Abstract
Cluster bean [Cyamopsis tetragonoloba (L.) Taub] is an important leguminous crop of India, accounting for nearly 80% of the global production. The seeds of this economically important crop are used in various industrial applications worldwide like, textile, cosmetics, pharmaceuticals, mining, petroleum, natural gas, oil industries etc. Despite such high economic value, the productivity of this crop is quite low. One such major constraint in the production of cluster bean is bacterial leaf blight disease caused by Xanthomonas campestris pv. Cyamopsidis. Yield losses of up to 58–68% have been reported in case of severe infestation. Therefore, there is a need to formulate suitable breeding strategies to overcome the yield losses. But the lack of complete understanding of underlying genomic base in cluster bean is a major constraint in developing high-yielding and resistant cultivars/varieties. Here we report the first linkage map construction at the level of interspecific polymorphism using F3-4 segregating population based on Cyamopsis’s the basis of microsatellite markers of cluster bean and other legume crops which can further advance the breeding programs. An interspecific cross was developed in our laboratory using a bacterial blight resistant wild species (C. serrata Schinz.) and a moderately susceptible with high gum content cultivar (HG563). A total of 177 SSRs comprising EST-SSRs from cluster bean and ESTs as well genomic SSRs from other legumes were used in molecular analysis and 38 SSRs were found polymorphic. Linkage analysis revealed that nine markers (CBN21, CBN16, CBN17, CTF234, CBN03, CBN11, CTN15, CTN14, and GDR22) formed a single linkage group spanning 190 cM of cluster bean genome. Four minor QTLs were identified in interspecific cross for plant height (qPH1.1), hundred seed weight (qHSW1.1), disease resistance (qPDI1.1), and gum content (qGC1.1) mapped on single linkage group at 39.41, 123.41, 112.41 and 102.41 cM, respectively. The LOD values for all four QTLs ranged from 3.3 to 15.5. (above the threshold LOD > 3.0). Marker interval of CTN15 and CTN14 spanning 35.4–190.0 cM distance harboured all the four QTLs. Interestingly, plant height QTL was found only 4.8 cM apart from CTN 15 SSR marker, which can be exploited further for cluster bean improvement programs in future.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
There are many native food legumes whose inherent potential is untapped and underexploited. Cluster bean [Cyamopsis tetragonoloba (L.) Taub.] belongs to family leguminosae and generally known as ‘Guar’ in India. Cluster bean is a drought-tolerant annual legume, having chromosome number 2n = 14 (Gresta et al. 2018; Kumar et al. 2017). The deep tap root system of guar helps in acclimatizing the water deficient crop system, thus making it suitable for growing in arid and semi-arid regions of India. The origin of Cyamopsis tetragonoloba is unknown since it has never been found in the wild (Whistler and Hymowitz 1979). It is reported to have developed from the African species i.e. Cyamopsis senegalensis (Gillett 1958). It was further domesticated in South Asia, where it has been cultivated for centuries (Mudgil et al. 2014). In India, crop is mainly grown in the dry habitats of Rajasthan, Haryana, Gujarat, Punjab, Uttar Pradesh, and Madhya Pradesh.
Cluster bean has undergone a noteworthy journey from poor man’s crop to industrial crop due to bounteous industrial applications of gum present in endosperm of seeds also known as ‘Guar gum’. Guar gum is a galactomannan polysaccharide, a natural thickener, and forms a gel in water. It has numerous industrial applications like oil and gas drilling, textile, paper, food industries, cosmetics, water treatment, mining, pharmaceuticals, etc. (Coveney et al. 2000; Mudgil et al. 2014; Undersander et al. 1991). Due to high industrial value and a steep spike in demand, guar has attracted farmers’ attention in Australia, Bangladesh, Myanmar, the USA, South Africa, Brazil, Congo, and Sri Lanka (Boghara et al. 2016; Kumar et al. 2017). India exports different cluster bean products to a large number of countries and dominates in exportation of guar gum worldwide. The country exported Rs. 381,880.16 MT of cluster bean gum to the world from April, 2019 to March, 2020 for the worth of Rs.3261.60 crores (APEDA, 2020) majorly to countries like the United States, China, Russia, Norway, and Germany.
Cluster bean being a profitable crop, yet encounters a lot of different yield constraints. Bacterial leaf blight (BLB) has been a major threat to this crop, particularly during the years of prolonged and late summer rains (Murwan and Abdallah 2008). Bacterial leaf blight of cluster bean is a seed-borne disease caused by Xanthomonas campestris pv. cyamopsidis (Mali et al. 1989; Mihail and Alcorn 1985) and was first reported in India as bacterial leaf spot by Patel et al. (1953). Later on, the same disease was identified as bacterial blight (Patel and Patel 1958; Srivastava and Rao 1963). Yield losses have been recorded up to 58–68% in severe infestation conditions (Gandhi and Chand 1985; Srivastava and Rao 1963). The high industrial demand for cluster bean (guar gum) in various industries has drawn attention of researchers to develop improved commercial genotypes.
Molecular markers are worthy of attention as of their significant roles in different areas of plant breeding like genetic diversity studies, phylogenetic analysis, deducing genetic polymorphism at particular loci or whole genome sequence level, genetic mapping, cloning of agronomical important trait governing genes based on map (Heffner et al. 2010; Jiang 2013; Morgante et al. 2002). The amalgam of breeding techniques and molecular biology has made deeper insight into traits possible, as the genome is saturated with more markers, one gets closer to a gene-controlling trait (Jaradat 2016; Xu 2010).
Despite of such an important crop, there is lack of complete understanding of underlying genomic base and no comprehensive microsatellite-based linkage map has been reported in cluster bean. This is first attempt on linkage map construction which may facilitate cluster bean breeding for the development of improved varieties of cluster bean.
Here we report construction of a linkage map using microsatellite markers of cluster bean along with other legumes using F3-4 segregating populations of interspecific (HG563 Cyamopsis serrata) cross developed in our laboratory. In interspecific cross, C. serrata highly resistant to bacterial leaf blight (BLB) and HG563, moderately susceptible with high gum content were used. Further, agronomic traits were utilized in order to identify QTLs for BLB resistance and gum content.
Materials and methods
Plant material
An interspecific cross between HG563 (high-yielding cultivated variety) and C. serrata (wild and BLB resistant; Fig. 1) was made in our laboratory and interspecific hybrids were recovered through embryo rescue method and validated at molecular level (Sombir 2013). F1 plants were grown to get F2 in the field of farm area of Forage Section, Department of Genetics and Plant Breeding, CCS Haryana Agricultural University, Hisar. F2-3 seeds were harvested from individual F2 plants and likewise, F3-4 was raised using F2-3 plants seeds, which were used for the present study along with parental genotypes. Segregating population (F3-4) of interspecific cross was raised at two locations i.e., research area of Forage Section, Department of Genetics and Plant Breeding, CCS Haryana Agricultural University, Hisar and Regional Research Station, Bawal, CCS Haryana Agricultural University, Hisar. All the plants were grown in natural conditions during the 2016–17 growing season. A total 64 lines of F3-4 population were used for further phenotypic and genotypic analysis.
Phenotypic observation
Phenotypic data was recorded on 64 lines derived from interspecific cross (HG563 × C. serrata) at both locations i.e., CCS Haryana Agricultural University, Hisar and Regional Research Station, Bawal, (after maturity) by taking five replications from each line and taking average of them. Phenotypic parameters like plant height, branches per plant, clusters per plant, pods per cluster, pods per plant, pod length, days to 50% flowering, days to maturity (The days were counted from the date of sowing to appearance up to 50 percent of flowering in each plant), 100 seed weight, and seed yield per plant were recorded. Statistical analysis of the data such as mean, range, standard error, and coefficient of variation was performed using OPSTAT software (Sheoran et al. 1998). The gum content was estimated by extraction of galactomannan followed by its purification, precipitation and precipitate was finally dissolved to measure gum content spectro-photometrically as described by Das et al. (1977) and further improved by Joshi et al. (2004).
Disease scoring
Observations for disease intensity of bacterial leaf blight were recorded in natural conditions using 0–5 disease scale given by Wheeler (1969). The percent disease index was calculated as:
Five plants from each replication selected randomly, were tagged and further assessed for percent disease index calculation (Table 1).
Genotyping
A total of 177 SSRs from cluster bean, common bean, adzuki bean, pigeon pea, and mung bean were used in the present study (Supplementary Table 1). EST SSRs from cluster bean used were11 EST SSRs (GDR1-11) related to disease resistance (Kaur 2014), 11 EST SSRs (GDR12-22) related to disease resistance (Aditi 2018), 36 EST SSRs related to galactomannan metabolism (CTN) from cluster bean (Wadhwa 2015) and 45 EST SSRs (CBN and CTF series; Kuravadi et al. 2014) synthesized from sequence available in the public domain. Remaining SSRs were taken from other legumes like 15 genomic SSRs from mung bean (MBSSRG series; Singh et al. 2013), 19 genomic SSRs from common bean (BM and PVBR series; Gaitán-Solís et al. 2002; Grisi et al. 2007, respectively), 20 EST SSRs from Adzuki bean (Az series; Chen et al. 2015) and 20 EST SSRs from Pigeon pea (ICPeM series; Raju et al. 2010). So, total EST SSRs are summed up to 143 out of 177, and remaining 34 SSRs were genomic (from mungbean and common bean).
SSR marker analysis
SSR markers were validated by polymerase chain reaction of genomic DNA in a 20 µl reaction mixture. Each reaction contained 50 ng of DNA template, 2.5 mM MgCl2 (G Biosciences), 250 µM of dNTPs mix (G Biosciences), 0.4 µM of primers (Integrated DNA Technologies, IDT), and 1.5 units of Taq DNA polymerase (G Biosciences). Polymerase chain reaction amplification was carried out in Benchtop thermocycler under following reaction conditions: initial denaturation at 94ºC for 3 min and 35 cycles of denaturation at 94ºC for 1 min, primer annealing at 46-52ºC depending upon primer pair for 1 min, primer extension at 72ºC for 2 min, final extension at 72°C for 10 min. The amplified products were resolved on 8% Polyacrylamide gel electrophoresis (PAGE) stained with Ethidium bromide (EtBr) (Sigma-Aldrich) and analysed in UV light and photographed using gel documentation system (Bio-Rad).
Polymorphism study and allele scoring
Validated SSR markers were further screened for polymorphism between parents of interspecific cross (HG 563 × C. serrata). Subsequently, polymorphic markers were genotyped in the 64 lines of F3-4 population by PCR amplification of genomic DNA as described above. After visualization of amplified products in PAGE, scoring was done based on presence (taken as 1) or absence (taken as 0) of band for each position in all plants. The size (in nucleotide base pairs) of the amplified bands was determined based on its migration relative to standard molecular marker (20 bp plus DNA ladder, Takara).
Linkage analysis
A genotyping data matrix was generated based on the scoring pattern observed in the segregating population with all the polymorphic microsatellite markers. The matrix generated was integrated with the markers and used as an input file in a publicly-available software MAPMAKER/EXP 3.0 (Lincoln et al. 1993) for construction of framework of linkage map. After reducing this dataset by merging perfectly linked markers with one another, linkage groups were assigned to chromosomes based on their ordering in the same group. Two-point, three-point, and multipoint analysis were used to determine the best order of marker loci within the linkage groups. The markers were grouped at LOD values of 4, 3, 2, and 1.5 to determine the best combination of markers within same linkage group and it was observed that at a LOD value of 1.5 and distance of 100 cM all the markers were arranged into 1 linkage group (LG). Correct order was checked using ‘ripple’ command in the Map maker. Distances between the loci were mapped in kosambi. The highest threshold LOD value of 1.5 was chosen based on as a uniform threshold for constructing the map. Map was constructed using software WinQTL cartographer, version 2.5.
QTL mapping
Windows QTL cartographer version 2.5 was used to map quantitative trait loci (QTL) using morphological data of 64 lines at both locations. The map positions (cM) of all the 9 linked polymorphic markers were obtained from linkage map of interspecific cross. SSR allelic data was scored according to the translation available in the manual for WinQTL cartographer. The number 2, was assigned to C. serrata; 0 to HG563; 1 to heterozygous alleles whereas -1 was assigned to the missing allele. QTL positions were assigned to the point of maximum LOD score above threshold value in the target regions. These LOD profiles were used to identify the most likely position for a QTL in relation to the linkage map, which was the position where the highest LOD value was obtained. An output excel file was created which included all the details of the QTL (i.e. chromosome location, map positions, additive effect, LR value, etc.). In addition, the proportion of observed phenotypic variance explained by each QTL and corresponding additive effect was estimated. The information in this file was used for localizing the QTL on linkage groups.
Results
Morphological traits evaluation in parents and segregating population
Morphological traits were recorded in segregating population including parents of mapping population at both locations i.e. Research area of Forage Section, Department of Genetics and Plant Breeding, CCS Haryana Agricultural University, Hisar and Regional Research Station, Bawal, CCS Haryana Agricultural University, Hisar (Supplementary Tables 2 and 3). Parents of interspecific mapping population were highly contrasting for all morphological traits which further exhibited the variation in segregating population for all traits as revealed by their mean value (Tables 2, 3).
Population was grown at research area of Forage Section, Department of Genetics and Plant Breeding, CCS Haryana Agricultural University, Hisar, and data was recorded. Plant height (PH) of parents of F3-4 population i.e. HG563 and C. serrata was 132.40 cm and 11.50 cm, respectively while plant height in their segregating population ranged between 85.00 and 150.00 cm with a mean value of 118.57 cm. likewise other morphological characters like branches per plant (BCH) in segregating population ranged between 7.00 and 16.00 with a mean value of 10.79 while C. serrata and HG563 had mean no. of branches per plant were 6.00 and 15.40 respectively. Clusters per plant (CL) ranged between 14.00 and 35.00, pods per cluster (P/CL) ranged between 3.60 and 7.72, pods per plant (P/Pl) ranged between 70.00 and 177.00, pod length (PL) ranged between 4.60 and 5.64 cm, Days to 50% flowering (FW) ranged between 37.00 and 43.21 days, days to maturity (DM) ranged between 87.60 and 100.40 days, 100 seed weight (HSW) ranged between 1.72 and 2.89 g while seed yield per plant (SY) ranged between 12.52 and 45.00 g.
In segregating population grown at Regional Research Station Bawal, PH varied between 89.00 and 151.00 cm with a mean value of 127.27 cm while C. serrata and HG563 had mean PH of 11.00 cm and 134.40 cm, respectively. Similarly, BCH ranged between 7.00 and 17.00, CL ranged between 37.00 and 59.40, P/CL ranged between 4.90 and 5.96, P/Pl ranged between 139.33 and 245.20, PL ranged between 4.75 and 5.27 cm, FW 35.25–43.80 days, DM ranged between 84.00 and 97.40 days, HSW ranged between 1.50 and 2.84 g, SY ranged between 17.30 and 42.10 g.
Screening of bacterial leaf blight in parents and segregating population under natural conditions
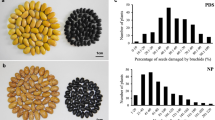
Along with parents, segregating population of interspecific cross (HG563 × C. serrata) was screened for bacterial leaf blight disease under natural conditions using 0–5 scale (Table 4) at research area of Forage Section, Department of Genetics and Plant Breeding, CCS Haryana Agricultural University, Hisar. No plant was found highly resistant to the disease. C. serrata was resistant while HG563 found susceptible to the disease. Three lines (2–8, 5–11, and 6–9) showed resistant reaction to the disease. Four lines (2–3, 3–2, 5–7 and 6–6) were found moderately resistant while nine lines (4–2, 5–2, 5–9, 5–10, 6–8, 6–13, 6–24, 13–3 and 13–5) were showed moderately susceptible reaction to the disease. Eighteen lines (2–1, 2–13, 3–8, 6–1, 6–4, 6–11, 6–17, 6–17, 6–18, 6–19, 6–23, 7–1, 7–2, 9–1, 9–4, 9–13, 13–2 and 13–4) showed susceptible reaction. Remaining 30 lines (1–2, 2–4, 2–5, 2–9, 2–11, 2–14, 3–1, 3–3, 4–3, 4–6, 5–1, 5–3, 5–8, 6–3, 6–7, 6–10, 6–13, 6–14, 6–16, 6–20, 6–21, 7–4,7–6, 9–2, 9–3, 9–5, 9–6, 11–1, 12–1 and 13–8) were found highly susceptible to the disease.
At Regional Research Station, Bawal also, no plant was found highly resistant (Table 5). C. serrata showed resistance to the disease while HG563 was found susceptible in a similar way to CCS HAU, Hisar location. Two lines (2–4 and 6–9) showed resistant reaction to the disease. Six lines (1–1, 2–8, 5–3, 6–3, 3–2, and 5–11) were found moderately resistant. Eight lines (1–1, 2–8, 3–2, 5–3, 5–11, 6–3, 3–2, and 5–11) showed moderately sensitive reactions to the disease. HG563 along with 11 lines (P2, 2–13, 2–14, 5–7, 5–8, 5–9, 5–10, 6–8, 6–12, 6–24, 7–1, and 9–13) showed sensitive reaction. Remaining 26 lines (2–9, 2–12, 3–3, 3–5, 4–2, 4–3, 4–6, 5–1, 6–6, 6–10, 6–18, 6–19, 6–20, 6–21, 6–23, 6–25, 7–4, 7–5, 7–6, 9–1, 9–2, 9–3, 9–6, 11–2, 13–3 and 13–5) were found highly sensitive to the disease.
Polymorphic markers for map construction
A total of 165 plants from 64 lines along with parents were taken for further molecular analysis using EST SSRs of cluster bean and SSRs from other legumes. In the present study, 177 SSRs were used for screening parental polymorphism, and out of these, 38 SSRs were found polymorphic in parents (Table 6) which were further screened in segregating population (Figs. 2, 3, 4, 5). So, in total 19.79% SSRs showed polymorphism among parents of interspecific cross. In case of SSRs from other legumes, mostly did not amplified the cluster bean genomic DNA except common bean primers from which 15.7% SSRs amplified and showed polymorphism among parents.
Linkage map construction
Out of 177 SSRs, thirty-eight markers were shown polymorphism among parents and subsequently used for genotyping of segregating population. At LOD 1.5, Only nine markers (CBN21, CBN16, CBN17, CTF234, CBN03, CBN11, CTN15, CTN14, and GDR22) out of 38 polymorphic markers were assigned on a single linkage group and remaining markers did not group to any of the linkage group and hence remain unlinked (Fig. 6). The interspecific map generated here spanned 190 cM distance of cluster bean genome.
QTL mapping for yield attribute, BLB disease traits, and gum content in cluster bean using F3-4 population (HG563 × C. serrata) under field conditions
Nine polymorphic SSR markers were mapped on one linkage group. Composite interval mapping (CIM) analysis by WinQTL Cartographer 2.5 revealed a total of 4 QTLs for yield attributes, BLB disease index, and gum content in F3-4 population. Out of these identified QTLs, 1 QTL for plant height (qPH1.1), 1 QTL for hundred seed weight (qHSW1.1), 1 QTL for gum content (qGC1.1), and 1 QTL for disease resistance (qPDI1.1) have been mapped on single linkage group at 39.41, 123.41, 102.41 cM and 112.41, respectively (Fig. 8). The LOD values for all four QTLs were 3.3, 12.5, 4.7, and 15.5, respectively (Above cut off of 3.0, Fig. 7). The R2% ranged between 0.1–1.3 suggesting that the identified QTLs were minor effect QTLs (Table 7). Plant height QTL was observed only 4.8 cM apart from CTN15 SSR marker which can be a useful marker for further molecular breeding approaches (Fig. 8).
Promising lines selected based on morphological traits and disease score
In interspecific cross three lines namely 2–8, 5–11, and 6–9 were selected on the basis agronomic data and BLB resistance data under field conditions recorded at research area of Forage section, Department of Genetics and Plant Breeding, CCS Haryana Agricultural University, Hisar. In same manner, two lines namely 2–4 and 6–9 were selected at Regional Research Station, Bawal, CCS Haryana Agricultural University, Hisar. These lines had seed yield per plant comparable to parent HG563 and showed disease resistance which suggests the transfer of disease-resistant alleles from parent C. serrata. Line no. 6–9 showed high seed yield and Bacterial leaf blight resistance at both locations i.e., Bawal and Hisar. This also showed early flowering (days to 50% flowering) than HG563 at both locations.
Discussion
Cluster bean [Cyamopsis tetragonoloba (L.) Taub] is an important Kharif legume (2n = 14) commonly known as cluster bean which is well adapted to Arid and Semi-Arid regions of the world. It is a multipurpose legume and the presence of galactomannan gum has made this crop industrially important (Reddy et al. 2014). The increased demand has put stress on cluster bean breeders and farmers for ameliorating its production and productivity. But cluster bean suffers from many yield-limiting factors and bacterial leaf blight is the major threat during the years of prolonged and late summer rains (Murwan and Abdallah 2008). Bai et al. (1997) reported that genetic studies indicate that resistance to common bacterial blight is different depending upon source of resistance and may be determined by both major and minor genes. Early maturing varieties should be used to increase yield and productivity. Thus, farmers are interested in cluster bean varieties that are resistant to diseases and are high yielding. In recent years, genetics and molecular biology techniques have come hand in hand for increasing our understanding of structural and functional aspects of plant genomes. Molecular markers are such a powerful tool of molecular plant breeding that can be used in various ways to assist plant breeding for selection of useful traits. Marker trait associations have been established for various agronomically useful traits in so many legumes like common bean, cow pea, chickpea, mung bean, pigeon pea, etc. However, such advancements are not visible in case of cluster bean making it an orphan crop and little genetic advancements have been made in cluster bean. Tanwar et al. (2017) reported for the first-time transcriptome characterization and SNP identification in cluster bean. Further, Rawal et al. (2017) constructed a database ‘ClustergeneDB’ for retrieval of unigenes and microsatellite markers. Genomic SSRs were developed via next-generation sequencing and validated in guar (Kumar et al. 2020). These studies will prompt the advancement in cluster bean improvement programs like construction of linkage maps, identification of QTLs linked to useful traits and will further aid in cost-effective breeding programs. In cluster bean, none of the cultivated germplasm has complete BLB resistance. So, to develop disease-resistant varieties, inheritance of disease resistance must be known, which can be used by breeders for precise selection of resistant genotypes. Anil et al. (2012) reported the inhibitory gene action for resistance to bacterial leaf blight disease in two crosses of cluster bean i.e. HG563 x PNB and HG75 x PNB. Segregation ratio of 13:3 suggested bacterial leaf blight resistance showing the non-Mendelian inheritance. In the present study, three lines of interspecific cross grown at Hisar location showed resistance and four lines were moderately resistant to disease and at Bawal, location, two lines were resistant and six lines were moderately resistant to BLB disease. Remaining lines were susceptible to the disease. So, segregating population was not following Mendelian inheritance for disease resistance. It suggests that epistatic interaction governs BLB resistance. Our study showed limitation that we cannot identify the genes involved in bacterial leaf blight resistance, so further investigation is required to identify genes involved in Bacterial leaf blight resistance.
For this study, interspecific cross made between HG563 (cultivated genotype) and C. serrata (wild relative) was used for widening the genetic base and creating greater variability in yield-related attributes in F3-4 population. Where C. serrata is a wild species that has some desirable attributes like drought tolerance, photo and thermo-insensitivity and disease resistance and HG563 is high gum content and high-yielding variety but susceptible to BLB. So desirable traits of both varieties can be combined in single line. Further, this population was derived from contrasting parents making them suitable for linkage map construction and QTL identification. In the present study, F3-4 populations of interspecific were evaluated for various morphological characters (plant height, no. of branches per plant, no. of clusters per plant, pods per cluster, pods per plant, pod length, days to 50% flowering, days to maturity, 100 seed weight and seed yield per plant) and were screened for bacterial leaf blight. Microsatellite analysis was done using EST SSRs from cluster bean and SSRs from other legumes. Segregating population showed large variation for plant height, branches per plant, clusters per plant, and pods per plant. In interspecific cross, plant height varied from 85 to 150 cm, pods per plant ranged from 70.00 to 177.00, cluster per plant ranged from 14.00 to 35.00 and seed yield ranged from 12.52 to 45.00 g at Hisar and likewise at Bawal also large variation observed for Plant height (89–151 cm) clusters per plant (37.00–59.40) and pods per plant (139.33–245.20). These results are in concurrence with study of Reddy et al. (2018) where they studied the variability in yield and yield attributed in 51 cluster bean genotypes. They also found large variation for plant height (36.9–121.5 cm), clusters per plant (9.6–24.8), and pods per plant (44–96.76). Seed yield per plant varied from 5.76 to 25.95 g in a study by Manvinnan et al. (2015) which is much less than what observed in the present study and this may be due to different geographical locations.
In the present study, low range of variation was recorded for hundred seed weight, pod length, and days to 50% flowering. In interspecific cross, hundred seed weight ranged from 1.72 to 2.89 g, pod length ranged from 4.60 to 5.64 cm and days to 50% flowering ranged from 37.00 to 43.21 at Hisar. In Bawal also, low-range variations were recorded for pod length (4.75–5.27 cm) days to 50% flowering (35.25–43.80) and hundred seed weight (1.50–2.84 g). Similarly, Manvinnan et al. (2015) in his study found low range of variation in hundred seed weight (3.32–4.99 g), pod length (5.26- 12.88 cm), days to 50% flowering (23.5–34.5), and pods per cluster (2.6–10.7). Reddy et al. (2018) also observed low range of variation for pod length (4–11.5), days to 50% flowering (38–48), and pods per cluster (3.1–5.6).
In this cross, 5 EST SSRs (GDR1, GDR6, GDR7, GDR10, and GDR11) and 5 genic SSRs (GDR15, GDR16, GDR18, GDR20, and GDR22) showed polymorphism which were directly related to disease resistance. These polymorphic EST SSRs were further used to screen segregating population. Like GDR1, SSR was reported to exhibit a distinct defence profile in maize (Liu et al. 2017), GDR6 showed upregulated in response to Phytophthora sojae infection and was found responsible for increased resistance in transgenic tobacco and soybean in response to P. nicotiana Breda and P. sojae respectively (Xu et al. 2014), GDR9 and GDR20 found to play role in fungal and bacterial disease resistance (Swada et al. 2004), GDR10 and GDR22 EST-SSRs showed putative function of SRC2 gene product which was found upregulated in in response in response to infection with the type II -non-host-pathogen Xanthomonas axonopodis pv. glycines 8 ra, which elicits a hypersensitive response (Kim et al. 2008), GDR11 showed homology with PR-1 protein in a study by Ali et al. (2017). Quantitative real-time PCR (qRT-PCR) results showed increased expression of BjPr1 gene in both local (infected) and distal (non-infected) leaves of Brassica juncea after infection whereas mechanical wounding showed expression only in local (wounded) leaves but not in distal (unwounded) leaves.
Linkage mapping
Linkage map was constructed in interspecific population using polymorphic markers. In interspecific cross, thirty-eight markers showed polymorphism and out of which nine markers were found suitable for mapping while remaining markers did not map to any linkage group. Interspecific polymorphic EST SSR markers (CBN21, CBN16, CBN17, CTF234, CBN03, CBN11, CTN15, CTN14 and GDR22) formed one linkage group covering distance190 cM.
This skeletal map was constructed based on interspecific microsatellite polymorphism. The EST SSRs used in the present study showed less polymorphism, in other studies, EST SSRs have also been reported to show less polymorphism in other crop plants because of greater DNA sequence conservation in transcribed regions unlike genomic SSRs (Chanbane et al. 2005; Cho et al. 2000; Corderso et al. 2001; Eujayl et al. 2001 and Russell et al. 2004) and less no. of markers used may be the reason of less genome coverage. In capsicum spp. first report of inter-specific partial linkage map was given by Arjun et al. (2018) in F2 population of interspecific cross. Only 28 markers were found polymorphic and screened in F2 population. Twenty-four SSRs mapped on three linkage groups (LGs): LG1, LG2, and LG3. Two fruit length-determining QTLs designated as paufl2.1 and paufl2.2 was identified on LG2.
QTL mapping is a highly effective approach for studying genetically complex forms of plant disease resistance. QTL analysis is helpful in finding the loci involved in disease resistance. QTL for disease resistance can further aid in selecting plants with disease resistance loci. In this study, in interspecific cross, four QTLs were identified for plant height (qPH1.1), hundred seed weight (qHSW1.1), disease resistance (qPDI1.1), and gum content (qGC1.1) which mapped on single linkage group at 39.41, 123.41, 112.41 and 102.41 cM, respectively. The LOD values for all four QTLs were 3.3, 12.5, 15.5, and 4.7, respectively. The R2% ranged between 0.1 and 1.3 suggesting that the identified QTLs were minor effect QTLs. QTL for plant height was found nearest to CTN 15 which is only 4.8 cM apart which can be advantageous for further cluster bean breeding programmes. Flanking markers for identified QTLs for plant height, disease resistance, hundred seed weight and gum content were CTN 14 and CTN 15, which were 154.6 cM apart. So, fine mapping is needed to find out flanking markers which will allow marker-assisted selection for targeted traits.
In the present study, promising lines were selected based on high yield and BLB resistance. In interspecific cross, line no. 6–9 showed better performance at both locations (Hisar and Bawal). These lines are being further evaluated for their use in cluster bean breeding programs.
Conclusions
An initiative was taken to conduct linkage studies and linkage map construction using F3-4 interspecific mapping population. Sixty-four polymorphic markers were used in linkage map construction, however, analysis showed nine markers forming only one linkage group. QTLs related to important traits such as plant height, disease resistance, and gum content were also identified on this linkage group. Interestingly, plant height QTL at 39.4 cM had close CTN 15 SSR at 34.5 cM, only 4.8 cM apart from QTL, seems quite promising for molecular breeding Interestingly, plant height QTL at 39.4 cM had close CTN 15 SSR at 34.5 cM, only 4.8 cM apart from QTL, seems quite promising for molecular breeding. The promising plants with alleles for BLB resistance (from C. serrata) and galactomannan metabolism (from HG563) have been identified which can be used for further evaluation and development of breeding lines with high BLB resistance and gum content. The study is the first report on linkage construction and QTL identification in cluster bean using 177 markers only, nonetheless shows the right step in this much-neglected crop in terms of molecular breeding when other crops have excellent genomic resources to work. Further efforts will definitely prove their worth in enriching genomic resources as well as providing molecular tools to plant breeders for its improvement.
References
Aditi (2018) EST-SSR and RGA polymorphism for diversity analysis and Bacterial Leaf Blight in cluster bean [Cyamopsis tetragonoloba (L.) Taub.]. Ph.D. thesis submitted to CCS Haryana Agricultural University, Hisar, Haryana
Ali S, Mir ZA, Tyagi A, Bhat JA, Chandrashekar N, Papolu PK, Rawat S, Grover A (2017) Identification and comparative analysis of Brassica juncea pathogenesis-related genes in response to hormonal, biotic and abiotic stresses. Acta Physiol Plant 39(12):1–15. https://doi.org/10.1007/s11738-017-2565-8
Anil PSK, Kadian P, Gandhi SK, Bhusal N (2012) Inheritance of bacterial leaf blight resistance in cluster bean. Forage Research 38(3):182–183
Arjun K, Dhaliwal MS, Jindal SK, Fakrudin B (2018) Mapping of fruit length related QTLs in interspecific cross (Capsicum annuum L. × Capsicum galapagoense Hunz.) of chilli. Breed Sci 68(2):219–226. https://doi.org/10.1270/jsbbs.17073s
Bai Y, Michaels TE, Pauls KP (1997) Identification of RAPD markers linked to common bacterial blight resistance genes in Phaseolus vulgaris L. Genome 40(4):544–551. https://doi.org/10.1139/g97-071
Boghara MC, Dhaduk HL, Kumar S, Parekh MJ, Patel NJ, Sharma R (2016) Genetic divergence, path analysis and molecular diversity analysis in cluster bean (Cyamopsis tetragonoloba L. Taub.). Ind Crop Prod 89:468–477. https://doi.org/10.1016/j.indcrop.2016.05.049
Chanbane K, Ablett GA, Cordeiro GM, Valkoun J, Henry RJ (2005) EST versus genomic derived microsatellite markers for genotyping wild and cultivated barley. Genet Resour Crop Evol 52(7):903–909. https://doi.org/10.1007/s10722-003-6112-7
Chen H, Liu L, Wang L, Wang S, Somta P, Cheng X (2015) Development and validation of ESTSSR markers from the transcriptome of adzuki bean (Vigna angularis). PLoS ONE 10(7):e0131939. https://doi.org/10.1371/journal.pone.0131939
Cho YG, Ishii T, Temnykh S, Chen X, Lipovich L, McCouch SR, Park WD, Ayres N, Cartinhour S (2000) Diversity of microsatellites derived from genomic libraries and GenBank sequences in rice (Oryza sativa L.). Theor Appl Genet 100:713–722. https://doi.org/10.1007/s001220051343
Coveney PV, Silva HD, Gomtsyan A, Whiting A, Boek ES (2000) Novel approaches to cross-linking high molecular weight polysaccharides: application to guar-based hydraulic fracturing fluids. Mol Simul 25:265–299. https://doi.org/10.1080/08927020008024503
Das B, Arora SK, Luthra YP (1977) A rapid method for determination of gum in guar (Cyamopsis tetragonoloba (L.) Taub.). in Proceedings of the 1st Institute for Conflict Analysis and Resolution Workshop (CAZRI '77), pp. 117–123, Jodhpur, India
Eujayl I, Sorrells M, Baum M, Wolters P, Powell W (2001) Assessment of genotypic variation among cultivated durum wheat based on EST-SSRs and genomic SSRs. Euphytica 119(1–2):39–43. https://doi.org/10.1023/a:1017537720475
Gaitán-Solís E, Duque MC, Edwards KJ, Tohme J (2002) Microsatellite repeats in common bean (Phaseolus vulgaris): isolation, characterization, and cross-species amplification in Phaseolus ssp. Crop Sci 42:2128–2136. https://doi.org/10.2135/cropsci2002.2128
Gandhi SK, Chand JN (1985) Yield losses in cluster bean due to bacterial blight caused by Xanthomonas campestris pv. cyamopsidis. Indian Phytopathol 38:516–519
Gillett JB (1958) Indigofera (Microcharis) in tropical Africa with the related genera Cyamopsis and Rhynchotropis. Kew Bull Addit Series 1:1–166
Gresta F, Avola G, Cannavo S, Santonoceto C (2018) Morphological, biological, productive and qualitative characterization of 68 guar (Cyamopsis tetragonoloba (L.) geotypes. Ind Crops Prod 114:98–107. https://doi.org/10.1016/j.indcrop.2018.01.070
Grisi MCM, Blair MW, Gepts P, Brondani C, Pereira PAA, Brondani RPV (2007) Genetic mapping of a new set of microsatellite markers in a refernce common bean (Phaseolus vulgaris) population BAT93 x Jalo EEP558. Genet Mol Res 6(3):691–706
Heffner EL, Lorenz AJ, Jannink JL, Sorrells ME (2010) Plant breeding with genomic selection: gain per unit time and cost. Crop Sci 50(5):1681–1690. https://doi.org/10.2135/cropsci2009.11.0662
Jaradat AA (2016) Breeding oilseed crops for climate change. In: Gupta SK (ed) Breeding oilseed crops sustain prod. Academic Press, Cambridge, pp 421–472
Jiang G (2013) Molecular markers and marker-assisted breeding in plants. In (Ed.), Plant breeding from laboratories to fields. IntechOpen, London, pp 45–83 https://doi.org/10.5772/52583
Joshi UN (2004) Advances in chemistry, biochemistry and industrial utilization of guar seed. In: Guar JV Singh, Dahiya BS (Eds.), p. 197, Indian Society of Forage Research, Hisar and Agricultural and Processed Food Products Export Development Authority (APEDA), New Delhi, India 229
Kaur A (2014) Molecular marker analysis for bacterial blight resistance in cluster bean (Cyamopsis tetragonoloba (L.) Taub.). M.Sc. thesis submitted to CCS Haryana Agricultural University, Hisar
Kim YC, Kim SY, Choi D, Ryu CM, Park JM (2008) Molecular characterization of a pepper C2 domain-containing SRC2 protein implicated in resistance against host and non-host pathogens and abiotic stresses. Planta 227(5):1169–1179. https://doi.org/10.1007/s00425-007-0680-2
Kumar S, Modi AR, Parekh MJ et al (2017) Role of conventional and biotechnological approaches for genetic improvement of cluster bean. Ind Crops Prod 97:639–648. https://doi.org/10.1016/j.indcrop.2017.01.008
Kumar S, Palve AS, Patel SK, Selvanayagam S, Sharma R, Rathore A (2020) Development of genomic microsatellite markers in cluster bean using next-generation DNA sequencing and their utility in diversity analysis. Curr Plant Biol 21:100134. https://doi.org/10.1016/j.cpb.2019.100134
Kuravadi NA, Tiwar PB, Tanwar UK, Tripathi SK, Dhugga KS, Gill KS, Randhawa GS (2014) Identification and characterization of EST-SSR markers in cluster bean (Cyamopsis spp.). Crop Sci 54(3):1097–1102. https://doi.org/10.2135/cropsci2013.08.0522
Lincoln SE, Lander SL (1993) Mapmaker/exp 3.0 and mapmaker/qtl 1.1. Technical report. Whitehead Institute of Medical Research, Cambridge, Mass
Liu Q, Liu H, Gong Y, Tao Y, Jiang L, Zuo W, Yang Q, Ye J, Lai J, Wu J, Lübberstedt T (2017) An atypical thioredoxin imparts early resistance to Sugarcane mosaic virus in maize. Mol Plant 10(3):483–497. https://doi.org/10.1016/j.molp.2017.02.002
Mali PC, Vyas SP, Lodha S (1989) Biochemical com- ponents of clusterbean genotypes in relation to bacterial blight. Indian Phytopathol 42:559–561
Manivannan A, Anandakumar CR, Ushakumari R, Dahiya GS (2015) Genetic diversity of cluster bean genotypes (Cyamopsis tetragonoloba (L.) Taub.) based on agro-morphologicaltraits. Bangladesh J Bot 44:59–65
Mihail JD, Alcorn SM (1985) Bacterial blight (Xanthomonas campestris pv. cyamopsidis) of guar in Arizona. Plant Dis 69:811. https://doi.org/10.1094/PD-69-811c
Morgante M, Hanafey M, Powell W (2002) Microsatellites are preferentially associate with nonrepetitive DNA in plant genomes. Nat Genet 30:194–200. https://doi.org/10.1038/ng822
Mudgil D, Barak S, Khatkar BS (2014) Cluster bean gum: processing, properties and food applications—a review. J Food Sci Technol 51(3):409–418. https://doi.org/10.1007/s13197-011-0522-x
Murwan KS, Abdalla AH (2008) Yield and yield components of forty-five cluster bean (Cyamopsis tetragonoloba) genotype grown in Sudan. Nile Basin Research Journal 11(4):48–54
Patel AJ, Patel MK (1958) A new bacterial blight in Cyamopsis tetragonoloba (L). Taub Curr Sci 27:258–259
Patel MK, Dhande GW, Kulkarni YS (1953) Bacterial leaf-spot of Cyamopsis tetragonoloba(L.) Taub. Curr Sci 22:183
Raju NL, Gnanesh BN, Lekha P, Jayashree B, Pande S, Hiremath PJ, Byregowda M, Singh NK, Varshney RK (2010) The first set of EST resource for gene discovery and marker development in pigeonpea (Cajanus cajan L.). BMC Plant Biol 10:45. https://doi.org/10.1186/1471-2229-10-45
Rawal HC, Kumar S, Mithra SVA, Solanke AU, Nigam D, Saxena S, Tyagi AVS, Yadav NR, Kalia P, Singh NP, Singh NK, Sharma TR, Gaikwad K (2017) High quality Unigenes and microsatellite markers from tissue specific transcriptome and development of a databse in cluster bean (Cyamopsis tetragonoloba, L. Taub). Genes 9:8–11. https://doi.org/10.3390/genes8110313
Reddy DS, Nagare PK, Reddaiah K, Reddy BR (2014) Effect of integrated nutrient management on growth, yield, yield attributing characters and quality characters in cluster bean (Cyamopsis tetragonoloba (L.) Taub.). The Ecoscan 4:329–332
Reddy DR, Saidaiah P, Reddy KR, Pandravada SR, Geetha A (2018) Genetic variability for growth, pod and quality attributes in germplasm of cluster bean [Cyamopsis tetragonoloba(L.) Taub.]. Legum Res 3917:1–5. https://doi.org/10.18805/LR-3917
Russell J, Booth A, Fuller J, Harrower B, Hedley P, Machray G, Powell W (2004) A comparison of sequence-based polymorphism and haplotype content in transcribed and anonymous regions of the barley genome. Genome Biol 47(2):389–398. https://doi.org/10.1139/g03-125
Sheoran OP, Tonk DS, Kaushik LS, Hasija RC, Pannu RS (1998) Statistical Software Package for Agricultural Research Workers. Recent Advances in information theory, Statistics and computer applications by D S Hooda and R C Hasija Department of Mathematics Statistics, CCSHAU, Hisar. Pp. 139–143
Singh N, Singh H, Nagarajan P (2013) Development of SSR markers in mung bean, Vigna radiata (L.) Wilczek using in silico methods. J Crop Weed 9(1):69–74
Sombir (2013) Interspecific hybridization in cluster bean (Cyamopsis tetragonoloba) using biotechnological approaches. M.Sc. thesis submitted to CCS Haryana Agricultural University, Hisar
Srivastava DN, Rao YP (1963) Seed transmission and epidemiology of the bacterial leaf blight disease of rice in north India. Indian Phytopathol 14:77–78
Swada K, Hasegawa M, Tokuda L, Kameyama J, Kodama O, Kohchi T, Yoshida K, Shinmyo A (2004) Enhanced resistance to blast fungus and bacterial blight in transgenic rice constitutively expressing OsSBP, a rice homologue of mammalian selenium-binding proteins. Biosci Biotechnol Biochem 68(4):873–880. https://doi.org/10.1271/bbb.68.873
Tanwar UK, Pruthi V, Randhawa GS (2017) RNA-Seq of Cluster bean (Cyamopsis tetragonoloba, L. Taub.) Leaves: de novo transcriptome assembly, functional annotation and development of genomic resources. Front Plant Sci 8(91):1–15. https://doi.org/10.3389/fpls.2017.00091
Undersander DJ, Putnam DH, Kaminski AR, Kelling KA, Doll JD, Oplinger ES, Gunsolus JL (1991) Guar. Alternative Field Crop Manual University of Wisconsin Cooperative Extension Service, University of Minnesota Extension Service, Center for Alternative Plant and Animal Products, 1991. https://hort.purdue.edu/newcrop/afcm/guar.html
Wadhwa N (2015) Analysis of simple sequence repeats and key enzymes for galactomannan content in cluster bean [Cyamopsis tetragonoloba (L.) Taub.] genotypes. Ph.D. thesis submitted to CCS Haryana AgriculturalUniversity, Hisar. pp 108
Wheeler BEJ (1969) An introduction to plant diseases. John & Wiley and Sons, New York
Whistler RL, Hymowitz T (1979) Cluster bean: agronomy, production, industrial use, and nutrition. Purdue University Press, West Lafayette
Xu Y (2010) Molecular plant breeding. CABI, Wallingford, p 736 (978-1-84593-392-0)
Xu P, Jiang L, Wu J, Li W, Fan S, Zhang S (2014) Isolation and characterization of a pathogenesis-related protein 10 gene (GmPR10) with induced expression in soybean (Glycine max) during infection with Phytophthora sojae. Mol Biol Rep 41(8):4899–4909. https://doi.org/10.1007/s11033-014-3356-6
Acknowledgements
Not applicable
Funding
This work was supported by a grant from the Director of Research, CCS Haryana Agricultural University, Hisar, India [C(a) BMB-6-Plan (Agri.)].
Author information
Authors and Affiliations
Contributions
NRY and RCY conceived and designed the research. P executed the experiments and performed the analysis. N RY and RCY reviewed the results. SKP and RP collaborated in the field experiments and reviewed the morphological data. P wrote the draft manuscript. NRY and RCY edited the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Priti, Panchta, R., Yadav, R.C. et al. Linkage mapping and identification of quantitative trait loci associated with bacterial leaf blight resistance and gum content in cluster bean using the interspecific population (HG563 × Cyamopsis serrata). Genet Resour Crop Evol 70, 1501–1516 (2023). https://doi.org/10.1007/s10722-022-01517-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10722-022-01517-0