Abstract
DNA barcoding allows the use of molecular markers to differentiate the species of an interest group. This is especially useful when morphological characters are insufficient due to high similarity between species. The genus Capsicum contains some species that are difficult to determine by taxonomic means, in particular the annuum complex. Thus, the objective of this study was to investigate the discriminatory ability of two molecular markers in Colombian Capsicum accessions, including three wild species belonging to the Andean clade of the genus. A total of 95 Capsicum accessions, representing eight species, were genotyped by high resolution melting analysis (HRM) using the Waxy and C2_At5g04590 markers. Waxy could discriminate the Andean clade species (C. rhomboideum, C. dimorphum and C. lycianthoides), C. baccatum, C. pubescens and C. chinense, while C2_At5g04590 could discriminate C. frutescens, C. annuum var. annuum and C. annuum var. glabriusculum. Hence, a combination of the two markers could be used for discrimination of the eight species including the wild variety of C. annuum: C. annuum var. glabriusculum. Most nucleotide substitutions and indels were found in the sequences of the three Andean species, indicating that the Andean clade has a high genetic diversity compared to the other species. The incorporation of more wild species and varieties in this study allowed to correct the power of both markers to discriminate Capsicum species, besides the registration of new haplotypes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Chilli is one of the world's most important crops, according to FAO, in 2021 more than 41 million tons were produced for consumption. All chili peppers belong to the genus Capsicum in the family Solanaceae, which currently includes 43 species (Barboza et al. 2022). However, only 5 have been domesticated and cultivated for commercial purposes: C. annuum L. var. annuum (Caa), C. chinense Jacq. (Cch), C. frutescens L. (Cfr), C. baccatum L. (Cba), and C. pubescens Ruiz & Pav (Cpu). The first three species form a complex of species called the annuum complex, which shows a great similarity not only in their morphological but also in their molecular characters and can show hybridization events (Baral and Bosland 2004; Carrizo García et al. 2016; Barboza et al. 2022). Therefore, some varieties of this complex resemble each other and make it difficult to establish the boundaries between species.
In this sense, molecular markers can be used as a complement to taxonomic identification through the so-called “DNA barcoding”. This methodology uses DNA regions that are conserved in a group of interest, but that vary sufficiently interspecifically to allow discrimination between species or another taxonomic group (Paz et al. 2011). This technique has been widely used in some animal groups such as insects and vertebrates using the COI (cytochrome c oxidase subunit I) mtRNA region with satisfactory results (Hebert et al. 2003a, b). It has also been applied to bacteria using the 16S rRNA region (Kim and Chun 2014). In plants, multilocus analysis (chloroplast and nuclear genes) has been used with better results (Jarret 2008; González et al. 2009). This methodology allows identification in situations where the key characters for taxonomy have not yet been developed or are absent: Early stages of individuals, when only fragments are available, or when there is a delay in obtaining the essential characters for taxonomy (time to flowering and/or fruiting in plants). It also allows identification in cases where morphological characters are insufficient or too ambiguous for discrimination.
In combination with DNA barcoding, the use of High-Resolution Melting (HRM) genotyping allows the identification of SNPs and indels in a sequence by their dissociation curves. Combined with DNA barcoding, it has the advantage of reducing the costs associated with sequencing, since only one or two samples are sequenced for each pattern of curves obtained, as opposed to the traditional barcoding approach where all samples in the study are sequenced. In addition, because it is a real-time PCR technique (Wittwer et al. 2003; Martino et al. 2010), it also reduces genotyping time. In plants, barcoding HRM analysis has been widely used for species and accession discrimination: Citrus (Distefano et al. 2012), Prunus avium (Ganopoulos et al. 2011), Lens culinaris (Bosmali et al. 2012), Melientha suavis, Sauropus androgynus, and Urobotrya siamensis (Thongkhao et al. 2020), among others.
In 2010, Jeong and coworkers developed a set of markers (nuclear and chloroplast) that allowed the discrimination of the five domesticated species of the genus Capsicum, including the annuum complex, as well as one wild species (C. chacoense) by the barcoding-HRM method. They found that Waxy and C2_At5g04590 (hereafter C2) amplifications were the most efficient marker combination for species discrimination. However, that study used accessions from World Vegetable Center (AVRDC) (Taiwan). These accessions have been selected to a certain extent and their genetic diversity has been reduced. Also, it does not include many wild species of the genus and does not represent much material coming from the American continent, where the genus Capsicum originated. Therefore, the objective of this work was to apply the barcoding-HRM technique with the markers developed by Jeong et al. (2010) in Colombian accessions of Capsicum, including some wild species of the Andean clade and domesticated landraces found in home gardens and marketplaces, which may represent new genetic variants for barcoding marker sequences.
Materials and methods
Plant materials
Capsicum accessions (seeds and/or plants) were collected from natural reserves, marketplaces, nurseries, home gardens and parks in different municipalities of the departments of Valle del Cauca and Nariño (Supplementary data 1). Seeds were germinated in a greenhouse at the Universidad del Valle, Colombia. The leaf tissue was collected and then preserved in silica gel until it was analyzed in the laboratory (Chase and Hills 1991). Then, adult plants were used by morphological identification using taxonomic characters (Barboza et al. 2022).
Genomic DNA extraction
All molecular analyses were performed at the Laboratorio de Biología Molecular de la Universidad del Valle. Genomic DNA was extracted from leaf tissue using the CTAB method of Doyle and Doyle (1987) modified according to Stewart (1997). Concentration and purity of DNA samples were measured using Nanodrop 2000 (Thermo Scientific, USA).
Barcoding and HRM analysis
Waxy and C2 markers were used for identified Capsicum species using the primers and PCR conditions described by Jeong et al. (2010) (Supplementary data 2). PCR consisted of a final volumen of 15 µL of 2X Supermix Melt Precision (BioRad), 0.7 µM of both primers, 5 µL of DNA (10 ng/µL) and 5.4 µL of distilled and deionized water. PCR and HRM analysis were performed in a CFX96 Touch™ Real-Time PCR thermocycler. HRM analysis was performed as follows: DNA was denatured at 95 °C for 30 s followed by 60 °C for 1 min. Then the temperature was gradually increased by 0.2 °C per cycle for 10 s from 65 to 95 °C. Fluorescence was sampled at each cycle. HRM data was analyzed in a Precision Melt Analysis™ v1.2 software. It was verified that the percentage confidence of cluster assignment for each sample was not less than 95%. Any sample with a lower percentage was repeated. One or two individuals per cluster (curve) were sequenced by the Sanger method using a specialized service (Macrogen Korea) to obtain the nucleotide sequence. As the annuum complex species (Ca, Cfr and Cch) are a group very difficult to distinguish by both morphological and molecular analysis, three additional individuals belonging to very well-known cultivars of each species were also sequenced to serve as a control. One individual of “ají Cayena” coded as C. annuum var. annuum cultivar, one of “ají Tabasco” coded as C. frutescens cultivar and one of “ají Habanero” coded as C. chinense cultivar.
Data analysis
Sequences were edited and cleaned using Sequencher 4.6 (Gene Codes Corporation 2006). After that, they were aligned using Muscle algorithm, UPGMA trees and different statistics as number of variable sites and unique polymorphism (singletons) were calculated in MEGA software version 11 (Tamura et al. 2021). Additionally, sequences from Waxy gen were compared with reported in NCBI using BLAST.
Results
Morphological identification
95 accessions were collected, representing 8 Capsicum species based on morphological characters (Supplementary data 1), 5 domestic species: C. annuum (Ca) including Caa and C. annuum var. glabriusculum (Cag), Cfr, Cch, Cba, Cpu) and 3 wild species from the Andean clade according to Carrizo García et al. (2016): C. rhomboideum (Dunal) Kuntze (Crh), C. lycianthoides Bitter (Cly) and C. dimorphum (Miers) Kuntze (Cdi). Some accessions were not identified morphologically because they are members of the annuum complex, and this complex has similar morphological characteristics among its species (Barboza et al. 2022). This species was determined only by molecular analysis.
Waxy marker
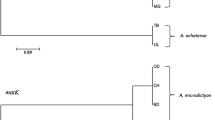
Six groups were defined by analyzing the HRM of the Waxy marker, Fig. 1 shows the normalized and differential HRM curves of these groups. The Andean species (Cly and Cdi) are each represented in a different group, as are the Cpu and Cba species. Crh species are not shown in that figure because their melting temperature (Tm) is very different from the rest of the species. Cch (including both cultivars and non-commercial accessions) could be easily discriminated in a group from the rest of the annuum complex (Cfr and Ca), while these last two could not discriminate each other and conformed a unique group. Sequence analysis of the fragment of 245 bp revealed 31 variable sites and 4 indels. Figure 2 shows the nucleotide composition of the Waxy marker fragment together with its variants among 20 sequenced individuals. The group of Andean species had most of the nucleotide substitutions, which made them differ enormously from the remaining species (Fig. 3). One of the most remarkable changes in this group was the unique deletion of 12 nucleotides from position 215–226 present in Crh (Fig. 2). The individuals of Cba and Cpu could also be distinguished from the rest of the species, while for the annuum complex only Cch could be distinguished from the other two species. Therefore, this marker could not be used to discriminate Ca and Cfr. Although Jeong et al. (2010) did not report their Waxy sequences on the NCBI database, a manual check of the haplotypes showed that the variants found in this work were the same as those of Jeong et al. (2010), except for the sequences associated with the Andean Capsicum group, which were not analyzed for them and therefore represent new haplotypes. A BLAST of the sequences showed almost always a high similarity (> 99%) with some sequences registered in the NCBI and with the same taxonomic assignment that we have assigned in this work. An exception was made for Cfr and Ca, which could not be discriminated with this marker.
Difference plot graph of the Waxy marker based on the high resolution melting (HRM) analysis, each curve represents a different Capsicum individual, each color represents a different cluster. C. annuum and C. frutescens (red), C. baccatum (blue), C. pubescens (purple), C. chinense (light blue), C. lycianthoides (pink), C. dimorphum (green)
C2 marker
Five different clusters (Fig. 4) were shown by HRM analysis. For the sequencing of the C2 marker, a fragment of 368 bp was obtained for 22 Capsicum sequences. A difference from the Waxy marker, which is shorter, only 9 variable sites were identified. These 9 nucleotide changes do not include insertions or deletions as it occurs for Waxy (Fig. 5), showing less nucleotide diversity for this marker. Andean samples were not present in this analysis as they do not amplify for this marker. C2 allowed to distinguish the Cch and Cpu species from the others, similar to the Waxy marker. Most of the variable sites of the sequence were present in this last species. Furthermore, it was possible to separate Ca into two groups, the domestic varieties (Caa) grouped together, and the wild varieties (Cag) grouped together with Cba (Fig. 6A–D). In contrast to Waxy, C2 was able to discriminate Cfr individuals from Ca, including some samples with a high morphological similarity to Cag individuals, which we called “wild Cfr” (Fig. 6E–F). Therefore, this marker was not able to discriminate between Cba and Cag, something that did not happen with the Waxy marker and that has not been reported by Jeong et al. (2010).
Difference plot graph from C2 marker based on HRM analysis, each curve represents a different individual, each color represents a different cluster. Capsicumannuum var. glabriusculum and C. baccatum (red), C. chinense (blue), C. annuum var. annuum (pink), C. pubescens (fluorescent green), C. frutescens (green)
Sequence comparison by BLAST was not possible because only one sequence of this marker is registered in NCBI. However, a manual check with the Jeong et al. (2010) sequences showed the same haplotypes for most species with the exception for Cag individuals, that were not studied by Jeong et al. (2010) and in this study converged in the same haplotype as Cba individuals.
Accessions B6 and KF were two samples that could not be identified by morphological characteristics because they belong to the high morphological similarity annuum complex, however, molecular barcoding showed that B6 belongs to the Cch species (Fig. 7) and KF belongs to the Cfr species (Fig. 2). Likewise, G17 was an accession that never developed a reproductive stage, so morphological identification was not possible. However, barcoding showed that this sample belonged to the Cpu species (Fig. 7).
Discussion
Often, the use of Colombian accessions in global Capsicum research is low (Lee et al. 2016; Taranto et al. 2016; Tripodi and Greco 2018; Colonna et al. 2019). This represents a gap in information, as the country has two centers of diversification of the genus, the Amazon and the Andes, and represents the point of entry of chilies to Centro America and North America (Carrizo García et al. 2016). The Waxy and C2 markers were developed for Jeong et al. (2010) and tested on commercial cultivars of Capsicum (domesticated species), with the exception of C. chacoense, which is a wild species. Although they work to discriminate these species, a test on wild accessions and other species of Capsicum, including Colombian material, were needed to confirm their discriminating ability. This study has shown that the use of Waxy and C2 markers over 95 accessions collected from the Colombian southwest, is usable for the discrimination of taxonomic species, including the Andean clade and semi-wild varieties, which have more genetic diversity than commercial crops.
The Andean clade of Capsicum represents the most ancestral group of the genus with eight species. Some common characteristics of the members include the absence of capsaicionoids in their fruits, yellow corolla for most members, and a karyotype 2n = 26 (Barboza et al. 2019). Phylogenetic analysis also distinguishes these species from the others, corresponding to a well-supported monophyletic clade (Carrizo García et al. 2016; Barboza et al. 2019). In this study, the analysis of three Andean species showed that the Waxy marker could discriminate them from each other as well as from the other species. In fact, most of the nucleotide substitutions and indels registered for Waxy in this study are attributed to the Andean species (Fig. 2), which shows the great genetic divergence between the Andean clade and the other species of the genus. This patronage was also observed by Jarret (2008), who evaluated the same marker in some species of Capsicum and reported that 54 of 73 substitutions were found in Crh, including the same indel of 12 nucleotides that we have found in this study. None of these species amplified for the C2 marker. However, we do not know if this lack of PCR amplification is due to mutations in the primer recognition area, so a more intense analysis needs to be done.
Ca is one of the Capsicum species that has two well-defined taxonomic varieties. A remarkable result was the differentiation of Ca varieties, Cag and Caa. On the one hand, Caa includes all commercial accessions that have undergone some degree of plant breeding and have medium or large fruit size, and on the other hand, Cag includes the wild accessions that have small fruit size. Jeong et al. (2010) included only Caa accessions in their study. Therefore, the discrimination of the two Ca varieties is a novel result of this study. In addition, Cag accessions share the same haplotype as Cba, so it is not possible to separate these two species with this marker, but the Waxy marker could do it. Thus, even after including wild accessions and Andean Capsicum species, the combined use of both markers to discriminate Capsicum species proposed by Jeong et al. (2010) is still valid. Waxy is useful for the discrimination of Cba and the Andean clade, and C2 is useful for the discrimination of Cfr and Cag individuals. Both could easily distinguish the Cpu and Cch species.
Molecular barcoding has the advantage of identifying an individual at any stage of this development, as we have shown for the G17 accession. This is different from taxonomic identification, which requires reproductive traits to make an effective categorization. However, DNA barcoding needs a good genetic database in order to make a correct identification, and although this study improves the database with new haplotypes for both markers, a better database, especially for C2, is needed to improve the discrimination of the barcoding.
Finally, by reducing the number of individuals to be sequenced, HRM analysis is a practical tool to reduce the costs associated with sequencing. Out of 95 samples, only 42 were sequenced because it is assumed that individuals with the same HRM curve have the same nucleotide sequence. Therefore, in order to have their nucleotide composition, only two or three individuals need to be sequenced for the curve. In addition, HRM curves could be stored for comparison with samples from future research, so only new curves would have to be sequenced.
Conclusion
The combination of Waxy and C2 markers is usable to discriminate efficiently the Capsicum species evaluated in this study, Waxy could separate Cba, Cch, Cpu and Andean clade (Cdi, Crh and Cly), while C2 could separate Caa, Cag and Cfr. In addition, this study registered a new haplotypic diversity with the genotyping of wild accessions (Cag, Cdi, Crh and Cly). This showed the great genetic diversity found in samples that have not been subjected to plant breeding and the importance of genotyping other Capsicum species and varieties to verify the discriminatory ability of Waxy and C2 markers.
Data availability statement
The data that support the findings of this study are available from the corresponding author, RAVV, upon reasonable request. Nucleotide sequences for both markers have been submitted to the NCBI database, accession numbers for Waxy marker: PP660239-PP660258 and for the C2 marker: PP660217-PP660238.
References
Baral JB, Bosland PW (2004) Unravelling the species dilemma in Capsicum frutescens and C. chinense (Solanaceae): a multiple evidence approach using morphology, molecular analysis, and sexual compatibility. J Am Soc Hort Sci 129:826–832. https://doi.org/10.21273/JASHS.129.6.0826
Barboza GE, Carrizo García C, Leiva González S, Scaldaferro M, Reyes X (2019) Four new species of Capsicum (Solanaceae) from the tropical Andes and an update on the phylogeny of the genus. PLoS ONE 14(1):e0209792. https://doi.org/10.1371/journal.pone.0209792
Barboza GE, García CC, de BemBianchetti L, Romero MV, Scaldaferro M (2022) Monograph of wild and cultivated chili peppers (Capsicum L., Solanaceae). PhytoKeys 200:1. https://doi.org/10.3897/phytokeys.200.71667
Bosmali I, Ganopoulos I, Madesis P, Tsaftaris A (2012) Microsatellite and DNA-barcode regions typing combined with High Resolution Melting (HRM) analysis for food forensic uses: a case study on lentils (Lens culinaris). Food Res Int 46(1):141–147. https://doi.org/10.1016/j.foodres.2011.12.013
Carrizo García C, Barfuss MH, Sehr EM, Barboza GE, Samuel R, Moscone EA, Ehrendorfer F (2016) Phylogenetic relationships, diversification and expansion of chili peppers (Capsicum, Solanaceae). Ann Bot 118(1):35–51. https://doi.org/10.1093/aob/mcw079
Chase MW, Hills HH (1991) Silica gel: an ideal material for field preservation of leaf samples for DNA studies. Taxon 40(2):215–220. https://doi.org/10.2307/1222975
Colonna V, D’Agostino N, Garrison E, Albrechtsen A, Meisner J, Facchiano A, Cardi T, Tripodi P (2019) Genomic diversity and novel genome-wide association with fruit morphology in Capsicum, from 746k polymorphic sites. Sci Rep 9(1):1–14. https://doi.org/10.1038/s41598-019-46136-5
Distefano G, Caruso M, La Malfa S, Gentile A, Wu SB (2012) High resolution melting analysis is a more sensitive and effective alternative to gel-based platforms in analysis of SSR–an example in Citrus. PLoS ONE 7(8):e44202. https://doi.org/10.1371/journal.pone.0044202
Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19:11–15
Ganopoulos I, Argiriou A, Tsaftaris A (2011) Microsatellite high resolution melting (SSR-HRM) analysis for authenticity testing of protected designation of origin (PDO) sweet cherry products. Food Control 22(3–4):532–541. https://doi.org/10.1016/j.foodcont.2010.09.040
Gene Codes Corporation (2006) Sequencher® (versión 4.6) [Software to analyzed DNA sequences]. http://www.genecodes.com. Accessed 10 Dec 2022
González MA, Baraloto C, Engel J, Mori SA, Pétronelli P, Riéra B, Roger A, Thébaud C, Chave J (2009) Identification of Amazonian trees with DNA barcodes. PLoS ONE 4(10):e7483. https://doi.org/10.1371/journal.pone.0007483
Hebert PD, Cywinska A, Ball SL, Dewaard JR (2003) Biological identifications through DNA barcodes. Proc R Soc Lond Ser B Biol Sci 270(1512):313–321. https://doi.org/10.1098/rspb.2002.2218
Hebert PD, Ratnasingham S, De Waard JR (2003) Barcoding animal life: cytochrome c oxidase subunit 1 divergences among closely related species. Proc R Soc Lond Ser B Biol Sci 270(suppl_1):S96–S99. https://doi.org/10.1098/rsbl.2003.0025
Jarret RL (2008) DNA barcoding in a crop genebank: the Capsicum annuum species complex. Open Biol J 1(1):35–42
Jeong HJ, Jo YD, Park SW, Kang BC (2010) Identification of Capsicum species using SNP markers based on high resolution melting analysis. Genome 53(12):1029–1040. https://doi.org/10.1139/G10-094
Kim M, Chun J (2014) 16S rRNA gene-based identification of bacteria and archaea using the EzTaxon server. Methods Microbiol 41:61–74. https://doi.org/10.1016/bs.mim.2014.08.001
Lee HY, Ro NY, Jeong HJ, Kwon JK, Jo J, Ha Y, Jung A, Han JW, Venkatesh J, Kang BC (2016) Genetic diversity and population structure analysis to construct a core collection from a large Capsicum germplasm. BMC Genet 17(1):1–13. https://doi.org/10.1186/s12863-016-0452-8
Martino A, Mancuso T, Rossi AM (2010) Application of high-resolution melting to large-scale, high-throughput SNP genotyping: a comparison with the TaqMan method. J Biomol Screen 15(6):623–629. https://doi.org/10.1177/1087057110365900
Paz A, González M, Crawford AJ (2011) Códigos de barras de la vida: introducción y perspectiva. Acta Biol Colomb 16(3):161–175
Stewart CN Jr (1997) Rapid DNA extraction from plants. In: Micheli MR, Bova R (eds) Fingerprinting methods based on arbitrarily primed PCR. Springer, Berlin, pp 25–28. https://doi.org/10.1007/978-3-642-60441-6_4
Tamura K, Stecher G, Kumar S (2021) MEGA 11: molecular evolutionary genetics analysis version 11. Mol Biol Evol. https://doi.org/10.1093/molbev/msab120
Taranto F, D’Agostino N, Greco B, Cardi T, Tripodi P (2016) Genome-wide SNP discovery and population structure analysis in pepper (Capsicum annuum) using genotyping by sequencing. BMC Genom 17(1):1–13. https://doi.org/10.1186/s12864-016-3297-7
Thongkhao K, Tungphatthong C, Phadungcharoen T, Sukrong S (2020) The use of plant DNA barcoding coupled with HRM analysis to differentiate edible vegetables from poisonous plants for food safety. Food Control 109:106896. https://doi.org/10.1016/j.foodcont.2019.106896
Tripodi P, Greco B (2018) Large scale phenotyping provides insight into the diversity of vegetative and reproductive organs in a wide collection of wild and domesticated peppers (Capsicum spp.). Plants 7(4):103. https://doi.org/10.3390/plants7040103
Wittwer CT, Reed GH, Gundry CN, Vandersteen JG, Pryor RJ (2003) High-resolution genotyping by amplicon melting analysis using LCGreen. Clin Chem 49(6):853–860. https://doi.org/10.1373/49.6.853
Acknowledgements
We are very grateful to Jhon Jairo Calderón for his support in collecting samples in the department of Nariño. To Leidy Laura Arias, Jorge Mario Ruiz, Mary Belcy Bonilla, Héctor Cifuentes, Mauricio Peñuela and José Cándelo for their guidance and support in collecting samples in the departments of Cauca and Valle del Cauca.
Funding
Open Access funding provided by Colombia Consortium. This project was funded by the internal call 2023 for the creation of a bank of eligible projects for research and artistic creation in the sciences, arts, humanities, technologies and innovation of the Universidad del Valle, Colombia (CI 71368) and the internal call for support to Ph.D. students (2021) of the Universidad del Valle, Colombia (CI71310).
Author information
Authors and Affiliations
Contributions
RAVV conceived and designed the experiment, analysed the data, prepared the figures and/or tables, wrote the original manuscript, revised and approved the final document. HC conceived and designed the experiment, revised and approved the final document, managed and supervised the project.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no declaration of interest statement.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Viáfara-Vega, R.A., Cárdenas-Henao, H. Identification of Capsicum species from Colombia by DNA barcoding and high resolution melting (HRM) analysis. Genet Resour Crop Evol (2024). https://doi.org/10.1007/s10722-024-02097-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10722-024-02097-x