Abstract
Markers linked to quantitative trait loci (QTL) must be validated in diverse genetic materials before they can be reliably used in molecular breeding programs. Here, 30 simple sequence repeat markers linked to QTL for grain iron content (GFeC), grain zinc content (GZnC), and grain protein content (GPC) were analyzed in 56 diverse dicoccum wheat genotypes. Seven markers were validated, including four (Xwmc617, Xbarc67, Xwmc283, Xgwm361) for grain iron content, one (Xbarc146) for both grain iron and protein contents; one more for grain protein (Xgwm408) and one for grain zinc content (Xgwm271). The segregating F2 population developed from the high-quality local landrace GPM DIC 87, and the high-yielding low-quality commercial cultivar (HW 1098) was used to revalidate the identified QTL-linked markers. As a result, Xgwm271 and Xbarc67 were re-validated in the F2 population, with values for phenotypic variation explained (PVE) of 56.50% and 72.78%, respectively. These two markers may serve as ideal candidates for molecular breeding programs to improve grain micronutrient contents. Additionally, the evaluation of the F2 population for agro-morphological, physiological, and quality traits, revealed large variability that can be useful for trait improvement. This population generated rare transgressive segregants with desirable allelic combinations that may be ideal for improving grain yield, grain quality, and physiological efficiency.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The consumption of staple foods that are low in essential minerals and vitamins leads to micronutrient deficiency in humans, which is also referred to as hidden hunger (Liu et al. 2019). Approximately two billion people worldwide are reportedly affected by malnutrition due to micronutrient dietary insufficiency, particularly of iron and zinc (Gupta et al. 2021; Von Grebmer et al. 2014). Further, children and women are the most vulnerable groups to micronutrient deficiencies (Darnton-Hill et al. 2005). Iron (Fe) is an essential micronutrient, and an insufficient Fe intake causes impaired cognitive ability, reduced immunity, birth weight, and severe anemia, resulting in maternal and child mortality. In turn, zinc (Zn) is another essential micronutrient required for various biological processes, and Zn deficiency causes retarded growth and reduced immunity to infectious diseases; furthermore, it increases infant mortality, and pregnancy and childbirth complications (Krebs et al. 2014).
Specifically, micronutrient deficiency is most prevalent in Africa, south of the Sahara, and the South Asian subcontinent (Black et al. 2013). Globally, a deficiency of at least one of three micronutrients (Fe, Zn, and vitamin A) prevails among 56% (372 million) and 69% (1.2 billion) of pre-school children and non-pregnant women at reproductive age, respectively. The highest number of micronutrient deficiencies affects preschool children living in South Asia (99 million), followed by those living in sub-Saharan Africa (98 million), East Asia, and the Pacific (85 million). In turn, more than half (57%) of non-pregnant women at reproductive age suffering from micronutrient deficiencies live in East Asia and the Pacific (384 million), or South Asia (307 million) (Stevens et al. 2022).
Grain protein is an important trait in wheat because of its nutritional significance, as it plays a key role in industrial processing and end-product quality. Specifically, wheat serves as a dietary cornerstone for nearly 2.5 billion people worldwide (Listman et al. 2019), a staple food for 30% of the global population (Lobell et al. 2011). Notably, wheat contributes substantially to the daily caloric intake, representing over one-fifth of the global dietary energy consumption. Given its pivotal role in global nutrition, wheat biofortification has emerged as a strategic approach to combating micronutrient malnutrition. Successful crop biofortification breeding programs rely on valuable germplasm diversity and a comprehensive understanding of genetic architecture. In particular, the status of grain micronutrients such as grain iron content (GFeC) and grain zinc content (GZnC) in the germplasm, advanced breeding lines, and mapping populations has been studied in wheat (Krishnappa et al. 2022; Rathan et al. 2022; Gopalareddy et al. 2015; Morgounov et al. 2007; Chhuneja et al. 2006; Cakmak et al. 2000; Monasterio and Graham 2000). Although efforts have been made to explore and utilize the existing germplasm diversity, the limited variability in modern wheat cultivars emphasizes the need for further exploration and incorporation of diverse genetic resources. A case in point, nutritionally rich hulled-wheat (T. turgidum ssp. dicoccum) is one of the oldest crop species, however, its cultivationis currently restricted to the mountainous regions of Europe and Asia. Nonetheless, dicoccum wheat is gaining importance because of its high nutritional and therapeutic values (Hammed and Simsek 2014; Lachman et al. 2012).
Although the enhancement of the nutritional status of modern wheat cultivars through conventional breeding approaches has been successful, and many high yielding biofortified cultivars have been released for commercial cultivation, breeding of nutrient-rich wheat cultivars through such conventional approaches is time consuming and curtailed for various reasons, including linkage drag, slow response to selection, and the quantitative nature of the relevant traits. Alternatively, the integration of molecular tools to the breeding scheme is a powerful and promising approach in the development of nutrient-rich wheat cultivars. Indeed, the development of molecular markers and their application in marker-assisted selection (MAS) in crop plants through the tagging of major genes, particularly for qualitative traits, have yielded many commercial cultivars of diverse crops (Krishnappa et al. 2024). Furthermore, mapping of quantitative trait loci (QTL) and genome-wide association study (GWAS) are the most efficient methods for dissecting complex quantitative traits. Thus, over the last decade, several QTL have been identified in different genetic backgrounds using various marker systems for quality traits in wheat (Gupta et al. 2021). However, except for the major QTL, Gpc-B1, most of the identified QTL are not used in MAS. Exceptionally, the wild emmer accession of Israel, i.e., FA15-3, is an extensively used genetic resource for many high-protein genes, including Gpc-B1, which can accumulate approximately 40% protein under sufficient nitrogen application (Avivi 1978). Specifically, the Gpc-B1 gene, identified on the 6BS chromosome, encodes the NAC transcription factor (NAM-B1), which has a pleiotropic effect and increases GFeC, GZnC, and grain protein content (GPC) (Distelfeld et al. 2007) through the remobilization of nutrients from source to sink organs by accelerating senescence (Uauy et al. 2006).
The major bottleneck in the utilization of QTL in cultivar development is the lack of validation of the identified QTL in different genetic backgrounds and production conditions through multi-environment evaluations. Furthermore, many investigations have several limitations, including limited mapping population size, low marker coverage, and a lack of robust phenotyping, altogether making it difficult to validate the identified QTL. Furthermore, there are very few validation research compared with mapping research. Molecular markers must be validated by testing for their presence in a range of cultivars and other important diverse genotypes for their utilization in MAS (Spielmeyer et al. 2003; Sharp et al. 2001). MAS-based transfer or pyramiding of QTL for quality traits in wheat has not been accomplished owing to the lack of validation. Hence, the objectives of this study were: i) to assess the genetic variability of agro-morphological, physiological, and quality traits in a segregating population derived from two highly diverse parents of tetraploid wheat; ii) to validate previously identified QTL for GFeC, GZnC, and GPC in a set of 56 diverse tetraploid wheat genotypes, and iii) to revalidate the validated QTL in the segregating F2 population derived from the HW 1098 × GPM DIC 87 biparental cross.
Materials and methods
Plant material and field experiments
The plant material used for the validation of previously identified QTL consisted of 56 dicoccum genotypes, including 34 germplasm lines, 13 breeding lines, and nine commercial cultivars. Details of the genetic material are provided in our previous study (Biradar et al. 2023). This diverse set of genotypes was tested at All India Coordinated Research Project (AICRP) on wheat, University of Agricultural Sciences, Dharwad, State of Karnataka, India (15°31′ N; 75° 07′ E; elevation: 678 m above mean sea level) for one year in 2020–21. Each genotype was sown in six rows (3 m in length) with two replicates in an alpha-lattice experimental design. The materials under study were planted under irrigated production conditions during the first fortnight of November 2020. To revalidate the validated QTL, a biparental F2 population was developed by crossing the micronutrient rich local collection, GPM DIC 87, with the commercial dicoccum cultivar, HW 1098 (high yielding cultivar with low GFeC and GZnC).This population was evaluated in 2020–21.
Estimation of grain micronutrients and protein
At physiological maturity, a random sample of 21–24 spikes was manually harvested in both the diversity panel of 56 genotypes and the F2 population, and threshed. Approximately 20 g of the grain sample from each entry was collected to estimate GFeC and GZnC contents. An Energy Dispersive X-ray Fluorescence (ED-XRF) instrument available at the ICAR-Indian Institute of Wheat and Barley Research, Karnal, India, was used to estimate GFeC and GZnC. Meanwhile, GPC was estimated by a nondestructive method using a near-infrared transmittance-based protein analyzer and expressed at 12.0% moisture level.
Agro-morphological and physiological parameters
Phenotypic characteristics were recorded for the F2 population at different stages of crop growth. Agro-morphological traits viz., days to 50% flowering (DFF), days to maturity (DM), plant height (PH), spike length (SL), number of spikelets per spike (SPS), number of grains per spike (GPS), number of productive tillers per plant (PTPP), 1000-grain weight (TGW), and grain yield (GY) were recorded. Similarly, physiological traits such as soil plant analysis development (SPAD) for chlorophyll content were recorded at three different stages, namely, booting (SPAD I), anthesis (SPAD II), and grain filling (SPAD III). Similarly, other physiological parameters, such as, the normalized difference vegetation index (NDVI), were recorded at three different crop growth stages: booting (NDVI I), anthesis (NDVI II), and grain-filling (NDVI III).
Genotyping
Thirty simple sequence repeat (SSR) markers (Table 1) available in the public domain and linked to the QTL for GFeC, GZnC, and GPC were screened for polymorphism among the set of 56 diverse genotypes. All 30 SSR primers were amplified; however, only nine markers were polymorphic among the genotypes and these nine polymorphic markers were used for genotyping the 56 genotypes. Further, seven markers (Xbarc146, Xwmc617, Xbarc67, Xwmc283, Xgwm361, Xgwm271, Xgwm408) validated in the set of 56 diverse genotypes were used to genotype the F2 population. Among these seven validated markers, only two (Xgwm271 and Xbarc67) showed both polymorphism and no segregation distortion in the F2 population; therefore, these two markers were further used for genotyping 200 genotypes.
Statistical analysis
The phenotypic coefficient of variation (PCV) and genotypic coefficient of variation (GCV), broad-sense heritability (h2 bs), and genetic advance over the mean (GAM) were computed using the formulas recommended by Burton and De Vane (1953), Hanson et al. (1956), and Johnson et al. (1955). Single-marker analysis (SMA) was conducted to assess the significance and contribution of SSR markers to the variation in grain micronutrient levels. Analysis of variance (ANOVA) and linear regression, as outlined by Haley and Knott (1992), were used in the single-marker analysis. These statistical methods were used to determine the significance of the markers and calculate the coefficient of determination (R2) for each marker. The R2 values served as indicators of the proportion of phenotypic variation associated with the linked markers, aiding in the understanding of their impact on the observed trait variations. All phenotypic and marker analyses were performed using SPSS v21.0 and Microsoft Excel.
Results
Phenotypic variation
The mean performance of the 56 genotypes suggested the presence of large variation for grain yield, GFeC, and GZnC in the genotypes under study. The top 20% of high yielding genotypes included five checks (DDK 1001, DDK 1025, Amruth, DWR 1006, and DDK 1029) and six germplasm lines (DDK 50404, DDK 50382, DDK 50422, DDK 50529, DDK 50533, and DDK 50377). Further, among the check cultivars, DDK 1001 was the best yielding genotype with high GFeC (41.2 mg/kg) and GZnC (39.0 mg/kg) contents, followed by DDK 1025 (GFeC: 40.8 mg/kg and GZnC: 39.1 mg/kg), Amruth (GFeC: 41.1 mg/kg and GZnC: 36.1 mg/kg), DWR 1006 (GFeC: 40.4 mg/kg and GZnC: 47.0 mg/kg), and DDK 1029 (GFeC: 44.7 mg/kg and GZnC: 32.4 mg/kg). Similarly, among the high yielding germplasm lines, DDK 50404 showed the highest yield, with a GFeC of 51.9 mg/kg and GZnC of 44.3 mg/kg, followed by DDK 50382 (GFeC: 55.7 mg/kg and GZnC: 43.4 mg/kg), DDK 50422 (GFeC: 49.7 mg/kg and GZnC: 36.0 mg/kg), DDK 50529 (GFeC: 46.1 mg/kg and GZnC: 33.6 mg/kg), DDK 50533 (GFeC: 43.7 mg/kg and GZnC: 29.2 mg/kg), and DDK 50377 (GFeC: 52.5 mg/kg and GZnC: 36.9 mg/kg).
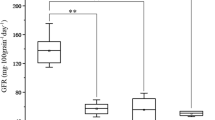
The genetic variability parameters of the F2 population for the agro-morphological, physiological, and quality traits analyzed are shown in Table 2. A wide range of variability was observed for all studied traits; i.e., DFF (61.0–101.0), DM (105.0–136.0), PH (52.6–110.0 cm), PTPP (4.0–31.0), SL (6.5–13.5 cm), SPS (14.0–28.0), GPS (21.0–54.0), TGW (18.5–57.5 g), GY (8.0–65.0 g), SPAD I (43.5–65.0), SPAD II (41.3–57.6), SPAD III (37.3–54.4), NDVI I (0.50–0.8), NDVI II (0.5–0.8), NDVI III (0.4–0.7), GFeC (35.0–66.8 mg/kg), GZnC (35.7–74.2 mg/kg), GPC (14.0%–18.6%). PCV and GCV were highest for GY (PCV, 68.4%; GCV, 46.8%), followed by PTPP (PCV, 54.2%; GCV, 46.6%), GPS (PCV, 21.9%; GCV, 21.9%), whereas the lowest PCV was observed for GPC (7.3%) and lowest GCV for SPAD III (5.7%). Similarly, broad-sense heritability was observed at more than 80.0% for eight traits, including the highest for TGW (90.3%), followed by SL (89.4%), SPS (88.5%), PH (86.2%), GZnC (86.1%), GFeC (83.9%), GPS (83.4%), and DFF (82.7%), whereas the lowest heritability values were recorded for GY (46.5%). Additionally, 10 traits recorded GAM of more than 20.0%, with the highest being for PTPP (82.5%), followed by GY (65.7%), GPS (37.8%), SPS (33.6%), SL (32.7%), TGW (32.2%), GZnC (29.8%), PH (25%), GFeC (23.7%), and DFF (22.9%), whereas, the lowest was observed for SPAD III (8.3%). The mean performance of the F2 population for the various agro-morphological, physiological, and quality traits is illustrated in the box plot in Fig. 1. Transgressive segregants that surpassed both parents were observed for all studied traits.
Box plots for agro-morphological, physiological, and quality traits in the second filial (F2) population of the cross HW1098 × GPM DIC 87. DFF days to 50% flowering, DH days to maturity, PH plant height (cm), PTPP productive tillers per plant, SL Spike length (cm), SPS spikelets per spike, GPS grains per spike, TGW thousand-grain weight (g), GY grain yield (g/plant), SPAD soil plant analysis development, NDVI I, II and III normalized difference vegetation index I (booting stage) II (anthesis stage) III (grain filling stage), GFeC, grain iron content (mg/kg), GZnC grain zinc content (mg/kg), GPC grain protein content (%)
Correlations
The correlation coefficients of agro-morphological, physiological, and quality traits in the second filial (F2) population of the cross HW1098 × GPM DIC 87 are shown in Table 3. GPC was significantly and positively correlated with four agro-morphological traits (DFF, DM, SL, and SPS) and two quality traits (GFeC and GZnC), whereas GPC was negatively correlated with TGW. In turn, GZnC, had a significant positive correlation with DFF, DM, SL, and GFeC. Another micronutrient, GFeC, showed a significant positive correlation with SPS, GPS, GZnC, and GPC. Further, the important physiological trait, NDVI, had a significant positive correlation with PH and GY at all three growth stages sampled; however, NDVI I (booting stage) had a significant negative correlation with DFF. Similarly, SPAD I (anthesis stage), was significantly and positively associated with SL, whereas SPAD I (booting stage) was significantly and negatively associated with PTPP. Meanwhile, GY had a significant positive association with PH, PTPP, SPS, GPS, and TGW, and with NDVI at anthesis and grain-filling. In turn, GPS was significantly and positively associated with DFF, DM, PH, SL, SPS, and GY. Another Spike-related trait, SL, was significantly and positively associated with DFF, DM, PH, SPS, GPS, SPAD II, GZnC, and GPC. Lastly, PH was significantly and positively associated with DM, SL, SPS, GPS, GY, and NDVI.
Validation of markers linked to GFeC, GZnC, and GPC
A list of markers linked to GFeC, GZnC, and GPC, validated in a set of 56 wheat genotypes, is shown in Table 4. Seven markers, including one linked to two traits, were validated in this study. Four markers, Xwmc617 (4A), Xbarc67 (3A), Xwmc283 (4A), and Xgwm361 (6B), were linked to GFeC, with a phenotypic variation explained (PVE) of 23.70%, 13.20%, 7.65%, and 7.47%, respectively. One marker, Xbarc146 (6A), was linked to both GFeC and GPC, with PVE of 23.94% and 7.11%, respectively; one SSR marker, Xgwm408 (5B) was linked to GPC with PVE of 11.09%. Similarly, one marker on 5B (gwm271) was linked to GZnC, with PVE of 7.11%.
Revalidation of markers linked to GFeC and GZnC
Identification of true hybrids and genotyping of the F 2 population
Two SSR markers, Xgwm271 and Xbarc67, were used to confirm the true hybrid nature of the HW 1098 × GPM DIC 87 cross. These two polymorphic markers were used to screen F1 plants to select true hybrids and avoid selfed progenies. The banding patterns of the parental genotypes and true hybrids are shown in Fig. 2. Subsequently, 12 plants verified as true F1s were subjected to selfing to generate F2 seeds, and the F2 population was further evaluated. Two informative SSR markers (Xgwm271 and Xbarc67) were used to genotype 200 plants from the F2 population of the HW 1098 × GPM DIC 87 cross. A representative banding pattern of the F2 population is shown in Fig. 3A and 3B.
Single marker analysis
The SMA was conducted on the F2 population derived from the HW1098 × GPM DIC 87 cross to assess the correlation between grain micronutrients and linked SSR markers. This analysis involved the computation of the F statistic and a simple regression coefficient, as outlined by Haley and Knott (1992). The extent to which phenotypic variance was accounted for by the markers was expressed as the phenotypic variance explained (PVE), and measured as R2. A comprehensive analysis leveraging genotypic and phenotypic data was conducted on each F2 individual resulting from the cross between HW 1098 and GPM DIC 87 to analyze GFeC and GZnC. Notably, the marker Xgwm 271 exhibited a significant association with GZnC, demonstrating a PVE of 56.63%. Similarly, the marker Xbarc 67 was found to be closely linked with GFeC, showing a significantly high PVE value of 72.78% (Table 5).
Discussion
Genetic improvement or response to selection for a particular trait depends mainly on heritability, selection intensity, and genetic variability, particularly additive genetic variance (Krishnappa et al. 2021; Singh and Narayanan 2013). The maximum variability measured as PCV and GCV, h2 bs, and GAM were observed for agro-morphological traits (except DM and GY), followed by quality traits (except GPC) and physiological traits. Although GCV was comparable for both GY (GCV: 46.8%) and PTPP (GCV: 46.8%), the GAM of the two traits is not comparable due to the influence of heritability, as the heritability of the GY was very low compared to the heritability of PTPP. Conversely, despite the higher heritability of grain micronutrients, the genetic advancement of the trait was low due to lower GCV and PCV. Therefore, the genetic improvement of a trait requires both high variability and heritability (Fyroj et al. 2020; Tazeen et al. 2009; Paul et al. 2006).
The population developed between contrasting parents enabled the identification of several transgressive segregants for most traits, predominantly grain quality traits (GFeC, GZnC, and GPC). These segregants were identified based on the superior performance of the progenies, surpassing the population mean in a desirable direction for each individual trait. Individuals who exhibit trait values that surpass those of the better parent indicate that the combination of alleles from the parental lines resulted in novel and potentially advantageous trait expression (Lephuthing et al. 2021; Rieseberg et al. 1999). Additionally, the occurrence of transgressive segregants is crucial in breeding programs, as it implies the availability of genetic variation that can be harnessed to select individuals with improved or unique trait combinations. Among quality traits and GY, a maximum of 50% were transgressive segregants for GPC, followed by 28.33% for GFeC, 13.33% for GZnC, and 10.0% for GY. A large number of transgressive segregants were observed for grain quality traits due to diverse parental allelic combinations, in which one parent was a high-yielding, well-adapted cultivar with a low micronutrient status and the other parent was a local germplasm collection with a very high micronutrient status. The efficient selection of nutrient-rich lines with favorable allelic combinations in further segregating generations would pave the way for the development of high-performing and nutritionally enhanced wheat cultivars. Historically, wheat-quality breeding has been slow and curtailed, compared with other economic traits, particularly grain yield (Krishnappa et al. 2019). Furthermore, quality evaluation of breeding material generally starts from advanced fixed lines, e.g., F7. During generation advancement, directional selection for high GY and disease resistance will eliminate much of the variability for grain quality traits, partially because of the difficulty in testing large segregating populations for quality parameters; the same holds true for physiological traits.
The significant positive correlations between GY and key morpho-physiological attributes (PH, PTPP, SPS, GPS, TGW, and NDVI) in the F2 population underscore the importance of these traits in influencing the overall GY. It is well established that GY and quality parameters are inversely related. Indeed, in this study, the association of GY with grain micronutrients was negative, although non-significant; similar to the case of another important quality trait, GPC, which had a positive but non-significant association with GY. Gene action and trait association also depend on the type of genetic material used. In the population used herein, the non-significant association between GY and quality traits makes this F2 population suitable for the simultaneous improvement of both GY and quality traits. Another important physiological parameter, NDVI, showed a significant positive correlation with GY. NDVI is an important and easily measurable physiological parameter that provides ground coverage quantification and crop nitrogen status; it is an important physiological tool that is highly correlated with grain yield, total biomass, and nitrogen status in wheat (Crain et al. 2012). Hence, the genetic potential of the genotypes for nitrogen use efficiency may influence the expression of related traits, such as NDVI and GPC (Krishnappa et al. 2023). Lastly, a significant and positive correlation was observed among quality traits (GFeC, GZnC, and GPC), suggesting a simultaneous improvement. Previous research has also reported the co-localization of genomic regions governing GFeC, GZnC, and GPC in wheat (Krishnappa et al. 2017; Uauy et al. 2006).
A set of nine polymorphic markers was used in the SMA in a set of 56 diverse genotypes to assess the linkage between those markers and traits. Of the nine polymorphic markers, seven (four markers for GFeC, one marker for GFeC and GPC, and one marker for GPC and GZnC) were identified. Among the seven markers above, Xbarc146, Xwmc617, and Xbarc67 were identified by Moradi et al. (2014), while the remaining four markers, Xwmc283, Xgwm361, Xgwm271, Xgwm408, were identified by Quarrie et al. (2006), Manish et al. (2014), Genc et al. (2009), and Fyroj (2017). To revalidate the validated makers, the F2 population was developed by comparing the parents HW 1098 and GPM DIC 87. Among the seven validated markers, one marker i.e., Xgwm271 linked to GZnC, was re-validated in the F2 population, with a PVE of 56.50%, and the second marker i.e., Xbarc67, linked to GFeC, was also re-validated, with a PVE of 72.78%. Previously, Krishnappa et al. (2018) validated three GZnC markers and two GFeC markers in a diverse set of 48 bread wheat genotypes.
Conclusion
The validation of molecular markers linked to QTL through MAS is an essential step in gene-pyramiding programs. Although several QTL have been identified in different investigations by using various marker systems and genetic materials, these novel QTL have seldom been validated in different genetic backgrounds. In this study, seven markers were validated in a set of 56 diverse dicoccum genotypes. Among these seven validated markers, two (Xgwm271 and Xbarc67) were revalidated in the F2 population, which are ideal candidates to use in MAS. In addition, the large variability generated between local landraces and popular cultivars will serve as potential genetic material for selecting high-yielding genotypes with high nutritional value. We believe our study makes a significant and unique contribution because it coupled conventional and molecular approaches to enhance the potential of plant breeding programs aimed to develop nutrient-rich, high-yielding wheat cultivars. Additionally, it re-validated two markers in a segregating population.
References
Avivi L (1979) High grain protein content in wild tetraploid wheat triticum dicoccoides Korn. in proceedings of the fifth international wheat genetics symposium, New Delhi, India, 23–28 pp. 372–380
Biradar SS, Fyroj U, Desai SA, Patil MK, Krishnappa G, Chethan CK, Sudha T (2023) GGE biplot analysis of biofortification traits in relation to grain yield in landraces of tetraploid wheat (Triticum turgidum ssp. dicoccum). Genet Resour Crop Evol 1–15. https://doi.org/10.1007/s10722-023-01766-7
Black RE, Victora CG, Walker SP, Bhutta ZA, Christian P, De Onis M, Ezzati M, Grantham-McGregor S, Katz J, Martorell R, Uauy R (2013) Maternal and child undernutrition and overweight in low-income and middle-income countries. The Lancet 382(9890):427–451
Burton GW, Dewane EH (1953) Estimating heritability in tall fescues (Festuca allamidiaceae) from replicated clonal material. Agron J 45:1476–1481
Cakmak I, Ozkan H, Braun HJ, Welch RM, Romheld V (2000) Zinc and Iron concentrations in seeds of wild, primitive and modern Wheat. Food Nutr Bull 21(4):401–403
Chhuneja P, Dhaliwal HS, Bains NS, Singh K (2006) Aegilops kotschyi and aegilops tauschii are the sources for high grain iron and zinc. Plant Breed 125:529–531
Crain J, Ortiz-Monasterio I, Raun B (2012) Evaluation of a reduced cost active NDVI sensor for crop nutrient management. J Sens 2012:582028
Darnton-Hill I, Webb P, Harvey PW, Hunt JM, Dalmiya N, Chopra M, Ball MJ, Bloem MW, de Benoist B (2005) Micronutrient deficiencies and gender: social and economic costs. Am J Clin Nutr 81(5):1198S-1205S
Distelfeld A, Cakmak I, Peleg Z, Ozturk I, Yazici AM, Budak H et al (2007) Multiple QTL-effects of wheat GpcB1 locus on grain protein and micronutrient concentrations. Plant Physiol 129:635–643
Fyroj U, Biradar SS, Desai SA, Rudra Naik V, Patil MK, Lakkanagoudar S, Ram S, (2020) Triticum dicoccum schubler wheat: a potential source for wheat bio-fortification program. Inter J Chem Stud 8(5):1417–1422
Fyroj U (2017) Genetic variability studies for grain nutrients, yield and yield attributes in tetraploid wheat. M. Sc. (Agri) Thesis, Univ. Agric. Sci., Dharwad, Karnataka (India)
Genc Y, Verbyla A, Torun A, Cakmak I, Willsmore K, Wallwork H, McDonald GK (2009) Quantitative trait loci analysis of zinc efficiency and grain zinc concentration in wheat using whole genome average interval mapping. Plant Soil 314:49–66
Gopalareddy K, Singh AM, Ahlawat AK, Singh GP, Jaiswal JP (2015) Genotype-environment interaction for grain iron and zinc concentration in recombinant inbred lines of a bread wheat (Triticum aestivum L.) cross. Indian J Genet Plant Breed 75:307–313
Von Grebmer K, Saltzman A, Birol E, Wiesman D, Prasai N, Yin S, Yohannes Y, Menon P, Thompson J, Sonntag A (2014) Synopsis: 2014 global hunger index: the challenge of hidden hunger Intl Food Policy Res Inst 83
Gupta PK, Balyan HS, Sharma S, Kumar R (2021) Biofortification and bioavailability of Zn, Fe and Se in wheat: present status and future prospects. Theor Appl Genet 134:1–35
Haley CS, Knott SA (1992) A simple regression method for mapping quantitative trait loci in line crosses using flanking markers. Heredity 6(4):315–324
Hammed AM, Simsek S (2014) Hulled wheat: a review of nutritional properties and processing methods. Cereal Chem 91:97–104
Hanson CH, Robinson HF, Comstock RE (1956) Biometrical studies on yield in segregating population of Korean lespedesa. Agron J 48:268–272
Johnson HW, Robinson HF, Comstock RE (1955) Estimation of genetic and environmental variability in soybeans. Agron J 47:314–318
Krebs NF, Miller LV, Michael Hambidge K (2014) Zinc deficiency in infants and children: a review of its complex and synergistic interactions. Paediatr Int Child Health 34(4):279–288
Krishnappa G, Singh AM, Chaudhary S, Ahlawat AK, Singh SK, Shukla RB, Jaiswal JP, Singh GP, Solanki IS (2017) Molecular mapping of the grain iron and zinc concentration, protein content and thousand kernel weight in wheat (Triticum aestivum L). PLoS ONE 12:e0174972
Krishnappa G, Singh AM, Ahlawat AK, Singh SK, Sharma P, Shukla RB, Singh GP (2018) Validation of QTLs for grain iron and zinc concentration in wheat (Triticum aestivum L). Indian J. Genet. Plant Breed. 78(03):378–381
Krishnappa G, Ahlawat AK, Shukla RB, Singh SK, Singh SK, Singh AM, Singh GP (2019) Multi-environment analysis of grain quality traits in recombinant inbred lines of a biparental cross in bread wheat (Triticum aestivum L.). Cereal Res Commun 47(2):334–344
Krishnappa G, Savadi S, Tyagi BS, Singh SK, Mamrutha HM, Kumar S, Mishra CN, Khan H, Singh G, Singh GP (2021) Integrated genomic selection for rapid improvement of crops. Genomics 113:1070–1086
Krishnappa G, Khan H, Krishna H, Kumar S, Mishra CN, Parkash O, Devate NB, Nepolean T, Rathan ND, Mamrutha HM, Srivastava P, Biradar S, Uday G, Kumar M, Singh G, Singh GP (2022) Genetic dissection of grain iron and zinc, and thousand kernel weight in wheat (Triticum aestivum L.) using genome-wide association study. Sci Rep 12:12444
Krishnappa G, Khan H, Krishna H, Devate NB, Kumar S, Mishra CN, Parkash O, Kumar S, Kumar M, Mamrutha HM, Singh GP (2023) Genome-wide association study for grain protein, thousand kernel weight, and normalized difference vegetation index in bread wheat (Triticum aestivum L.). Genes 14(3):637
Krishnappa G, Mamrutha HM, Rathan ND, Khan H, Mishra CN, Kumar V, Reddy KV, Pandey V, Khobra R, Singh C, Yashavanthakumar KJ (2024) Micronutrient biofortification in wheat: status and opportunities. In wheat science (pp. 285–301). CRC Press
Lachman J, Orsak M, Pivec V, Jiru K (2012) Antioxidant activity of grain of einkorn (Triticum monococcum L.), emmer (Triticum dicoccum schuebl [Schrank]) and spring wheat (Triticum aestivum L.) varieties. Plant Soil Environ 58:15–21
Lephuthing MC, Tolmay VL, Baloyi TA, Hlongoane T, Oliphant TA, Tsilo TJ (2021) Relationship of grain micronutrient concentrations and grain yield components in a doubled haploid bread wheat (Triticum aestivum) population. Crop Pasture Sci 73:116–126
Listman M, Ordonez R (2019) Ten things you should know about maize and wheat. CIMMYT Batan, Mexico
Liu J, Bihua W, Ravi PS, Govindan V (2019) QTL mapping for micronutrients concentration and yield component traits in a hexaploid wheat mapping population. J Cereal Sci 88:55–64
Lobell DB, Schlenker W, Costa-Roberts J (2011) Climate trends and global crop production. Science 333(6042):616–620
Manish K, Mishra VK, Gupta PK, Yadav PS, Kumar H, Joshi Arun AK (2014) Introgression of the high grain protein gene Gpc-B1 in an elite wheat variety of Indo-gangetic plains through marker assisted backcross breeding. Curr Plant Biol 1:60–67
Monasterio I, Graham R (2000) Breeding for trace minerals in wheat. Food Nutr Bull 21:392–396
Moradi N, Badakhshan H, Mohammadzadeh H, Zakeri MR, Mirzaghaderi, (2014) Assessment of genetic diversity and identification of SSR markers associated with grain iron content in Iranian prevalent wheat genotypes. J Plant Mol Bred 2:64–73
Morgounov A, Gomez-Becerra HF, Abugalieva A, Dzhunusova M, Yessimbekova M, Muminjanov H, Zelenskiy Y, Cakmak OL, I, (2007) Iron and zinc grain density in common wheat grown in central asia. Euphytica 155(2):193–203
Paul AK, Islam MA, Hasan MJ, Chowdhury MMH (2006) Genetic variation of some morpho-physiological characters in Triticum durum wheat. Int J Sust Agril Tech 2(8):11–14
Quarrie S, Quarrie PS, Radosevic R, Rancic D, Kaminska A (2006) Dissecting a wheat QTL for yield present in a range of environments: from the QTL to candidate genes. J Exp Bot 57:2627–2637
Rathan ND, Krishna H, Ellur RK, Sehgal D, Velu G, Ahlawat AK, Krishnappa G, Jaiswal JP, Singh JB, Saiprasad SV, Ambati D, Singh SK, Bajpai K, Mahendru-Singh A (2022) Genome-wide association study identifies loci and candidate genes for grain micronutrients and quality traits in wheat (Triticum aestivum L). Sci Rep 12:7037
Rieseberg LH, Archer MA, Wayne RK (1999) Transgressive segregation, adaptation and speciation. Heredity 83(4):363–372
Sharp PJ, Johnston S, Brown G, Mcintosh RA, Pallotta M, Carter M, Bariana HS, Khatkar S, Lagudah ES, Singh RP, Khairallah M, Potter R, Jones MGK (2001) Validation of molecular markers for wheat breeding. Aust J Agri Res 52:1357–1366
Singh K, Ghai M, Garg M, Chhuneja P, Kaur P, Schnurbusch T, Keller B, Dhaliwal HS (2007) An integrated molecular linkage map of diploid wheat based on a T. boeoticum × T. monococcum RIL population. Theor Appl Genet 115:301–312
Singh P, Narayanan SS (2013) Biometrical techniques in plant breeding. In. Kalyani publishers, India
Spielmeyer W, Sharp PJ, Lagudah ES (2003) Identification and validation of markers linked to broad-spectrum stem rust resistance gene Sr2 in wheat (Triticum aestivum L.). Crop Sci 43:333–336
Stevens GA, Beal T, Mbuya MNN, Luo H, Neufeld LM (2022) Global micronutrient deficiencies research group micronutrient deficiencies among preschool-aged children and women of reproductive age worldwide: a pooled analysis of individual level data from population-representative surveys. Lancet Glob Health 10(11):1590–1599
Tazeen M, Nadia K, Farzana NN (2009) Heritability, phenotypic correlation and path co-efficient studies for some agronomic characters in synthetic elite lines of wheat. J Food Aric Environ 7(3):278–282
Uauy C, Distelfeld A, Fahima T, Blechl A, Dubcovsky J (2006) A NAC gene regulating senescence improves grain protein, zinc and iron content in wheat. Science 314:1298–1301
Xu Y, Diaoguo A, Dongcheng L, Aimin Z, Hongxing X, Bin L (2012) Molecular mapping of QTLs for grain zinc, iron and protein concentration of wheat across two environments. Field Crops Res 38:57–62
Author information
Authors and Affiliations
Contributions
SSB, RK, RRH and SAD conceptualized the research plan. GK and RK carried out data analysis. MKP, LJ, UF, and KKM carried out the experimentation. GK and SR analysed grain iron, zinc and protein content. GK wrote the original draft and all authors reviewed and approved the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
All the authors declare no conflict of interest.
Ethical statement
This investigation does not involve any research with animals at any stage of experimentation.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kumar, R., Biradar, S.S., Patil, M.K. et al. Validation of quantitative trait loci for biofortification traits and variability research on agro-morphological, physiological, and quality traits in dicoccum wheat (Triticum dicoccum Schrank.). Genet Resour Crop Evol (2024). https://doi.org/10.1007/s10722-024-01973-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10722-024-01973-w