Abstract
Panicle size is one of the most important agronomic traits highly associated with grain yield in rice. For quantitative trait locus (QTL) analysis and breeding utilization of panicle size in rice, a large panicle rice line YR1 was used as a donor to cross and backcross with a commercial cultivar Ningjing 1 (NJ1), and developed backcross populations. In a BC2F2 population, a total of 3 QTL associated with panicle size were found on chromosome 1 and 8. Two of the QTLs, qPBN1 and qGNP1, were mapped to an interval on chromosome 1, where a previously cloned gene GN1a was located. qPBN8, a major QTL for primary branch number on chromosome 8, was finally narrowed to a 51.8 kb region, containing reported gene OsSPL14 regulating branch number and plant architecture. Through sequence and expression comparison, the qPBN8 was considered to be the allele of OsSPL14. The investigation of yield structure in six pairs of near-isogenic lines revealed that the effects of qPBN8 on grain yield were related to the length of vegetative period. The qPBN8 allele of large panicle was suitable for late-maturing varieties to improve yield.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Grain yield is one of the most valuable traits in crop production. Increasing rice yield per unit area is particularly important in the global situation that crop yields on limited arable land are provided to a growing population (Takai et al. 2018). Therefore, cultivating high-yield varieties is one of the most economical and effective approach to solve rice yield. Different environments and cultivation methods require different varieties to achieve high yield. In the high humidity environment where most rice grows, large-panicle varieties can effectively improve field climate, reduce disease and lodging incidence (Peng et al. 2008). Moreover, direct-seeded rice is becoming a global tendency of labor-saving rice production (Kumar and Ladha 2011), and claim suitable varieties that has the characteristics of large-panicle, thick stem, less tillers or panicles and rapid seedling (Liu et al. 2015).
It has been recognized that panicle size is the major factors for rice yield. Panicle traits includes four factors: panicle length (PL), primary branch number (PBN), secondary branch number (SBN), and spikelet number per panicle/grain number per panicle (SPP/GNP) (Peng et al. 2014). Primary and secondary branches bear spikelets. The number of spikelets is an important determinant of grain yield (Bai et al. 2016). SBN and GNP are positively correlated (Luo et al. 2009; Mei et al. 2006). During panicle development, the inflorescence meristem is an important regulator of grain number formation (Li et al. 2013; Sasaki et al. 2017). The inflorescence meristem initiates primary branch meristems. The primary panicle branches also produce a certain number of secondary branch meristems, which generate secondary panicle branches or differentiate into spikelets. Ultimately, the primary and secondary branch meristems differentiate into terminal spikelets (Tabuchi et al. 2011; Xing and Zhang 2010).
In recent years, many quantitative trait loci (QTLs) for grain number have been identified. Some genes or loci related grain number in rice have been cloned. Such as, Gn1a which encodes a cytokinin oxidase/dehydrogenase (CKX), was the first isolated QTL for grain number (Ashikari et al. 2005). APO1 positively controls spikelet number by suppressing the precocious conversion of inflorescence meristems to spikelet meristems (Ikeda et al. 2007). Ghd7 was reported to function in rice growth, development and environmental response, thus regulating rice grain yield, plant height and heading date (Weng et al. 2014; Xue et al. 2008). Artificial selection for the PROSTRATE GROWTH 1 (PROG1) mutant during rice domestication led to the transition from the plant architecture of wild rice to that of domesticated rice, resulting in erect growth, greater grain number and higher grain yield (Jin et al. 2008; Tan et al. 2008). DEP1 enhanced meristematic activity, increased number of grains per panicle and a consequent increased in grain yield (Huang et al. 2009). WFP/IPA1 regulates primary branch number and ideal plant architecture (Jiao et al. 2010; Miura et al. 2010). SPIKE/NAL1 can increase spikelet number without changing grain quality or growth period (Fujita et al. 2013). PAY1 affects plant architecture and grain yield in rice (Zhao et al. 2015). NOG1, which encodes an enoyl-CoA hydratase/isomerase, increases the grain yield of rice by enhancing grain number per panicle without a negative effect on the number of panicles per plant or grain weight (Huo et al. 2017). Molecular characterization of genes controlling rice grain yield will not only strengthen our understanding of regulatory mechanisms of these traits, but also be valuable for high-yield breeding in rice.
In this study, QTL analysis was carried out in BC2F2 population. A major QTL (qPBN8) was detected on chromosome 8. The target region was narrowed by map-based cloning strategy with BC2F4 population. Based on sequencing and expression analysis, the predicted ORF3 for qPBN8 encoding the plant-specific transcription factor OsSPL14, was proved to be not only responsible for branch number (Miura et al. 2010), but also for ideal plant architecture (Jiao et al. 2010).
Materials and methods
Plant materials
YR1 is an F8 line with erected large panicles, and selected from the selfing of a hybrid rice Yongyou 11. Ningjing 1 (NJ1) is a commercial japonica cultivar that is widely planted in the Lower Yangtze River of China. YR1 was crossed with NJ1, and F2 plants with large panicles were continuously backcrossed with NJ1, so that the beneficial trait of large panicle was introduced into NJ1 background. A BC2F2 population of 164 plants was used for preliminary QTL mapping of panicle size. Subsequently, the heterozygotes from BC2F2 population were selected by marker assisted selection (MAS) to generate segregating BC2F3 populations for QTL confirmation. A total of 1374 BC2F4 plants were used for fine mapping. In addition, heterozygous plants from F2 and BC1F2 with different other agronomic traits were successively self-pollinated and keep residual heterozygous qPBN8 by MAS to F6 and BC1F5. At last each residual heterozygote was formed a pair of pure lines (near-isogenic line, NIL) in F8 and BC1F7, whose qPBN8 come from two different parents, and each pair of NIL has its own genetic background. These NILs were used to evaluate the qPBN8 effects on grain yield.
Field experiments and trait evaluation
Rice materials were grown in the field at the Experimental Station of Nanjing Agricultural University (Jiangsu Province, China). Field management was carried out in keeping with the local standard methods (Cheng et al. 2014). At maturity, panicles on the main stems were harvested, and then dried naturally after harvesting and stored at 45 °C for 3 days before testing (Zhu et al. 2011). Panicle length (PL), primary branch number (PBN), secondary branch number (SNB), spikelets number per panicle (SPP) and seed setting rate (SSR) were evaluated as the mean measurement from the three main-stem panicles. Heading date is described by the number of days from seeding to initial heading, and initial heading is 10% of the panicles in population extracted from the sheathes.
Molecular marker development
Simple sequence repeat (SSR) markers were designed as described on the Gramene database (http://www.gramene.org/). Insert and deletion (InDel) markers were newly designed on the RiceVarMap v2.0 database (http://ricevarmap.ncpgr.cn/v2/).
Linkage and QTL analysis
A total of 119 polymorphic SSR markers and InDel markers evenly distributed on 12 chromosomes were used to identify the genotypes of 164 BC2F2 individuals in this study. The genomic DNA of each plant was extracted from fresh rice leaves according to a modified CTAB protocol. PCR was performed as following: 95 °C for 3–5 min, followed by 32–35 cycles of 95 °C for 15 s, 55–60 °C for 20 s, 72 °C for 40 s, and a final elongation step at 72 °C for 5 min. The PCR products were analyzed on 6–8% agarose gels, which depend on PCR product length. The genetic map construction and QTL analysis were conducted by the composite interval mapping method using the IciMapping4.1.0 (www.isbreeding.net/) software. We selected the LOD threshold of 3 and walking speed of 1.0 cM for QTL identification (Fang et al. 2016).
Fine mapping
To further narrow down qPBN8 locus, new polymorphic molecular markers were developed (Table 1). For the convenience of scoring the phenotype, numbers of primary branch were selected as the target trait for fine mapping. BC2F4 individuals were used to screen recombinant events between InDel markers S98 and S21, flanking qPBN8. Nucleotide sequence alignment of candidate genes were performed between YR1 and NJ1.
RNA extraction and expression analysis
Total RNA was extracted from young panicles with the RNA extraction kit (Bioteke). The complementary DNA (cDNA) was synthesized with random oligo nucleotides utilizing a reverse transcription system (vazyme). Quantitative real-time PCR (20 μl reaction volume) was conducted using 1 μl of cDNA, 0.5 μl of each primer and 10 μl of AceQ qPCR SYBR Green Master Mix (vazyme, http://www.vazyme.com/) in a Roche480 real-time PCR detection system. The rice Actin gene (Os03g0718100) was used as the internal control. Each sample was performed in triplicate. A relative gene expression level was calculated using the comparative Ct method (Livak and Schmittgen 2001). The gene-specific primers were shown on Table 1.
Grain yield evaluation of qPBN8 NILs
Six pairs of qPBN8 NILs with different heading date were seeded in nursery trays containing nutrient matrix. After 30 days, the NILs were transplanted to experimental plot in randomized complete block design with three replications. Each replication had forty plants in a density of 25 cm (4 lines) × 13.3 cm (10 plants). Twenty plants, excluding marginal effects, were collected to evaluated yield structure, when the grain matured. The agronomy traits were surveyed included heading date, panicle number per plant, spikelet number per panicle, seed setting rate, thousand grain weight and grain yield per plant.
Results
Phenotypic variation
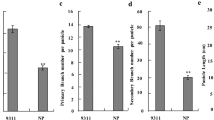
The two parents, YR1 and NJ1, displayed significant differences in the panicle traits examined (Fig. 1, Table 2). The main panicle of the commercial cultivar NJ1 has approximately 16.7 cm in length, 164.3 spikelets, 13.3 primary branches, 21.7 secondary branches, and 93.1% seed setting rate. However, that of YR1 has approximately 19.3 cm in length, 421.0 spikelets, 23.0 primary branches and, 76.0 secondary branches, and 89.3% seed setting rate. Moreover, YR1 heading date is about 30 days earlier than NJ1 (Table 2). A great differences existed between two parents in panicle traits, which provided an abundance of trait variation for population development and QTL mapping. The frequency distribution of PL, PBN, SBN, SPP and SSR in BC2F2 population are shown in Fig. 2. The panicle length and the primary branch are close to a normal distribution, and the positive super-parent individuals of primary branch are relatively more. The variation of the number of secondary branches and spikelets per panicle is large, showing a skew distribution, and the number of positive super-parents is small. The seed setting rate is skewed, and most individuals have a high seed setting rate.
Quantitative trait loci mapping
To uncover the genetic factors of panicle size, QTL analysis was performed in a BC2F2 population of 164 individuals. 119 Polymorphic markers distributed on 12 chromosomes were used to identify the genotypes of BC2F2 individuals (Fig. 3). For five panicle traits as mentioned above, only three major QTL were identified on chromosomes 1 and 8 (Table 3, Fig. 3). qSBN1 and qGNP1 controlling secondary branch number (SBN) and spikelets per panicle (SPP) respectively, shared the same interval between markers RM8105 and RM1220 on chromosome 1, explaining 30.6% and 25.4% of phenotypic variance with an additive effect of 11.0 secondary branches and 34.7 spikelets, respectively. In this interval, a reported grain number locus Gn1a was located (Ashikari et al. 2005). qPBN8, a QTL for primary branch number (PBN), was located between InDel markers S4 and S21 on chromosome 8. This QTL explained 39.6% of phenotypic variance with an additive effect of 3.6 primary branches in the BC2F2 population. YR1 allele had dominant and positive effects on primary branches. No QTL was identified controlling PL and SSR. In addition, one QTL of heading date was located on chromosome 10, explaining 7.3% of phenotypic variance with an additive effect of 5.4 heading date.
For verification of the qPBN8 loci, the BC2F2 plants with heterozygous qPBN8 and homozygous NJ1 qSBN1 locus were developed to BC2F3 populations. 134 plants of a BC2F3 line were used for qPBN8 re-mapping. The results showed that the qPBN8 was narrowed to an interval between markers S98 and S21, explaining 43.4% phenotypic variation with an additive effect of 4.4 primary branches (Fig. 4, Table 3). A QTL for the spikelet number qSPP8 was detected in this interval, and explained 18.3% of phenotypic variation with an additive effect of 27.4 spikelets. This shows that increasing the number of primary branches can increase the number of spikelets.
Fine mapping of qPBN8
In 1374 BC2F4 plants, a total of 56 recombinant events were detected between InDel markers S98 and S21. Through phenotype analysis of the crucial recombination plants and their next generation, qPBN8 was delimited to a 51.8 kb region between the marker S30 and S10 (Fig. 5). There are six annotated genes in the 51.8 kb region, referring to Rice Genome Annotation Project. ORF1 encodes Os8bglu27-beta-glucosidase. ORF2 encodes G-patch domain containing protein. ORF3 encodes the plant-specific transcription factor OsSPL14. ORF4 might encode hypothetical protein. ORF5 and ORF6 might encode retro-transposon protein (Table 4). Previous study demonstrated that ORF3 (LOC_Os08g39890) regulates primary branch number and ideal plant architecture (Jiao et al. 2010; Miura et al. 2010). Therefore, ORF3 was selected as the candidate gene for further analysis.
Fine mapping of qPBN8, a location of qPBN8 on rice chromosome 8, b high-resolution linkage analysis of qPBN8 locus. Recombinants numbers are given between markers. The hollow, black and grille bars represent genotypes of NJ1, YR1 and heterozygote, respectively. Serial numbers indicated the key recombinants. GT is the derived genotype of recombinant plant by the phenotype of its progeny line, N (NJ1), Y (YR1), H (heterozygote). PP (± SD) is average value (± standard error) of phenotype of progeny line. c Open reading frames (ORFs) within mapped interval of the qPBN8 on rice genome annotation project
Sequence and expression comparison of OsSPL14
Sequence analysis revealed that there was no difference in the coding region of OsSPL14 between two parents. The expression level of OsSPL14 at the 1–2 mm panicle stages was about ninefold higher in YR1 than in NJ1. But there is no difference at the 5–10 cm panicle stages (Fig. 6).
The yield effect of qPBN8
In six pairs of NILs, all the lines with YR1 qPBN8 allele had fewer tiller, more spikelets, lower seed setting rate and slightly larger grain, showing the basic characteristics of qPBN8. NILs of qPBN8 were derived from different F2 and BC1F2 heterozygotes. Their genetic background were very different especially in vegetative growth. The days from seeding to heading ranged from 69 to 115 days. The yield of NIL was related to the length of vegetative period. Between early-maturing NILs, the grain yield of the NIL with YR1 allele of qPBN8 was significantly lower than that with NJ1-qPBN8. However, between late-maturing NILs, the yield of YR1-qPBN8 NILs was significantly higher than that of NJ1-qPBN8 NILs (Table 5).
Discussion
Increasing yield is one of the most important goals in rice breeding. Grain number per panicle is a major component of rice yield that is typically controlled by many quantitative trait loci (QTLs). The identification of QTLs controlling grain number per panicle in rice would be valuable for the breeding of high-yielding rice (Hu et al. 2018; Zhu et al. 2011). Many QTLs for yield related traits have been verified and used for developing high yield rice (Miura et al. 2011). Panicle traits have been the vital factors for rice yield. It includes four factors: panicle length, primary branch number, secondary branch number, and spikelet number (Cheng et al. 2007; Peng et al. 2014). Increasing the value of each panicle characteristic would be an effective method for improving grain yield. To develop varieties with a large number of grain is a major direction in super rice breeding. Most of QTLs controlling GNB/SPP and SBN were located in cluster or closely linked on chromosomes, and the directions of their additive effects were consistent, which explained the genetic basis of significant correlations between their phenotypic characters (Luo et al. 2009). Therefore, one of the methods to increase yield is to pyramid the favorable alleles in the present high-yielding cultivar backgrounds (Takai et al. 2018).
In this study, panicle traits were focused on and genetic analysis was carried out in BC2 population derived from donor parent YR1 and recurrent parent NJ1. The two QTLs, qSBN1 and qGNP1, were mapped to the same interval of GN1a (Ashikari et al. 2005). One QTL (qPBN8) for primary branch were fine-mapped and targeted to IPA1/WFP. The YR1 alleles of QTLs on chromosomes 1 and chromosome 8 increased the number of grain per panicle, primary branch and secondary branch. The additive effect of three QTLs came from YR1. These results suggested that larger panicles can be achieved by pyramiding the favorable alleles of these QTLs. qPBN8 was finally localized to 51.8 kb interval containing six annotated genes. Of them, ORF3 has been reported not only regulating branch number and plant architecture but also promoting both yield and disease resistance by sustaining a balance between growth and immunity (Jiao et al. 2010; Miura et al. 2010; Wang et al. 2018). There was no difference in nucleotide sequence. The expression level of OsSPL14 was significantly higher in YR1 than NJ1 at the initial development stage of panicle (Fig. 6). And the primary branch number of YR1 was larger than NJ1. Therefore, it was considered that candidate of qPBN8 is OsSPL14 and plays an important role in controlling primary branch number and grain number.
In six paired NILs of qPBN8, the large panicle lines showed fewer tiller, more spikelets, lower seed setting rate and slightly larger grain. The yield of NILs was related to the length of vegetative period. The earlier the heading, the lower the yield. Between late-maturing NILs, the yield of YR1-qPBN8 NILs was significantly higher than that of NJ1-qPBN8 NILs. Between early-maturing NILs, however, the grain yield of the NIL with YR1-qPBN8 allele was unexpectedly lower than that with NJ1-qPBN8. This means that the yield effect of the large panicle allele of YR1-qPBN8 depended more on the length of vegetative period. The main reason for the result might be that the yield effect of more spikelets in early-maturing YR1-qPBN8 NIL was difficult to compensate for its effect of fewer panicles and lower seed setting rate. Therefore, the qPBN8 large panicle genotype was suitable for late-maturing varieties to increase yield.
Larger panicles have been an important target for improvement of rice grain yield because of its relationship with grain number. The utilization of favorable QTL panicles related, for example, GN1A, IPA1/WFP, SPIKE/NAL1, has increased the rice yield. Three QTL for the panicle traits were detected, which all were most likely to be GN1a and IPA1. The additive effect of three QTL all came from YR1. Our study suggest that larger panicles might be possible with the combination of the alleles of these QTLs. Therefore, pyramiding of elite QTLs will certainly benefit for breeding high-yield varieties by marker-assisted selection for rice in the future.
References
Ashikari M, Sakakibara H, Lin SY, Yamamoto T, Takashi T, Nishimura A, Matsuoka M (2005) Cytokinin oxidase regulates rice grain production. Science 309(5735):741–745
Bai X, Huang Y, Mao D, Wen M, Zhang L, Xing Y (2016) Regulatory role of FZP in the determination of panicle branching and spikelet formation in rice. Sci Rep 6:19022
Cheng FM, Zhang QF, Zhu HJ, Zhao NC, Wang F, Chen KM, Zhang GP (2007) The difference in amylose content within a panicle as affected by the panicle morphology of rice cultivars. J Cereal Sci 46(1):49–57
Cheng J, Wang L, Du W, Lai Y, Huang X, Wang Z, Zhang H (2014) Dynamic quantitative trait locus analysis of seed dormancy at three development stages in rice. Mol Breed 34(2):501–510
Fang NY, Wang RS, He WW, Yin CF, Guan CH, Chen H, Zhang HS (2016) QTL mapping of panicle blast resistance in japonica landrace heikezijing and its application in rice breeding. Mol Breed 36(12):171
Fujita D, Trijatmiko KR, Tagle AG, Sapasap MV, Koide Y, Sasaki K, Kobayashi N (2013) NAL1 allele from a rice landrace greatly increases yield in modern indica cultivars. Proc Natl Acad Sci USA 110(51):20431–20436
Hu ZJ, Cao LM, Sun XJ, Zhu Y, Zhang TY, Jiang L, Luo XJ (2018) Fine mapping of a major quantitative trait locus, qgnp7(t), controlling grain number per panicle in African rice (Oryza glaberrima S.). Breed Sci 68(5):606–613
Huang X, Qian Q, Liu Z, Sun H, He S, Luo D, Fu X (2009) Natural variation at the DEP1 locus enhances grain yield in rice. Nat Genet 41(4):494–497
Huo X, Wu S, Zhu Z, Liu F, Fu Y, Cai H, Sun C (2017) NOG1 increases grain production in rice. Nat Commun 8(1):1497
Ikeda K, Ito M, Nagasawa N, Kyozuka J, Nagato Y (2007) Rice ABERRANT PANICLE ORGANIZATION 1, encoding an F-box protein, regulates meristem fate. Plant J 51(6):1030–1040
Jiao Y, Wang Y, Xue D, Wang J, Yan M, Liu G, Li J (2010) Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat Genet 42(6):541–544
Jin J, Huang W, Yang J, Shi M, Zhu M, Luo D, Lin H (2008) Genetic control of rice plant architecture under domestication. Nat Genet 40(11):1365–1369
Kumar V, Ladha JK (2011) Direct seeding of rice: recent developments and future research needs. Adv Agron 111:297–413
Li S, Zhao B, Yuan D, Duan M, Qian Q, Tang L, Li C (2013) Rice zinc finger protein DST enhances grain production through controlling Gn1a/OsCKX2 expression. Proc Natl Acad Sci USA 110(8):3167–3172
Liu HY, Hussain S, Zheng MM, Peng SB, Huang JL, Cui KH, Nie LX (2015) Dry direct-seeded rice as an alternative to transplanted-flooded rice in Central China. Agron Sustain Dev 35(1):285–294
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2 − ΔΔCT method. Methods 25(4):402–408
Luo X, Tian F, Fu Y, Yang J, Sun C (2009) Mapping quantitative trait loci influencing panicle-related traits from Chinese common wild rice (Oryza rufipogon) using introgression lines. Plant Breed 128(6):559–567
Mei HW, Xu JL, Li ZK, Yu XQ, Guo LB, Wang YP, Luo LJ (2006) QTLs influencing panicle size detected in two reciprocal introgressive line (IL) populations in rice (Oryza sativa L.). Theor Appl Genet 112(4):648–656
Miura K, Ikeda M, Matsubara A, Song XJ, Ito M, Asano K, Ashikari M (2010) OsSPL14 promotes panicle branching and higher grain productivity in rice. Nat Genet 42(6):545–549
Miura K, Ashikari M, Matsuoka M (2011) The role of QTLs in the breeding of high-yielding rice. Trends Plant Sci 16(6):319–326
Peng SB, Khush GS, Virk P, Tang QY, Zou YB (2008) Progress in ideotype breeding to increase rice yield potential. Field Crops Res 108(1):32–38
Peng YL, Gao ZY, Zhang B, Liu CL, Xu J, Ruan BP, Qian Q (2014) Fine mapping and candidate gene analysis of a major QTL for panicle structure in rice. Plant Cell Rep 33(11):1843–1850
Sasaki K, Fujita D, Koide Y, Lumanglas PD, Gannaban RB, Tagle AG, Ishimaru T (2017) Fine mapping of a quantitative trait locus for spikelet number per panicle in a new plant type rice and evaluation of a near-isogenic line for grain productivity. J Exp Bot 68(11):2693–2702
Tabuchi H, Zhang Y, Hattori S, Omae M, Shimizu-Sato S, Oikawa T, Sato Y (2011) LAX PANICLE2 of rice encodes a novel nuclear protein and regulates the formation of axillary meristems. Plant Cell 23(9):3276–3287
Takai T, Nakano H, Yoshinaga S, Kondo M (2018) Identification of a novel QTL for the number of spikelets per panicle using a cross between indica- and japonica-type high-yielding rice cultivars in Japan. Plant Breed 137(2):109–117
Tan L, Li X, Liu F, Sun X, Li C, Zhu Z, Sun C (2008) Control of a key transition from prostrate to erect growth in rice domestication. Nat Genet 40:1360
Wang J, Zhou L, Shi H, Chern M, Yu H, Yi H, Li Y (2018) A single transcription factor promotes both yield and immunity in rice. Science 361(6406):1026–1028
Weng X, Wang L, Wang J, Hu Y, Du H, Xu C, Zhang Q (2014) Grain number, plant height, and heading date7 is a central regulator of growth, development, and stress response. Plant Physiol 164(2):735–747
Xing Y, Zhang Q (2010) Genetic and molecular bases of rice yield. Annu Rev Plant Biol 61:421–442
Xue WY, Xing YZ, Weng XY, Zhao Y, Tang WJ, Wang L, Zhang QF (2008) Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat Genet 40(6):761–767
Zhao L, Tan L, Zhu Z, Xiao L, Xie D, Sun C (2015) PAY1 improves plant architecture and enhances grain yield in rice. Plant J 83(3):528–536
Zhu JY, Zhou Y, Liu YH, Wang ZD, Tang ZX, Yi CD, Liang GH (2011) Fine mapping of a major QTL controlling panicle number in rice. Mol Breed 27(2):171–180
Acknowledgements
This work was supported by the Natural Science Foundation of Jiangsu Province (Grant No. BK20151427).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Shang, F., Chen, L., Meng, X. et al. Fine mapping and grain yield analysis of a major QTL controlling primary branch number in rice (Oryza sativa L.). Genet Resour Crop Evol 67, 421–431 (2020). https://doi.org/10.1007/s10722-019-00857-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10722-019-00857-8