are not completely clear in barley. Moreover, the type of water stress may be implicated with the selection of drought-tolerant genotypes under laboratory conditions, which is a crucial step for developing stress tolerant breeding lines as well as performing comparative transcriptomics analyses. The main objective of this study, thus, was to evaluate the effects of shock-like dehydration and slowly developing drought on the mRNA transcript levels of the major drought responsive genes, in parallel to the physiological changes in barley leaves. To achieve this, we have identified two contrasting genotypes from barley cultivars of Turkey origin by assessing the leaf water loss rate (WLR) and relative water content (RWC), and measured the expression levels of several antioxidant-related genes (HvGST6, Cu–Zn/SOD, HvAPX, HvCAT2) and six additional genes related to water stress response comparatively in both cultivars.
Materials and methods
Plant material
Seeds of germplasm material were obtained from The Central Research Institute for Fields Crops (Ankara, Turkey), The Aegean Agricultural Research Institute (Izmir, Turkey) and The Trakya Agricultural Research Institute (Edirne, Turkey) and repropagated in the field before screening. Hordeum vulgare L. cultivars Martı (MR), and Erginel90 (ER), were selected as two contrasting genotypes for their water stress tolerance in a screening with ten breeding barley cultivars (Bolayır, Bornova92, Erginel90, Martı, Kaya, Serife Hanım, Suleyman Bey, Tarm-92, Tokak 157/37, Vamık Hoca) (Table S1). The experiment was arranged in a complete randomized block design with 12 containers including one replicate of each of the cultivar. Screening was performed by assaying leaf RWC and WLR after 10 days of water stress (down to 38 % SWC) as described by Suprunova et al. (2004).
Water stress treatments and sampling
Shock drought (ShD) of selected contrasting cultivars, MR and ER, was performed as described by Ergen et al. (2009) with minor modifications. Seeds were surface sterilized with 0.6 % NaOCl and germinated in vials containing perlite. Seedlings of a similar developmental stages (Zadocks scale 12; 70–90 cm) were transferred to 5 l containers filled with half-strength Hoagland’s solution (Hoagland and Arnon 1950) and grown under controlled conditions (25 °C, 40–50 % relative humidity and 8 h light/16 h dark photoperiod with a light intensity of 600 μMol m−2 s−1) while the nutrient solution was renewed in every 3–4 days. ShD was performed 10 days after transfer to hydroponics (3-leaf stage). Plants were removed from containers and placed on filter papers inside the plant growth chamber, while control plants were kept in fresh hydroponic solutions under the same conditions. Fully expanded leaf segments from three plants per cultivar were harvested after 0.5, 1, 4, and 8 h from both control and ShD treated plants during light period (between 8.00 and 16.00). Leaves were directly frozen in liquid nitrogen and storen at −80 °C. 0 h represents the time point when plants were taken out of containers.
For slow drought (SDD) treatment, barley seeds were surface sterilized with 0.6 % NaOCl and germinated on filter papers. Five-day-old seedlings were transferred to pots containing rich organic soil. Eight pots per cultivar were used and pots were arranged in two random blocks in plant growth chambers (EKOCHL 1500, Angelantoni, Massa Martana, Italy), to grow under controlled conditions (25 °C, 40–50 % relative humidity and 8 h light/16 h dark photoperiod with a light intensity of 600 μMol m−2 s−1) with changing their position in the block every other day. Soil water content (SWC) in the pots was adjusted to 70–75 % by weighing and plants were kept well-watered until the 3-leaf stage. Dehydration was performed by withholding the water while closely monitoring the development of water stress by continuous measurement of the relative SWC [SWC = ((FW − DW)/(MW − DW)) × 100 %; FW, fresh weight; DW, dry weight; MW, weight of the soil at 100 % of field capacity] (Teulat et al. 1997). Fully expanded leaf segments from three plants per cultivar were harvested when SWC of stressed plants were decreased to 50 and 35 % in approximately 26 and 55 days respectively, and directly frozen in liquid nitrogen. SWC of control plants was kept at 70 %. Samples from control plants were collected at the same time points.
Assaying leaf WLR
WLR for ShD experiment was calculated according to Ristic and Jenks (2002). The FW values of first leaves were recorded as T0 (0 time point). Weight records were then taken by periods of 0.5, 1, 4, and 8 h (T0.5, T1, T4 and T8). Leaf samples were incubated at 80 °C for 24 h and dry weights (DW) were recorded. WLR values were calculated according to the following formula:
All measurements were performed as three biological replicates.
Ion leakage, osmolality, and biomass measurements
For measurement of ion leakage in cell walls, five samples were taken from elongated parts of the leaves at different time points. Samples were then washed three times with distilled water, capped and placed in a thermostatically controlled chamber maintained at 25 °C in the dark. After 16 h, 60 μl of the sample was used for conductivity measurement (E1) using a conductivity meter (HORIBA Scientific, NJ, USA). The tubes were then autoclaved for 20 min and the measurement was repeated to record E2 values. Ion leakage was calculated as E1/E2 × 100.
Leaf sap osmolality was measured by a semi-micro osmometer (Knauer K-7400, Germany). Two leaf samples from the same time point were collected and stored at −20 °C overnight. Cell sap was obtained by centrifugation at 4 °C for 30 min at 15,000×g. Leaf sap osmolality was determined using 150 µl sample and expressed as mosmol kg−1. Biomass (dry matter) of 12–15 water-stressed plants (SWC final 35 %) and control plants of contrasting cultivars was measured by weighing the whole plants after drying at 80 °C for 24 h.
Determination of H2O2 content
H2O2 was determined as putative hydroperoxides in leave segments of the plants using PeroxiDetect KIT (Sigma PD1) according to the manufacturer’s procedure. To briefly elaborate, leaf samples (five from each cultivar and treatment) were weighed, frozen in liquid nitrogen and homogenized in a cold mortar. After the addition of 100 µl of cold distilled water, samples were centrifuged at 4 °C at 18,000×g for 5 min. Working Color Reagent (200 μl) was added to 20 µl of supernatant and the mixture was incubated at room temperature for 30 min. Final accumulation of Fe3–xylenol orange complex was determined by spectrophotometry (µQuant, Bio-Tek Inc., VT, USA) at 560 nm. H2O2 content was determined using a hydrogen peroxide standard curve.
Total RNA extraction and PCR analyses
Total RNA was isolated with the TRIZOL® reagent (Invitrogen, Grand Island, NY, USA) according to manufacturer’s instructions and treated with DNase I (Fermentas, Vilnius, Lithuania) in a final volume of 100 μl. For both cultivars and for all stress applications, three biological replicates of two pooled plants were used for total RNA isolation. RNA quality was controlled by gel electrophoresis and quantified by spectrophotometer.
The change in the expressions of selected antioxidant enzymes, namely Cu/Zn SOD (copper-zinc superoxide dismutase), HvGST6 (glutathione S-transferase), HvAPX (ascorbate peroxidase) and HvCAT2 (catalase 2) were assessed by RT-PCR analyses where α-TUB (alpha-tubulin) was used for loading control. Primers used in RT-PCR and qRT-PCR analyses were designed by NCBI Primer-BLAST program. Details of the primers are given in Table S1. RT-PCR was performed in a total volume of 50 μl containing 500 ng RNA, 2× AccessQuick™ Master Mix (Promega, Madison, USA), 0.5 µM of forward and reverse primers and 5 U AMV reverse transcriptase at 45 °C for 50 min followed by enzyme inactivation at 94 °C for 2 min. PCR was performed as 35 cycles of 94 °C for 30 s, 60 °C for 30 s and 72 °C for 30 s with a final extension step for 10 min. RT-PCR was done in triplicates and amplified samples were analyzed on 1 % agarose gel.
For qRT-PCR analysis, first strand cDNA synthesis was performed using 4 µg of total RNA, 100 U of SuperScriptII RT (Invitrogen) and 1 µg of Oligo(dT)20 primer in a final volume of 40 μl. LightCycler 480 II (Roche Diagnostics, Mannheim, Germany) was used for the qRT-PCR analysis. The amplifications were performed in a total volume of 10 μl containing 4 μl of cDNA (1:4 diluted), 1 U of GoTaq DNA polymerase (Promega), 0.2 mM dNTP, 2.5 mM MgCl2, 0.2 μM of each primer, and 1 μl of EvaGreen (Biotium). PCR was performed at 95 °C for 5 min and 40 cycles of 95 °C for 20 s, 60 °C for 20 s, 72 °C for 20 s followed by a melting curve analysis with a temperature gradient of 0.11 °C s−1 from 95 to 65 °C. Standard curves were prepared from a dilution series of vectors containing cDNAs as follows; each cDNA was amplified by PCR from control RNA samples and ligated to pTZ57R/T vector by InsTAclone PCR Cloning Kit (Thermo Scientific). The resulting vectors were diluted with the copy numbers arranged from 109 to 104. Raw data was handled with the LightCycler 480 II Software (Roche, version 1.5.0), and target gene expression was normalized using the barley actin (HvACT) gene expression values for each sample.
Statistical analysis
The data were analyzed by SPSS software version 21 (Chicago, IL, USA) using one-way or two-way analysis of variance (ANOVA). Significance of differences between treatment means was calculated by Tukey’s studentized range (HSD) test. Data obtained from physiological responses of two barley cultivars (MR and ER) under ShD and SDD treatments were analyzed with two-way ANOVA at 95 % CI level with Bonferroni’s posthoc test. To compare gene expression values, t test assuming homogeneous variances was performed in Excel, with 0.05 CI level. The correlations between the four physiological parameters and expressions of three genes were calculated for ShD treatment by the Spearman’s rank correlation coefficient using Microsoft Excel.
Results
Effects of ShD and SDD treatments on plant physiology
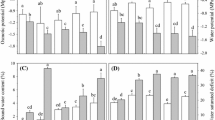
Figure 1 summarizes ion leakage, H2O2 content, osmolality, and WLR of contrasting cultivars in response to ShD at different time points and SDD at different SWC. During ShD treatment, the percentage ion leakage of ER rapidly increased in the first half an hour and started to decrease at the 8th hour, whereas that of MR showed a stable response during the first 4 h of ShD and started to increase at the 8th hour, emphasizing the contrasting response of selected barley cultivars to water deficit (Fig. 1a). The difference in the percentage ion leakage of ER and MR cultivars was 5.8-fold at t = 0.5 (Fig. 1a). In contrast to ShD treatment, ion leakage in cell walls of contrasting cultivars in SDD treatment was relatively stable in all SWC data points, and the increase in ion leakage with decreasing SWC was similar in contrasting cultivars (Fig. 1b). The measurement of H2O2 content of contrasting cultivars in response to ShD and SDD treatments indicated dehydration and cultivar dependent differences. Accumulation of H2O2 in the leaves of MR became significant at 0.5 h (Fig. 1c) but was stabilized at subsequent hours. H2O2 accumulation in ER was developed after 8 h of dehydration and the difference between the cultivars was more pronounced at this time point. The H2O2 content values were similar in both ShD and SDD treatments; however, the accumulation of H2O2 in response to SDD treatment was less pronounced in both cultivars. It is important to note that a significant increase in H2O2 content in MR at t = 0.5 during ShD treatment was observed in response to severe drought (35 % SWC) during SDD treatment (Fig. 1d). Osmolality of both cultivars was increased by 4th hour during ShD treatment with a significant difference between the cultivars (Fig. 1e). Data revealed that control plants grown in standard soil conditions showed a higher osmolality, therefore higher osmotic potential, compared to the ones grown in hydroponic conditions (Fig. 1e, f). During severe stress (35 % SWC), osmolality was measured slightly lower than 50 and 70 % of SWC conditions, as an average 1146 mosmol kg−1 in both genotypes.
Physiological responses of two barley cultivars (MR and ER) under ShD and SDD treatments. a, b Ion leakage; b, c H2O2 content; e, f osmolality; g epidermal water loss rate (WLR) under ShD treatment according to Ristic and Jenks (2002). Data represent mean ± SD of at least 12 plants per cultivar and analyzed with two-way ANOVA at 95 % CI level with Bonferroni’s posthoc test. P values for significance level were indicated with above asterisks (*P < 0.05; **P < 0.01; ***P < 0.001)
The changes in WLR of selected cultivars with respect to time after ShD are shown in Fig. 1g. In both cultivars, WLR values significantly decreased in the first half an hour of ShD (t = 0.5) (P ≤ 0.05 and P ≤ 0.005 in MR and ER, respectively) compared to other time points. The comparison of WLR values at different time points indicated significant difference between contrasting cultivars only at 4th hour of ShD (P = 0.041) (Fig. 1g).
Expression patterns of stress-responsive genes in response to ShD and SDD
All antioxidant enzymes tested were actively transcribed in the leaf tissues of MR and ER in response to both treatments (Fig. 2). The differences in the expression patterns of some antioxidant enzymes in response to different treatments indicated cultivar- and time-dependent responses. During ShD treatment, Cu/Zn-SOD expression was gradually increased with time and showed a strong similarity in contrasting cultivars (Fig. 2a). Transcript levels of HvGST6 and HvCAT2 were increased by time in MR, contrary to ER in which expression of both genes were decreased after 1 h (Fig. 2a). Differences between HvAPX expressions were also clear between two cultivars, where HvAPX was almost constitutively expressed in MR (Fig. 2a). While the expression pattern of Cu/Zn-SOD in MR was similar in ShD and SDD treatments, its expression was increased in ER plants by SDD (Fig. 2b). Similarly, HvGST6 and HvCAT2 expressions were induced by time in ER in response to SDD, contrary to the decrease in expression level observed in response to ShD (Fig. 2). Time dependent accumulation of HvCAT2 transcription in MR during ShD treatment was not detectable in response to SDD (Fig. 2). HvAPX transcript levels were consistent in MR in response to both treatments; however, in contrast to ShD treatment, the gene expression level of HvAPX induced with the decrease in SWC in the sensitive cultivar ER.
Quantitative RT-PCR data provided more evidence on the differences of transcript accumulations of other stress-responsive genes with respect to stress treatment and cultivar. Results indicated that the primer pairs corresponding to HvMT2, HvBAS1, HvABA7, HvACT, HvLHCB, HvDRF1 and HvWRKY12 genes worked ideally for development of the qRT-PCR assay to monitor differential expression of these genes under two different water stress treatments showing single product specific melting temperatures (data no shown). Of the six genes studied, the highest expression level was observed in HvMT2 and the lowest in HvDRF1 and HvWRKY12 (Fig. 3). As in the case of semi-quantitative PCR, the expression patterns of stress-responsive genes showed no clear similarities between ShD and SDD treatments (Fig. 3). In general, transcript levels normalized with actin expression were much higher in ShD than SDD with the exception of HvDRF1 and HvWRKY12 which showed a contrary response (Fig. 3). It is important to note that upregulation of HvBAS1, HvMT2 and HvDRF1 was significant in MR cultivar during the first half hour of ShD treatment, which was shown to be a critical time-point in physiological studies (Fig. 1). In terms of HvABA7 expression, genotypes showed significant difference in the ShD treatment and an increase was observed at 8th hour in MR (Fig. 3). Relative comparison of transcriptional regulation of these genes in control and stressed plants by a heat-map also showed that HvABA7 was up-regulated in both varieties at 8th hour (Fig. 3b). The expression level of HvWRKY12 gene showed slight similarity in contrasting cultivars in response to both treatments with a rather stable transcript amount in both cultivars during ShD time points and a decrease in response to 50 % SWC and an increase in 35 % (Fig. 3). Heat-map analysis also confirmed the upregulation of TFs only in SDD treatment (Fig. 3b).
a Relative transcription levels of water stress responsive genes in the leaves of MR and ER plants under ShD and SDD treatments. Transcript levels were measured by qRT-PCR in HvBAS1, HvMT2, HvLHCB, HvABA7, HvDRF1 and HvWRKY12 normalized to HvACT. Error bars represent standard deviations of three biological replicates with two technical repeats. Asterisks indicate statistical significance at *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001 of the differences between MR and ER according to t test. b qRT-PCR data in a represented in a heat map
Evaluation of genotypes and treatments under ShD and SDD
Exposure of shock dehydration caused a gradual increase in leaf osmolyte accumulation, WLR, ion leakage, H2O2 concentration and the expression of some genes during ShD treatment. A high significant correlation between osmolality and electrolyte leakage was observed in MR during ShD, while there was no such correlation in ER (Table S3). Correlation between osmolality and HvABA7 expression was high in both genotypes, but significant only in tolerant genotype, MR. There was positive but not significant correlation between HvMT2 and HvBAS1 expressions in ShD (Table S3). In MR, a significant positive correlation was found between H2O2 accumulation and epidermal water loss. Ion leakage was correlated with WLR in ER.
During SDD treatments, plants grown in control (70 % SWC) and severe stress (35 % SWC) conditions were visually compared (Fig. 4). MR is a lodging-tolerant cultivar under standard growth conditions and partly lost this phenotype during water stress imposition. Both genotypes showed wilting in the leaf tips and suppressed growth under 35 % SWC conditions (Fig. 4). The mean RWC decreased in both cultivars in response to SDD; however, the difference in the mean RWC of contrasting cultivars was significant only at severe drought stress (SWC 35 %) (P ≤ 0.05). Biomass was significantly reduced by SDD in both cultivars without a genotypic difference (Fig. 4b).
a Appearance of MR and ER plants during SDD treatments. b Relative water content (% RWC) and biomass measurements in two barley cultivars (MR and ER) in SDD treatment. Twelve plants per treatment were used for RWC. Dry weight biomass values are given as mean ± SD of six whole plants per cultivar. Different letters indicate significant differences at P ≤ 0.05 using least square means
Discussion
A significant relationship between osmotic adjustment and drought tolerance as well as RWC and soluble sugar content was previously shown in wheat and barley (Teulat et al. 1997; Suprunova et al. 2004; Shao et al. 2009). Based on this, the selection of contrasting cultivars was performed by screening ten Turkish barley genotypes with respect to RWC % and WLR in a slow-developing drought stress treatment (Table S2). Accordingly, two 6-rowed cultivars, namely Martı (MR) and Erginel90 (ER), showing contrasting leaf RWC and WLR values were selected for further analyses. The comparison of change in WLR of two cultivars in response to ShD indicated a sharp decrease in the first half hour of water deficit (t = 0.5), which showed that the development of water stress, i.e. water loss, was relatively fast and almost the same in both cultivars (Fig. 1g). The RWC % values of cultivars in response to SDD, on the other hand, were higher even in severe drought stress (average 85.7 % for MR and 71.6 % for ER) with a significant difference between them (Fig. 4b). Even without further watering in a slow-developing water stress experiment in pots, it is possible that there is still some soil moisture left, thus, plants that have more tolerance to water deficit might use this water more efficiently than sensitive ones. Therefore, only slight decrease in RWC observed in SDD experiment may be related higher water use efficiency of the barley plants grown under 35 % SWC. At the same plants, decrease in osmolality may be due to different solute partitioning between leaves at severe drought conditions. Previous studies suggest the main difference between contrasting genotypes might be the early sensing and activation of defense mechanisms in response to water deficit by drought tolerant plants (Ergen et al. 2009). In the case of ShD treatment, however, both plants were subjected to the same conditions where there was no more water in the environment (they were taken out of hydroponics). Therefore, rapid- and slow-developing water stress conditions used in this study should be different with respect to their effect on plant stress response mechanisms as suggested in literature (Talame et al. 2007).
Figure 1a summarizes the change in ion leakage of contrasting cultivars in response to ShD and SDD treatments. Ion leakage from the plant cells may be an indication of the level of cell membrane injury induced by drought stress (Bajji et al. 2002). The comparison indicated that, similar to WLR in ShD experiment, 0.5 h was critical time point for increase in ion leakage in sensitive cultivar, and the difference in ion leakage between contrasting cultivars were more pronounced. Lower values of ion leakage (6.4–30.6 %, ShD, MR) might even indicate a possible protection of membrane integrity (Babu et al. 2004; Kocheva et al. 2014). The ion leakage in SDD treatment showed a similar change pattern with the decrease in SWC in both barley cultivars with contrasting responses to drought (Fig. 1b). It appears that ion leakage might provide reliable background for the selection of contrasting genotypes with their water tolerance only in rapid-developing water stress conditions. It is important to note that a significant difference in percentage ion leakage between contrasting cultivars might also develop in response to SDD; however, during a SDD treatment, it is not easy to select the correct time points to detect such a difference.
Hydrogen peroxide (H2O2), a nonradical reduction product of oxygen, has a key role in the programmed cell death and other biological events including growth, development and environmental stress response (Gadjev et al. 2008; Li et al. 2011; Richards et al. 2014). Figure 1c, d summarizes the change in the cellular H2O2 concentration in response to ShD and SDD treatments. The comparison of values from contrasting cultivars in ShD and SDD treatments indicated that H2O2 concentration was relatively independent of the type of water deficit, i.e. time for stress development. The semi-quantitative RT-PCR results of several known H2O2 detoxifying enzymes are given in Fig. 2. In most cases, especially in ShD treatment, the expression patterns of antioxidant enzymes were completely opposite in MR and ER. For example, the expression of antioxidant enzymes HvGST6 (glutathione S-transferase 6) and HvCAT2 (catalase 2) started earlier in ER; however, their expression were either decreased (for HvGST6) or completely stopped (for HvCAT2) with time. The expression of the same genes in MR was in very low amounts during the first hours of ShD treatment, while the decrease in WLR was the highest. Their accumulation was observed after 0.5 h of rapid water deficit. In contrast, H2O2 content was gradually increased in sensitive cultivar, which may be caused by late expression of antioxidant gene expression (Fig. 1c). The comparison of expression of antioxidant enzymes in response to SDD treatment showed that H2O2 increase in MR at 35 % SWC was parallel to the decrease in HvCAT2 expression, while there was more transcript accumulation of this gene in ER. It is important to note that transcriptional data are not necessarily correlated with the enzyme activities (Furlan et al. 2014), and therefore our results provide only transcriptional indication of the involvement of the examined genes in ShD and SDD treatments.
We have used the HvACT gene for reference in our qRT-PCR analyses. Since experimental validation is suggested for the utility of reference genes for each study (Bustin et al. 2009); we have tested α-tubulin and GAPDH (glyceraldehyde-3-phosphate dehydrogenase) other than actin (HvACT) in our preliminary experiments with MR and ER. We have observed differential expression with GAPDH in leaf samples in several conditions tested (Data not shown). Conversely, the expression of HvACT and α-tubulin was relatively stable and not affected from water deficit treatments. Since actin was previously reported as one of the optimal reference genes in barley leaf under low-temperature stresses (Janska et al. 2013), we have preferred the expression of this gene for normalization. Our opposite results with GAPDH, once more, showed that commonly used reference genes should be tested for each cultivar and stress treatment to obtain reliable and repeatable results. As most of the stress-responsive genes show a diurnal rhythm of expression (Habte et al. 2014), we performed the sampling during the light period for both genotypes. As suggested by Ozturk et al. (2002) and Talame et al. (2007), the effect of rapid-developing water deficit on stress response mechanisms was more pronounced compared to slow-developing water deficit.
The normalized expression values of most of the drought responsive genes quantified in this study were several folds higher in response to ShD when compared to SDD treatment (Fig. 3). In addition, the comparison of normalized expression values of HvABA7, an important indicator of ABA-signaling and reported to be highly induced in salinity-stressed barley (Grillo and Leone 1996; Walia et al. 2006); HvBAS1, a peroxiredoxin gene upregulated by combined drought and heat stress in barley (Rollins et al. 2013); and HvMT2, a metallothionein-like protein type 2 that was reported to be highly expressed in barley leaf and roots after exposure to 150 mM NaCl (Ozturk et al. 2002), indicated water stress regime and cultivar dependent responses (Fig. 3). Interestingly, induction of HvBAS1 and HvMT2 in barley roots by combined stress of salinity and heat shock were also recently reported (Faralli et al. 2015), and the transient induction of these two genes in response to 50 % decrease in SWC supported the early sensing of soil water loss by the tolerant cultivar and activation of defense mechanisms in response to water deficit (Ergen et al. 2009). Upregulation of chlorophyll a-b binding protein of LHCII type III was previously shown in response to heat stress (Rollins et al. 2013) and HvLHCB expression was not shown to be significantly induced by osmotic stress and drought treatment in barley (Habte et al. 2014). Therefore as expected, both ShD and SDD treatments used in this study did not cause a significant increase in HvLHCB transcription level (Fig. 3). The most interesting observation was related with the normalized expression levels of two drought or abiotic stress related transcription factors functioning on ABA-dependent stress response pathway (Xue and Loveridge 2004; Rushton et al. 2010), namely HvDRF1 and HvWRKY12. The expression levels of these two genes were several folds higher in response to SDD compared to ShD treatment, where the effect of water loss was more than that of SDD (Fig. 3). It is known that after the induction of the stress responsive pathways, the expression of most transcription factors either is lost or stays at very low levels. Therefore, the low amounts of expression of these two transcription factors in response to ShD might be related without been able to detect the accumulation during induction of signaling, especially in more drought tolerant cultivar MR, since a significant increase in both genes were observed in earlier time points. These results showed the importance of selection of water stress regimes when evaluating gene specific transcript amounts or in comparison of total transcriptome.
In conclusion, our time-course dehydration experiment showed that in both MR and ER cultivars, there were positive correlations between osmolality and HvABA7 expression as well as H2O2 accumulation and leaf WLR. Both solute accumulation and HvABA7 gene expression could be coincided by abscisic acid accumulation in cell during shock dehydration (Table S3). Similarly, the role of abscisic acid accumulation in response to osmotic stress during cellular dehydration was reported in plants (Fujita et al. 2011), however the direct correlation between the expression of ABA-responsive gene and solute accumulation is less known in barley. Our data clearly proven that water stress regime directly influences the expression of drought responsive genes and its effects were most obvious on the expression of antioxidant system genes (HvCAT2, HvAPX, HvGST6, HvMT2, HvBAS1) in barley cultivars with contrasting response to drought conditions. Even though qRT-PCR data seems complex compared the semi-quantitative PCR; the comparison of detection limits clearly represents the shock-like effects of rapid-developing water deficit on plant drought stress response mechanisms. Transcriptional data of individual genes obtained under comparative water stress conditions would contribute to better understanding of stress perception and response in barley that is an important cereal grass for functional genomics studies.
References
Abebe T, Melmaiee K, Berg V, Wise RP (2010) Drought response in the spikes of barley: gene expression in the lemma, palea, awn, and seed. Funct Integr Genomics 10:191–205
Apel K, Hirt H (2004) Reactive oxygen species: metabolism, oxidative stress and signal transduction. Annu Rev Plant Biol 55:373–399
Atienza SG, Faccioli P, Perrotta G, Dalfino G, Zschiesche W, Humbeck K, Stanca AM, Cattivelli L (2004) Large scale analysis of transcripts abundance in barley subjected to several single and combined abiotic stress conditions. Plant Sci 167:1359–1365
Babu RC, Zhang JX, Blum A, Ho THD, Wu R, Nguyen HT (2004) HVA1, a LEA gene from barley confers dehydration tolerance in transgenic rice (Oryza sativa L.) via cell membrane protection. Plant Sci 166:855–862
Bajji M, Kinet J, Lutts S (2002) The use of the electrolyte leakage method for assessing cell membrane stability as a water stress tolerance test in durum wheat. Plant Growth Regul 36:61–70
Balazadeh S, Kwasniewski M, Caldana C, Mehrnia M, Zanor MI, Xue G-P, Mueller-Roeber B (2011) ORS1, an H2O2-responsive NAC transcription factor, controls senescence in Arabidopsis thaliana. Mol Plant 4:346–360
Bandurska H, Niedziela J, Chadzinikolau T (2013) Separate and combined responses to water deficit and UV-B radiation. Plant Sci 213:98–105
Bedada G, Westerbergh A, Müller T, Galkin, E, Bdolach E, Moshelion M, Fridman E, Schmid KJ (2014) Transcriptome sequencing of two wild barley (Hordeum spontaneum L.) ecotypes differentially adapted to drought stress reveals ecotype-specific transcripts. BMC Genomics 15:995
Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT (2009) The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem 55(4):611–622
Cattivelli L, Rizza F, Badeck FW, Mazzucotelli E, Mastrangelo AM, Francia E, Marè C, Tondelli A, Stanca AM (2008) Drought tolerance improvement in crop plants: an integrated view from breeding to genomics. Field Crop Res 105:1–14
Chaves MM, Maroco JP, Pereira JS (2003) Understanding plant responses to drought from genes to the whole plant. Funct Plant Biol 30:239–264
Chen W, Chao G, Singh KB (1996) The promoter of a H2O2-inducible, Arabidopsis glutathione S-transferase gene contains closely linked OBF- and OBP1-binding sites. Plant J 10:955–966
Cushman JC, Bohnert HJ (2000) Genomic approaches to plant stress tolerance. Curr Opin Plant Biol 3:117–124
de Mezer M, Turska-Taraska A, Kaczmarek Z, Glowacka K, Swarcewicz B, Rorat T (2014) Differential physiological and molecular response of barley genotypes to water deficit. Plant Physiol Biochem 80:234–248
Ergen ZN, Thimmapuram J, Bohnert HJ, Budak H (2009) Transcriptome pathways unique to dehydration tolerant relatives of modern wheat. Funct Integr Genomics 9:377–396
Faralli M, Lektemur C, Rosellini D, Gürel F (2015) Effects of heat shock on salinity tolerance in barley (Hordeum vulgare L.): plant growth and stress-related gene transcription. Biol Plant 59(3):537–546
Fujita Y, Fujita M, Shinozaki K, Yamaguchi-Shinozaki K (2011) ABA-mediated transcriptional regulation in response to osmotic stress in plants. J Plant Res 124:509–525
Furlan AL, Eliana B, Carmen TM, Castro S, Dietz K-J (2014) Antioxidant enzyme activities and gene expression patterns in peanut nodules during a drought and rehydration cycle. Funct Plant Biol 41:704–713
Gadjev I, Stone JM, Gechev TS (2008) Programmed cell death in plants: new insights into redox regulation and the role of hydrogen peroxide. Int Rev Cell Mol Biol 270:87–144
Gechev TS, Hille J (2005) Hydrogen peroxide as a signal controlling plant programmed cell death. J Cell Biol 168:17–20
Grillo S, Leone A (1996) Physical stresses in plants: genes and their products for tolerance. Springer, Berlin
Grzesiak M, Filek M, Barbasz A, Kreczmer B, Hartikainen H (2013) Relationships between polyamines, ethylene, osmoprotectants and antioxidant enzymes activities in wheat seedlings after shortterm PEG- and NaCl-induced stresses. Plant Growth Regul 69:177–189
Guo P, Baum M, Grando S, Ceccarelli S, Bai G, Li R, von Korff M, Varshney RK, Graner A, Valkoun J (2009) Differentially expressed genes between drought-tolerant and drought-sensitive barley genotypes in response to drought stress during the reproductive stage. J Exp Bot 60:3531–3544
Habte E, Müller LM, Shtaya M, Davis SJ, von Korff M (2014) Osmotic stress at the barley root affects expression of circadian clock genes in the shoot. Plant Cell Environ 37:1321–1337
Hoagland DR, Arnon DI (1950) The water-culture method for growing plants without soil. Calif Agric Exp Stn Circ 347:1–32
Janska A, Hodek J, Svoboda P, Zamecnik J, Prasil IT, Vlasakova E, Milella L, Ovesna J (2013) The choice of reference gene set for assessing gene expression in barley (Hordeum vulgare L.) under low temperature and drought stress. Mol Genet Genomics 288(11):639–649
Kocheva KV, Georgiev GI, Kochev VK (2014) An improvement of the diffusion model for assessment of drought stress in plant tissues. Physiol Plant 150:88–94
Li J, Wang X, Zhang Y, Jia H, Bi Y (2011) cGMP regulates hydrogen peroxide accumulation in calcium-dependent salt resistance pathway in Arabidopsis thaliana roots. Planta 234:709–722
McDonald AJS, Davies WJ (1996) Keeping in touch: responses of the whole plant to deficits in water and nitrogen supply. Adv Bot Res 22:229–300
Miller G, Suzuki N, Ciftci-Yilmaz S, Mittler R (2010) Reactive oxygen species homeostasis and signalling during drought and salinity stresses. Plant Cell Environ 33:453–467
Mir RR, Zaman-Allah M, Sreenivasulu N, Trethowan R (2012) Integrated genomics, physiology and breeding approaches for improving drought tolerance in crops. Theor Appl Genet 125:625–645
Ozturk ZN, Talame V, Deyholos M, Michalowski CB, Galbraithe DW, Gözükırmızı N, Tuberosa R, Bohnert HJ (2002) Monitoring large-scale changes in transcript abundance in drought- and salt-stressed barley. Plant Mol Biol 48:551–573
Ramanjulu S, Bartels D (2002) Drought- and desiccation-induced modulation of gene expression in plants. Plant Cell Environ 25:141–151
Rezaei MK, Shobbar ZS, Shahbazi M, Abedini R, Zare S (2013) Glutathione S-transferase (GST) family in barley: identification of members, enzyme activity, and gene expression pattern. J Plant Physiol 170:1277–1284
Richards SL, Wilkins KA, Swarbreck SM, Anderson AA, Habib N, Smith AG, McAinsh M, Devies JM (2014) The hydroxyl radical in plants: from seed to seed. J Exp Bot 66:37–46
Ristic Z, Jenks MA (2002) Leaf cuticle and water loss in maize lines differing in dehydration avoidance. J Plant Physiol 159:645–651
Rollins JA, Habte E, Templer SE, Colby T, Schmidt J, von Korff M (2013) Leaf proteome alterations in the context of physiological and morphological responses to drought and heat stress in barley (Hordeum vulgare L.). J Exp Bot 64:3201–3212
Rushton PJ, Somssich IE, Ringler P, Shen QJ (2010) WRKY transcription factors. Trends Plant Sci 15:247–258
Shao HB, Chu LY, Jaleel CA, Manivannan P, Panneerselvam R, Shao MA (2009) Understanding water deficit stress-induced changes in the basic metabolism of higher plants—biotechnologically and sustainably improving agriculture and the ecoenvironment in arid regions of the globe. Crit Rev Biotechnol 29:131–151
Suprunova T, Krugman T, Fahima T, Chen G, Shams I, Korol A, Nevo E (2004) Differential expression of dehydrin genes in wild barley, Hordeum spontaneum, associated with resistance to water deficit. Plant Cell Environ 27:1297–1308
Talame V, Ozturk NZ, Bohnert HJ, Tuberosa R (2007) Barley transcript profiles under dehydration shock and drought stress treatments: a comparative analysis. J Exp Bot 58:229–240
Teulat B, Monneveux P, Wery J, Borries J, Souyris I, Charrier A, This D (1997) Relationships between relative water content and growth parameters under water stress in barley: a QTL study. New Phytol 137:99–107
Walia H, Wilson C, Wahid A, Condamine P, Cui X, Close TJ (2006) Expression analysis of barley (Hordeum vulgare L.) during salinity stress. Funct Integr Genomics 6:143–156
Xu Z, Zhou G, Shimizu H (2010) Plant responses to drought and rewatering. Plant Signal Behav 5:649–654
Xue GP, Loveridge CW (2004) HvDRF1 is involved in abscisic acid-mediated gene regulation in barley and produces two forms of AP2 transcriptional activators, interacting preferable with a CT-rich element. Plant J 37:326–339
Yang S, Huang C, Wu Z (2006) Stomatal movement in response to long distance-communicated signals initiated by heat shock in partial roots of Commelina communis L. Sci China Ser C Life Sci 49:18–25
Acknowledgments
This work was supported by The Scientific Research Projects Coordination Unit of Istanbul University, Project Number BAP 4712. The authors thank to the staff of Dr. Maria von Korff’s lab for experimental advice on stress applications and qRT-PCR.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Table S1
Primer sequences used for gene expression analysis by semiquantitative RT-PCR and qRT-PCR (DOC 42 kb)
Table S2
Percent change in relative water content (RWC) and water loss rate (WLR, gh−1 g−1 dry weight) in ten barley cultivars at the end of 10 days of dehydration. Values are given as mean ± SD (n = 5). Different letters indicate significant differences at P ≤ 0.05 using least square means (DOCX 13 kb)
Table S3
Bivariate correlation coefficients (R) between physiological parameters and gene expressions of HvBAS1, HvMT2 and HvABA7 during shock-like dehydration (ShD) treatment, 0–8 h period, in MR and ER genotypes. IL: Ion leakage, Osmol: osmolality. Significance (*P < 0.05, **P < 0.01) (DOCX 17 kb)
Rights and permissions
About this article
Cite this article
Gürel, F., Öztürk, N.Z., Yörük, E. et al. Comparison of expression patterns of selected drought-responsive genes in barley (Hordeum vulgare L.) under shock-dehydration and slow drought treatments. Plant Growth Regul 80, 183–193 (2016). https://doi.org/10.1007/s10725-016-0156-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10725-016-0156-0