Abstract
Akebia trifoliata (Thunb.) Koidz. is a perennial woody liana that belongs to Akebia Decne. Being high-value medicinal and economic crops, different parts of the plants are utilized for producing medicines, fruit, tea, wine, edible oil, etc. A. trifoliata is mainly wild, slow growth and low reproduction. However, the fragmented habitats caused by increasingly human activities led to a sharp decline yield of wild resources. Therefore, the domestication and cultivation of this plant could be the solution. Study on the genetic diversity among individuals or populations are the basis of making its conservation strategies and germplasm selection. In this study, 242 A. trifoliata accessions of 12 populations, collected from Qinba mountain area of China, were assessed using ISSR and SRAP markers, which generated 99.4% and 98.7% polymorphism, respectively. Both ISSR and SRAP revealed the high level of genetic diversity (H = 0.31, I = 0.47, ISSR; H = 0.33, I = 0.50, SRAP), and also revealed high genetic differentiation (Gst = 0.36 for ISSR, Gst = 0.34 for SRAP) and gene flow (Nm = 1.76 for ISSR, Nm = 1.44 for SRAP). Genetic variation observed within population was higher than of among population. The Mantel Test showed that the genetic variation among populations could be explained by the geographical distances (r2 = 0.1797, p = 0.020, ISSR marker; r2 = 0.2793, p = 0.010, SRAP marker;) The results of Structure analysis and PCoA also supported clustering model. Finally, based on ISSR and SRAP data, a core collection of 56 accessions by the LDSS method was selected. Collectively, our results lay the foundation for making A. trifoliata’s conservation strategies and germplasm selection.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Akebia trifoliata (Thunb.) Koidz. is a perennial woody liana that belongs to the genus Akebia Decne. Stems are grayish brown, verruculose and laxly lenticellate. leaflets 3, ovate, ovate-oblong, or broadly ovate, papery, subleathery, or leathery, base truncate, rounded, cordate, or cuneate, margin sinuate, shallowly lobed, subentire, or entire, apex obtuse to narrowly rounded and slightly emarginate, cuspidate. Racemes axillary on short branches, Male flowers: 15–30. Sepals 3 or 4, pale purple to purple, Stamens 6, pistillodes 3. Female flowers: 1–2. Sepals 3, purplish brown, dark purple, or purplish black, Carpels 4–9. Fruit oblong, straight or slightly curved. Seeds ovoid. The flowering period is from April to May, and the fruiting period is from July to August. The plant reproduces by seeds under natural conditions. Akebia trifoliata, growing in semideciduous forest margins, open forest along valleys, scrub on hillsides, by streams; 200–2100 m. The species are among the most economically important plants in China, and different parts of plants are used for producing medicine, fruit, tea, wine, edible oil, etc. (Flora of China 2001; Deng et al. 2018). The stems and fruits of this plant have been used thousands of years as Traditional Chinese Medicine (TCM), for the treatment of reducing fever, causing diuresis, and soothing the liver (National Pharmacopoeia Committee 2015). Most importantly, for the past few years, numerous pharmacological studies demonstrated that the active ingredients of A. trifoliata have good antineoplasmic activities (Lu et al. 2018; Ouyang et al. 2018). Therefore, A. trifoliata possess high medicinal value and industrial use.
The genus Akebia Decne. contains 4 species and 2 subspecies, distributed in China, Japan, and Korea in East Asia. There are 3 species and 2 endemics in China: Akebia quinata (Houttuyn) Decaisne, Akebia longeracemosa Matsumura, Akebia trifoliata (Thunb.) Koidz, Akebia trifoliata ssp. australis (Diels) T. Shimizu and Akebia trifoliata ssp. longisepala H. N. Qin. are subspecies of Akebia trifoliata (Flora of China 2001). Akebia trifoliata is one of the most commonly used species as medicinal herb resource, which contains more residual resources compared with other species of Akebia Decne. (Li et al. 2010). A. trifoliata is mainly distributed in Hebei, Shanxi, Shandong, Henan, Shaanxi, and southeastern of Gansu province, and provinces along the Yangtze River Basin in China (Flora of China 2001). The Qinba mountain area is the core distribution area of A. trifoliata. The Qinba mountain area refers to the Daba Mountain, Qinling Mountain, and its adjacent areas on the upstream of the Han River, the largest anabranch of the Yangtze River. It spans six provinces and cities, including Shaanxi, Gansu, Henan, Hubei, Sichuan, and Chongqing, and its main body is located in southern Shaanxi (Li et al. 1991).
A. trifoliata is mainly wild, and has slow growth and low reproductive capacity. Ever-increasing habitat fragmentation caused by continually human activities made it difficult to meet the increasing demand and sustainable use (Li et al. 2010). Therefore, the domestication and cultivation of this plant was emergency needed. Most reports of A. trifoliata focused on the chemical composition analysis, pharmacological research, and domestic cultivation system (Mimaki et al. 2003; Xiong et al. 2006; Wang et al. 2014, 2015; Tan 2016; Xu et al. 2016; Wu et al. 2017). However, rare studies paid attention to genetic characteristic and improvement, and to our knowledge, there has been no research on the establishment of core collections Kitaoka et al. (2009) reported the differences of Akebia plants growing in various wild habitats in Japan using ITS sequences. Huang et al. (2014), Zhang et al. (2016a, b), Li et al. (2018a) and Li et al. (2018b) collected the accessions from narrow regions, and Li's research focused on another species, Akebia quinata(Houtt) Decne. of Akebia Decne. (Li et al. 2009). In the early stage, our research group analyzed A. trifoliata accessions of 6 populations collected from Qinling Mountain in Shaanxi Province using 10 AFLP markers (Zhang et al. 2016a, b). Therefore, it is necessary to evaluate the genetic diversity and differentiation, and explore the population structure of all populations of A. trifoliata collected from the main distribution region, Qinba mountain area.
It is difficult to use morphological characteristics to assess variations between different populations, because morphology changes according to the plant growth conditions (Wang et al. 2009). Molecular marker technology at the DNA level is not affected by environmental factors and developmental stages; moreover, it is widely used in the study of genetic relationship. Both the Inter-Simple Sequence Repeat (ISSR) and Sequence Related Amplified Polymorphism (SRAP) are extensively used dominant DNA markers for their abundant observable loci, high polymorphism, reliable identification ability, and particularly for simple technique and low cost. They have been widely utilized in many previous studies for purpose of exploring the genetic diversity and differentiation (Peng et al. 2015; Chen et al. 2017; Zheng et al. 2017).
An Australian scholar, Frankel (1984), proposed the concept of core germplasm for the first time, which is to represent the genetic diversity of the species to the maximum with the smallest number of resources. Selection of core collections can be based on molecular data or phenotypic characteristics (Thachuk et al. 2009). Regarding the selection of the core subset, several algorithms have been proposed. Some based on genetic distance, such as the Modified Rogers (MR, Wright 1969) and Cavalli Sforza and Edwards (CE, Cavalli-Sforza and Edwards 1967), and others based on genetic diversity calculations, such as Shannon’s diversity index (SH, Shannon 1948) or the expected proportion of heterozygous loci (HE).
In the present study, 10 ISSR markers and 10 SRAP primer combinations were utilized for evaluating the genetic diversity among 242 A. trifoliata accessions of 12 populations that grow naturally in the Qinba mountain area of China. Our objectives were: (1) to estimate the genetic structure of collected populations in the Qinba mountain area, (2) to evaluate the genetic diversity of accessions we collected, (3) to detect the validity of the two markers in genetic diversity assessment, (4) to establish a core collection of A. trifoliata in the Qinba mountain area, and (5) to lay the foundation for making A. trifoliata’s conservation strategies and germplasm selection.
Materials and methods
Plant materials and DNA extraction
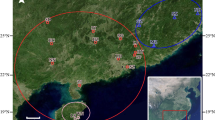
According to references (Zhang et al. 2013; Li et al. 2007) and the author’s investigation about the geographical distribution and resource status of wild A.trifoliata using quadrat method in the Qinba mountain area, there were 242 wild accessions belong to 12 populations from Shaanxi, Sichuan, Gansu, Henan, Shanxi and Hubei Province were selected in the current study. The number of individuals in each population was basically consistent with the resource reserve. More details about populations and the sampling localities were shown in Table 1 and Fig. 1. Plants with spacing greater than 50 m were selected at random, and 10–15 young leaves with no pests in the middle or lower part were collected. The materials collected in this study were identified by plant taxonomy professor of Shaanxi Normal University Xiaohui Zhang, and determined to be A. trifoliata of Akebia Decne. (Lardizabalaceae). Then, young leaves were dried on silica gel and stored at − 20 °C. Genomic DNA was isolated by the modified CTAB protocol. The quality and concentration were estimated by gel electrophoresis with 0.8% agarose and a nucleic acid-protein analyzer.
ISSR-PCR amplification
ISSR amplification was conducted in 25 μL volumes, which contained 20 ng DNA template, 1 × PCR Buffer (without Mg2+), 2.5 mM Mg2+, 0.2 mM dNTPs, 0.4 μM primers, and 2.0U Taq DNA polymerase (TaKaRa, China). The PCR cycling program was set to initial denaturation at 94 °C for 10 min, followed by 40 cycles of denaturation at 94 °C for 1 min, annealing at (Tm-2)°C for 1 min, extension at 72 °C for 2 min, and a final extension at 72 °C for 10 min. A total of 10 primers (Table 2, https://wenku.baidu.com/view/62eea8f1fc4ffe473268ab76.html) out of 100 which amplified the DNA well with clear and reliable polymorphic bands were finally selected for subsequent amplification. The amplified products were detected by gel electrophoresis with 2% agarose and further analyzed.
SRAP-PCR analysis
The system of SRAP was set as follows: (25 μL) 20 ng DNA template, 1 × Reaction Buffer, 2.5 mM Mg2+, 0.2 mM dNTPs, 2.0U Taq DNA polymerase (TaKaRa, China), and 0.4 μM primer. The PCR cycling program was set to initial DNA denaturation at 94 °C for 5 min; 5 cycles of denaturation at 94 °C for 1 min, annealing at 37 °C for 1 min, extension at 72 °C for 2 min; followed by 35 cycles of denaturation at 94 °C for 1 min, annealing at 54 °C for 1 min, extension at 72 °C for 2 min, and a final extension at 72 °C for 10 min. Eventually, 10 primer pairs (Table 3, https://www.docin.com/p-286948618.html) out of 100 were selected for subsequent amplification. The amplified products were electrophoresed with 2% agarose.
Statistical analysis
POPGENE 1.32 was used to calculate genetic parameters, such as the observed number of alleles (Na), effective number of alleles (Ne), Nei's gene diversity (H), Shannon's Information index (I), diversity within population (Hs), total gene diversity (Ht) and gene flow (Nm). A dendrogram was constructed using MEGA 7.0 based on genetic distances among individuals with Unweighted Pair Group Method with Arithmetic average (UPGMA). Principle Coordinate Analysis (PCoA) and the Analysis of Molecular Variance (AMOVA) were carried out by GenAlEx 6.2 software. A Mantel test based on Nei genetic distance and geographical distance was also conducted.
The STRUCTURE 2.3.4 software was utilized to estimate the population genetic structure of 12 populations. The initial K (presumed number of subpopulations) was set from 1 to 12, and 10 repetitions were performed for each K (100,000 Length of Burnin Period and 1,000,000 Number of MCMC Reps after Burnin). Then, the △K was calculated according to LnP (D) of each K (In addition to K = 1 and 12) based on the operation results to determine the most appropriate K.
On the basis of cluster analysis of ISSR and SRAP markers by NTsys2.10e software, the core collection of A. trifoliata was constructed using the LDSS method (Least distance stepwise sampling) (Hu et al. 2000). After each sampling, GenAlEx 6.2 software was used to analyze the genetic diversity of the core collection, and then to evaluate the representativeness of the preliminary core collection.
Results
Genetic diversity analysis
In this experiment, 10 ISSR primers generated 171 bands, among which 170 (99.4%) were polymorphic. Percentage of polymorphism revealed by primer UBC825 was 92% and the rest were 100%. The number of polymorphic bands generated by these primers varied from 10 to 24, with an average of 17 bands per primer. See in Table 2, the average number of alleles (Na) produced by per primer was close to 2, and the minimum number of alleles was amplified by primer UBC825 (1.92). The average effective number of alleles was 1.51, with a maximum and a minimum of 1.68 (primer UBC856) and 1.36 (primer UBC811), respectively. Among these ISSR primers, primer 891 revealed maximum H (0.37) and I (0.54). The mean values of total gene diversity (Ht), diversity within the population (Hs) and gene flow (Nm) detcted by ISSR primers were 0.31, 0.18, and 1.76, respectively.
10 SRAP primer combinations amplified 237 bands, of which 234 (98.7%) were polymorphic. Among these primer pairs, M5E2, M4E3, and M12E6 revealed the lowest percentage of polymorphism (95%). The number of polymorphic bands generated by these primer pairs varied from 20 to 31, with an average of 23.4 bands per primer combination. See in Table 3. The average values of Na and Ne produced by these primer pairs were 1.99 and 1.56, respectively. Of these, the primer combination M3E6 revealed the maximum of H (0.38) and I (0.55). The mean values of total gene diversity (Ht), diversity within population (Hs), and gene flow (Nm) were 0.33, 0.21, and 1.44, respectively.
Cluster analysis
Based on ISSR data, the genetic distance matrix between two populations in 12 populations was calculated (Table 4). According to the genetic distance, A. trifoliata genotypes were separated into two main clusters: Cluster I and Cluster II (Fig. 2). Cluster I was further grouped into two subgroups. One subgroup included all individuals of population WY, and the other subgroup contained all accessions of population MJ, JL, HXX, and SXX. Cluster II also comprised of two subgroups. The first subgroup contained all accessions from population FP except individual FP7, and the second subgroup included all accessions from population ZAX, ZAY, TB, NS, SN, HX and individual FP 7.
Genetic distance matrix between two populations based on SRAP data was shown in Table 5. Similarly, 242 A. trifoliata accessions were also divided into two main clusters: Cluster I and Cluster II (Fig. 3). Cluster I contained all accessions from population SXX, which was from Xiaxian, Shanxi Province. Cluster II was further divided into two subgroups, of which one subgroup contained all accessions from population ZAX, NS, SN, HX, FP, ZAY, and TB, and the another subgroup included all individuals from population JL, WY, HXX, and MJ. Individuals from the same population were generally divided into the same subgroup.
In view of the different results of the two markers clustering, we used Mantel test to analyze the correlation between ISSR and SRAP genetic distance matrix, the analysis results of ISSR and SRAP were highly and significantly correlated at the population level(R2 = 0.4188, p = 0.010) (Fig. 4). Therefore, ISSR and SRAP are highly consistent and reliable in analyzing the genetic diversity and population structure of A. trifoliata. Referred to Yan’s method (2019), UPGMA clustering was carried out using combined ISSR and SRAP data. 242 accessions were divided into two main clusters: Cluster I and Cluster II (Fig. 5). Unlike the clustering results based on ISSR or SRAP, all individuals from the same population completely clustered on the same branch based on the combined data. Cluster I was further grouped into two subgroups, one subgroup included population JL and MJ, and another subgroup contained population HXX and SXX. Cluster II also comprised of two subgroups, the first subgroup contained population WY, and the second subgroup included all populations from Shaanxi Province.
Principal coordinates analysis (PCoA) and analysis of molecular variance (AMOVA)
Principal coordinate analysis (PCoA) and analysis of molecular variance (AMOVA) were carried out for further study of genetic relationships among A. trifoliata accessions. In the ISSR analysis, the first two PCoA axes could explain 28.46% and 20.11% of the genetic variation, respectively. The PCoA showed that 12 populations were divided into three groups. Similar to the result of cluster analysis, different populations from Shaanxi Province gathered together. The population SXX and HXX were divided into another group. The third group were population JL, MJ and WY, respectively (Fig. 6). AMOVA analysis was conducted to evaluate diversity components within and among population. The results showed that 59% of the variation was within population, while 41% of the variation was among population. The gene differentiation coefficient was 0.407 (p = 0.010), indicating that the difference was significant. In addition, the result of the Mantel test showed that Nei genetic distance had a certain correlation with geographical distance in ISSR analysis (R2 = 0.1797, p = 0.020) (Fig. 7).
In the SRAP analysis, the first two axes in the PCoA plot accounted for 28.39% and 18.47% of the genetic variation, respectively. The PCoA showed that these populations of A. trifoliata were also divided into three groups. The results of PCoA were consistent with that of the cluster analysis based on SRAP data (Fig. 8). AMOVA results revealed that 60% of the variation was attributed to diversity within population and 40% of the variation was associated with differences among population. And the gene differentiation coefficient was 0.396 (p = 0.010 < 0.05), indicating that the difference was significant. Similarly, the Nei genetic distance was also related to geographical distance based on SRAP data (R2 = 0.2793, p = 0.010) (Fig. 9).
In addition, PCoA was also carried out based on the combined ISSR and SRAP data. The first two axes in the PCoA plot explained 9.12% and 5.47% of the genetic variation, respectively. The PCoA showed that 12 populations were divided into three groups. Similar to the result of ISSR. 7 populations from Shaanxi Province gathered together, the population SXX and HXX were included in another group, the third group consist of population JL, MJ and WY. (Fig. 10). The results of AMOVA revealed 60% and 40% of the variation within and among population, respectively. The gene differentiation coefficient was 0.400 (p = 0.010), indicating that the difference was significant. Therefore, ISSR and SRAP are highly consistent and reliable in analyzing the genetic diversity and population structure of A. trifoliata.
Population structure analysis
In order to evaluate the reliability of cluster analysis and PCoA, the genetic structure of the population was also calculated by the Structure software. The maximum value of △K appeared at K = 2, indicating that 12 populations were divided into two groups, the results of structure analysis on ISSR or SRAP markers were basically the same. For ISSR, the structure analysis were basically consistent with the cluster analysis, except for population WY, some of which were assigned to another group (Fig. 11A). However, for SRAP, the structer results was quite different from cluster analysis (Fig. 11B). In the PCoA plot, population SXX, JL, WY, HXX, and MJ were also clearly separated from the other seven populations.
Preliminary construction of core collection of A. trifoliata
Based on the clustering results of 242 A. trifoliata germplasms, the core collection was constructed using LDSS. According to Yonezawa (1995), 20–30% (p = 0.2–0.3) of the total amount of original genetic material is considered to be better representative. In both two markers, a minimum of 56 A. trifoliata was selected after four samplings, and they retained about 23% of original accessions. The genetic diversity parameters of the core collections constructed after each sampling were shown in Tables 6 and 7. Therefore, these 56 genetic materials can be used as the core resources of the original germplasm resources. The clustering results of these 56 germplasms were shown in Figs. 12 and 13. Of the two core collections, 21 accessions were exactly the same.
Discussion
Genetic diversity of A. trifoliata
Studies on molecular markers of A. trifoliata have been reported previously, however, accessions were limited and most used single markers (Huang et al. 2014; Zhang et al. 2016a; Li et al. 2018a, 2018b). The limitations of sample collection resulted in the conclusion probably under-represented. In the present study, a large number of germplasms were collected from Qinba mountain area, China, the main distribution area of A. trifoliata, therefore, the results of this study were reliable and representative. In this study, the percentage of polymorphism revealed by ISSR (99.4%) and SRAP markers (98.7%) was high, indicating that both markers were effective. Meanwhile, it also suggested that both markers can effectively analyze and evaluate A. trifoliata germplasm resources. However, in our previous studies, 10 AFLP markers revealed considerably low polymorphism (59.7%) among 6 populations (Zhang et al. 2016a). The results may be attributed to differences in molecular marker and the limitations of sample collection since all accessions were collected from the Qinling mountain in Shaanxi Province and the sample size was relatively small.
In this work, ISSR and SRAP markers revealed higher level of H and I (ISSR: H = 0.31, I = 0.47; SRAP: H = 0.33, I = 0.50) compared to other studies (Liu et al. 2015; Kumar and Agrawal 2017). This phenomenon indicated that ISSR and SRAP were successful in identifying the germplasm resources of A. trifoliata. Also, genetic polymorphism of A. trifoliata resources in Qinba mountain area was quite abundant, protecting wild resources is particularly important to improve the current situation of A. trifoliata.
Genetic structure of A. trifoliata populations
This study revealed high level of genetic differentiation (ISSR: Gst = 0.36; SRAP: Gst = 0.34. classified Gst > 0.15 as high) with both two markers. Several factors could take responsibility for the high population differentiation, such as mode of pollen and seed dispersal, geographical isolation, breeding system, population fragmentation, environmental heterogeneity and genetic drifts (Harish et al. 2014; Shekhawat et al. 2018). Li et al. (1991) reported that the flower of A. trifoliata had the characteristics of both insect-mediated flowers and wind-media flowers. Its flower characteristics determine its pollination methods, which in turn can explain high genetic differentiation.
The gene flows (Nm) among the A. trifoliata populations were 1.76 (ISSR) and 1.44 (SRAP), respectively. Gene flow refers to the movement of genes within and between populations (Grant 1991). Genetic drift can lead to the genetic differentiation between populations. It is generally accepted that in the case of Nm < 1, Genetic drift maybe the main reason for the differentiation of genetic structure among populations. In the case of Nm > 1, gene flow can prevent Genetic drift, reduce genetic diversity. In this research, both markers obtained higher gene flow, indicating that higher gene exchange occurred between populations, this gene exchange prevented the genetic structure differentiation caused by genetic drift among the populations of A. trifoliata.
AMOVA results suggested that the genetic variation within population (ISSR: 59%; SRAP: 60%) was higher than among population (ISSR: 41%; SRAP: 40%) for both markers. This indicated a relatively high level of heterogeneity within the natural population of A. trifoliata in Qinba mountains area (Wang et al. 2019). Factors such as mating system, population size, selection, somatic mutations, multiple founder events, etc. may be used to explain the causes of high levels of genetic diversity within population (Kumar and Agrawal 2019; Gomes et al. 2013; Rumbou et al. 2016).
The results of cluster analysis based on ISSR, SRAP markers are not consistent, individuals of the population ZAY, TB were not clustered together on ISSR, this is not uncommon. The possible reason for this phenomenon might be that different markers targeting different segments of the genome, a single marker cannot fully reflect the relationship between populations. Since both ISSR and SRAP markers are dominant markers, Mantel test show the correlation between ISSR and SRAP was highly and significantly at the population level, so we combined the two markers for analysis to avoid the limitation of single marker.
In this research, the Mantel test based on ISSR or SRAP data indicated that there was a significant correlation between Nei genetic distance and geographical distance. The cluster analysis of both markers showed that 7 populations from Shaanxi Province always gathered in the same branch, this suggested that the clustering results based on genetic distance were consistent with the geographic location. In addition, the clustering results of the 5 populations outside Shaanxi Province in different clustering methods were not exactly the same. In cluster analysis base on SRAP and combine of two markers, WY population groups together with 7 populations of Shaanxi province. We speculate that it might be related to the geographic distances, similar habitat. WY situated at the Northeast of Sichuan, very close to the south of Shaanxi, and located in the south of Qinling mountains with ZAX, ZAY, NS, SN, FP populations, smaller geographic distances make gene communication easier, reduced genetic variation. In addition, clustering results based on combined ISSR and SRAP data could cluster individuals from the same population on the same branch, but clustering results based on single markers were not, so we combined the two markers for analysis to avoid the limitation of single marker. The combined analysis of two markers is more comprehensive than that of single marker.
SXX is a noteworthy population. In clustering and PCoA analysis, the analysis results of two single markers are quite different, while in double-marker combination analysis, the two results support each other. SXX located at the northern of Qinling mountains, geographically far away from other populations, there is greater genetic variation between SXX and other populations. Due to geographical barriers, SXX has the greater genetic distance from SN,TB populations located in the central region of Qinling Mountains. HXX, like SXX, is also located in the north of Qinling mountains and has a similar habitat. Therefore, even though HXX is closer to the south of Shaanxi, the genetic distance between them is not large and there is a small genetic variation, so they tend to cluster together.
Core collection of A. trifoliata
A core collection of A. trifoliata was constructed, which could represent the genetic diversity of 242 original accessions. In the future genetic breeding work, we can focus on the 21 identical germplasm materials. By establishing core collections based on ISSR and SRAP data, A. trifoliata germplasm resources in Qinba mountain area of China with higher genetic diversity were collected and preserved. The establishment of the core collection of A. trifoliata in Qinba mountain area of China had improved the management and utilization efficiency of germplasm resources, and provided a reference basis for the establishment of the core collection of A. trifoliata in other regions.
Conclusion
In this study, we used ISSR and SRAP markers, for the first time, to evaluate A. trifoliata germplasm in Qinba mountain area of China. The results revealed a high level of genetic diversity, genetic differentiation, gene flow, and genetic variation within population. The results of population structure analysis and PCoA substantial supported to the clustering results of the populations. Finally, we established a core collection of 242 A. trifoliata germplasm based on ISSR data and SRAP data. A total of 56 accessions remained after samplings, which constituted the core collection of 242 A. trifoliata. The polymorphisms of core collection constructed based on ISSR and SRAP data were relatively high, being 98.25% and 97.05%, respectively. In summary, our study evaluated the genetic diversity of the collected accessions and compared the effectiveness of ISSR and SRAP markers in germplasm identification. Most importantly, we laid the foundation for germplasm conservation and selection strategies of A. trifoliata.
Availability of data and materials
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
References
Cavalli-Sforza LL, Edwards AWF (1967) Phylogenetic analysis: models and estimation procedures. Evolution 21(3):550–570
Chinese Flora Commission (2001) Flora of China 6: 440–454. https://www.iplant.cn/info/Akebia%20trifoliata?t=foc
Chen Y, Peng Z, Wu C, Ma Z, Ding G, Cao G, Ruan S, Lin S (2017) Genetic diversity and variation of Chinese fir from Fujian province and Taiwan, China, based on ISSR markers. PLoS ONE. https://doi.org/10.1371/journal.pone.0175571
Deng AH, Li HY, Wang Y, Xie P, Cheng LN, Peng CW (2018) Study on fabrication of Akebia trifoliata fruit tea. Guangzhou Chem Ind 46(01):120–123
Frankel OH (1984) Genetic perspectives of germplasm conservation. Genetic manipulation: impact on man and society, vol 61(3). Cambridge University Press, Cambridge, pp 161–170
Gomes F, Costa R, Ribeiro MM, Figueiredo E, Canhoto JM (2013) Analysis of genetic relationship among Arbutus unedo L. genotypes using RAPD and SSR markers. J For Res 24:227–236. https://doi.org/10.1007/s11676-012-0302-0
Grant V (1991) The evolutionary process: a critical study of evolutionary theory. Columbia University Press, New York
Hu J, Xu HM, Zhu J (2000) Constructing core collection of crop germplasm by multiple clusters based on genotypic values. J Biomath 15:103–109
Huang PB, Chen SH, Cao L, Wang F (2014) AFLP fingerprinting analysis of Akebia trifoliata. Lishizhen Med Mater Med Res 25(4):871–873
Harish GAK, Phulwaria M, Rai MK, Shekhawat NS (2014) Conservation genetics of endangered medicinal plant Commiphora wightii in Indian Thar Desert. Gene 535(2):266–272. https://doi.org/10.1016/j.gene.2013.11.018
Kitaoka F, Kakiuchi N, Long C, Itoga M, Yoshimatsu H, Mitsue A, Atsumi T, Mouri C, Mikage M (2009) Difference of ITS sequences of Akebia plants growing in various parts of Japan. J Nat Med 63(3):368–374
Kumar J, Agrawal V (2017) Analysis of genetic diversity and population genetic structure in Simarouba glauca DC. (an important bio-energy crop) employing ISSR and SRAP markers. Ind Crops Prod 100:198–207. https://doi.org/10.1016/j.indcrop.2017.02.035
Kumar J, Agrawal V (2019) Assessment of genetic diversity, population structure and sex identification in dioecious crop, Trichosanthes dioica employing ISSR SCoT and SRAP Markers. Heliyon. https://doi.org/10.1016/j.heliyon.2019.e01346
Li WL (1991) Studies on Lardizabalaceae in Qinba Mountains. Chin Wild Plant 01:19–23
Li JR, Li JG, Shui SQ (1991) Studies on flowering Biology of Akebia trifoliata kiodz. J Northwest A F Univ (Nat Sci Edn) 19(4):33–37
Li XH, Xiao YP, Xie J (2007) Investigation on the morphological diversity of the germplasm resources of Akebia trifoliata (Thunb) Koidz. J Shaanxi Normal Univ (Nat Sci Edn) 35(04):88–93
Li L, Yao XH, Chen XZ, Huang HW (2009) Development and characterization of microsatellite loci in Chinese medicinal plant Akebia trifoliata ssp. Australis and cross-species amplification in closely related taxa. Conserv Genetics 10(4):959–962. https://doi.org/10.1007/s10592-008-9666-2
Li L, Chen XZ, Yao XH, Tian H, Huang HW (2010) Geographic distribution and resource status of three important Akebia species. Journal of Wuhan Botanical Research 28(04):497–506
Li TJ, Dong J, Liao L, Jin HG, Han XJ, Wen F, Xu LL (2018a) Isolation and characterization of microsatellite markers for Akebia trifoliata. Guihaia 38(09):1117–1124
Li HY, Wang XH, Qin DJ, Xiang JQ, Yang YK, Chen FF, Wan HY, Zhou M, Zhu L (2018b) Analysis on the RAPD phylogenetic relationship of Akebia trifoliata (Thunb.) Koidz in Hunan and Hubei provinces. Hubei Agric Sci 57(23):148–152
Liu J, Wang ZR, Li C, Bian YB, Xiao Y (2015) Evaluating genetic diversity and constructing core collections of Chinese Lentinula edodes cultivars using ISSR and SRAP markers. J Basic Microbiol 55(6):749–760. https://doi.org/10.1002/jobm.201400774
Lu WL, Yang T, Yang QJ, Fang ZQ, Pan ZQ, Liang C, Jia DW, Peng PK (2018) Akebia trifoliata (Thunb.) Koidz Seed Extract inhibits human hepatocellular carcinoma cell migration and invasion in vitro. J Ethnopharmacol 234:204–215. https://doi.org/10.1016/j.jep.2018.11.044
Mimaki Y, Kuroda M, Yokosuka A, Harada H, Fukushima M, Sashida Y (2003) Triterpenes and Triterpene Saponins from the stems of Akebia trifoliata. Chem Pharm Bull 51:960–965. https://doi.org/10.1248/cpb.51.960
National Pharmacopoeia Committee (2015) Chinese Pharmacopoeia. China Medical Science and Technology Press, Beijing, pp 297
Ouyang JK, Dong LM, Xu QL, Wang J, Liu SB, Qian T, Yuan YF, Tan JW (2018) Triterpenoids with α-glucosidase inhibitory activity and cytotoxic activity from the leaves of Akebia trifoliata. RSC Adv 8(70):40483–40489
Peng X, Ji QY, Fan SW, Zhang YJ, Zhang JJ (2015) Genetic diversity in populations of the endangered medicinal plant Tetrastigma hemsleyanum revealed by ISSR and SRAP markers: implications for conservation. Genet Resour Crop Evol 62(7):1069–1078. https://doi.org/10.1007/s10722-014-0210-6
Rumbou A, von Bargen S, Demiral R, Langer J, Rott M, Jalkanen R, Buttner C (2016) High genetic diversity at the inter-/intra-host level of Cherry leaf roll virus population associated with the birch leaf-roll disease in Fennoscandia. Scand J For Res 31:546–560
Shannon CE (1948) A mathematical theory of communication. Bell system technical journal 27(3):379–423. https://doi.org/10.1002/j.1538-7305.1948.tb01338.x
Shekhawat JK, Rai MK, Shekhawat NS, Kataria V (2018) Start codon targeted (SCoT) polymorphism for evaluation of genetic diversity of wild population of Maytenus emarginata. Ind Crops Prod 122:202–208. https://doi.org/10.1016/j.indcrop.2018.05.074
Tan C (2016) Plant propagation and cultivation techniques of Akebia trifoliata in Qinba mountain area. Agric Technol 36(18):183
Thachuk C, Crossa J, Franco J, Dreisigacker S, Warburton M, Davenport GF (2009) Core hunter: an algorithm for sampling genetic resources based on multiple genetic measures. BMC Bioinformat 10(1):243. https://doi.org/10.1186/1471-2105-10-243
Wang HZ, Wu ZX, Lu JJ, Shi NN, Zhao Y, Zhang ZT, Liu JJ (2009) Molecular diversity and relationships among Cymbidium goeringii cultivars based on inter-simple sequence repeat (ISSR) markers. Genetica 136:391–399. https://doi.org/10.1007/s10709-008-9340-0
Wang J, Xu QL, Zheng MF, Ren H, Lei T, Wu P, Zhou ZY, Wei XY, Tan JW (2014) Bioactive 30-noroleanane triterpenes from the pericarps of Akebia trifoliata. Molecules 19:4301–4312. https://doi.org/10.3390/molecules19044301
Wang J, Ren H, Xu QL, Zhou ZY, Wu P, Wei XY, Cao Y, Chen XX, Tan JW (2015) Antibacterial oleanane-type triterpenoids from pericarps of Akebia trifoliata. Food Chem 168:623–629. https://doi.org/10.1016/j.foodchem.2014.07.105
Wang XD, Li XH, Zhang ZW, Wang KJ (2019) Genetic diversity and genetic structure of natural populations in an extremely narrowly distributed perennial species Glycine tabacina (Labill.) Benth. on the southeast islands in China. Genetic Resour Crop Evolut 66(5):989–1008. https://doi.org/10.1007/s10722-019-00764-y
Wright S (1969) Evolution and the genetics of populations. Theory gene Freq vol 2.
Wu ZB, Xu XH, Yang L, Xian XM (2017) Discussion on planting technique of Akebia trifoliata in Nanchuan District of Chongqing City. South China Agric 11(28):78–79
Xiong DS, Wang JY, Li ZH, Zhao RH (2006) Operational rules for standardized production of Akebia trifoliata. Mod Chin Med 8(5):37–40
Xu QL, Wang J, Dong LM, Zhang Q, Luo B, Jia YX, Wang HF, Tan JW (2016) Two new pentacyclic triterpene saponins from the leaves of Akebia trifoliata. Molecules. https://doi.org/10.3390/molecules21070962
Yan WP, Li JL, Zheng DJ, Friedman C, Wang HF (2019) Analysis of genetic population structure and diversity in Mallotus oblongifolius using ISSR and SRAP markers. PeerJ 7:e7173. https://doi.org/10.7717/peerj.7173
Yonezawa K (1995) Sampling strategies for use in stratified germplasm collections. Core collections of plant genetic resources.
Zhang Z, Jian JY, Wang Z (2016) AFLP analysis on genetic diversity of Akebia trifoliate in qinling mountains of Shaanxi. Chin Tradit Herbal Drugs 47(21):3890–3895
Zhang X, Ma YH, Zhong WM (2016) Analysis of genetic relationship and genetic diversity of Akebia trifoliata in Guizhou. Acta Bot Boreali Occident Sin 36(11):2192–2197
Zhang YJ, Dang HS, Yang LL, Wei GY, Wang Y (2013) Geographical distribution and resource survey of wild medicinal plant Akebia trifoliata subsp. trifoliata. Chin Wild Plant Resour 32(3):58–62
Zheng Y, Xu S, Liu J, Zhao Y, Liu J (2017) Genetic diversity and population structure of Chinese natural bermudagrass [Cynodon dactylon (L.) Pers.] germplasm based on SRAP markers. PLoS ONE. https://doi.org/10.1371/journal.pone.0177508
Funding
This work was supported by the Key Program of Science and Technology of Shaanxi Province, China (Project No. 2013K14-03-01).
Author information
Authors and Affiliations
Contributions
Conceived and designed the experiments: ZZ; Conducted the experiments: ZZ, YN, YZ, SD, WZ; Data analysis: QY; Contributed reagents/materials/analysis tools: ZZ; Writing—original draft: QY; Writing—review and editing: ZZ; Funding: ZW.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Consent for publication
The manuscript is approved by all authors for publication.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Zhang, Z., Yang, Q., Niu, Y. et al. Diversity analysis and establishment of core collection among Akebia trifoliata (Thunb.) Koidz. in Qinba mountain area of China using ISSR and SRAP markers. Genet Resour Crop Evol 68, 1085–1102 (2021). https://doi.org/10.1007/s10722-020-01051-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10722-020-01051-x