Abstract
Purification of amylases from digestive tracts of three freshwater fish species with Q-Sepharose Fast Flow and Sephacryl S-200 columns displayed two isoforms of amylases from Osteochilus hasselti (O1, O2) and three isoforms of those from both Hampala dispar (UB, H1, H2) and Puntioplites proctozystron (P1, P2, P3). The optimum pH values displayed at 7.0 and 8.0, while the optimum temperatures revealed at 40 and 50 °C. Almost isoenzyme activities were activated by NaCl and CaCl2, whereas EDTA and SDS strongly inhibited all enzymatic activities. Verification with an atomic absorption spectrophotometry exhibited the presence of Ca2+ ions in the range of 0.02–13.53 ppm per mg protein indicating that amylases are Ca2+ dependent. Molecular weight analysis revealed 12 to 147 kDa. The UB, O1, and H2 amylases with appropriate molecular masses of 64, 49, and 25 kDa validated with LC-MS/MS were selected. Three certain enzymes revealed high stability in a sample buffer after five cycles of freeze-thawing process upon storage at − 20 °C for 12 weeks. No protein degradation was observed on polyacrylamide gel, and the enzymes still displayed sharp and clear bands on zymograms. The result suggested that the purified fish amylases, which expressed high activities and stabilities, were potentially used as enzyme molecular weight markers for zymography.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Amylases are glycoside hydrolases that catalyze the hydrolysis of glycosidic linkage in starch molecules to produce a small fragment of saccharides. According to catalytic function, amylases are divided into three types consisting of α-amylase, β-amylase, and γ-amylase. α-Amylase (EC 3.2.1.1) is an endo-amylase family that randomly cleaves at internal α-1,4-glycosidic bonds in starch molecules, ultimately yielding malto-oligosaccharide or glucose. β-Amylase (EC 3.2.1.2) is an exo-amylase that catalyzes the hydrolysis of second α-1,4-glycosidic linkages from the non-reducing end releasing two glucose units. γ-Amylase (EC 3.2.1.3) is an exo-amylase that catalyzes the hydrolysis of both α-1,6- and α-1,4-glycosidic linkages from the non-reducing end yielding glucose (Saini et al. 2017; Kaplan and Guy 2004). Among all types, α-amylase is widely produced from living organisms such as microorganisms (Wu et al. 2018; Sodhi et al. 2005), fungi (Xian et al. 2015; Abdulaal 2018), plants (Stanley et al. 2005; Asatsuma et al. 2005), and animals (MacDonald et al. 1980; Hagenbüchle et al. 1980). Calcium plays an important role for the catalytic function and the stabilization of an α-amylase structure by interacting with negatively charged amino acid residues, such as glutamic acid and aspartic acid (Rogers 1985; Svensson 1994). In fish, this type of enzyme is normally synthesized and secreted by the digestive tract and plays an essential role in the biochemical process of carbohydrate digestion, especially in herbivorous and omnivorous (Gioda et al. 2017; Al-Tameemi et al. 2010). In previous studies, the amylase enzymes in different fish species exhibit the different characteristics and isoenzymes patterns in terms of number and molecular sizes (Champasri and Champasri 2017; Fernández et al. 2001; Ji et al. 2012; Champasri et al. 2021). Moreover, characterization of isoenzymes from each species regarding optimum temperature, optimum pH, thermostability, and effects of metal ions has become important and helps to understand more about their properties for determining their related applications such as hydrolysis of malt starch for alcohol production by the thermostable α-amylase from Bacillus sp. PS-7 (Sodhi et al. 2005) and double-enzymatic sugar-making process in starch industry by the acidic α-amylase from Alicyclobacillus sp. A4 (Bai et al. 2012).
Substrate zymography is an electrophoresis technique widely used for studying in-gel hydrolytic enzymes (Vandooren et al. 2013). Specific substrate of enzymes can be probably co-polymerized with acrylamide gel before electrophoresis or soaked with acrylamide gel after electrophoresis. The location of an enzyme, which can hydrolyze the substrate in gel, is visualized as a clear band against a dark background of overspreading substrate-specific dye. Many enzymes such as proteases (Champasri and Champasri 2017; Van de Louw et al. 2002; Jones 2014; Banerjee et al. 2016), amylases (Champasri and Champasri 2017; Champasri et al. 2021; Banerjee et al. 2016), cellulases (Champasri et al. 2015; Cano-Ramírez et al. 2017), and endopeptidases (Wong and Blaise 2020) have been studied by using this method. Determination of the molecular weights of hydrolytic enzymes requires comparing their electrophoretic movements on zymograms with those of protein molecular weight markers on another gel stained with Coomassie brilliant blue. Using different gels stained with different dyes and destained with different reagents may sometimes cause an error of estimation due to the distance shift of the protein band. The commercial kit of enzyme molecular weight markers separated and detected on single zymographic gel has not been available. Our previous studies have found that each fish species has exhibited several isoenzymes of amylases and proteases with different molecular masses (Champasri and Champasri 2017; Champasri et al. 2021). In addition, the enzymes have been stable in wide pH and temperature ranges with residual activities more than 80% after incubation at 25 to 50 °C, especially for amylases (Champasri et al. 2021). Therefore, in this study, we aimed to (i) purify the amylases extracted from digestive tract of three fish species, (ii) determine the molecular weights of the purified enzymes, (iii) characterize the biochemical properties of the purified enzymes, and (iv) select the enzyme with the desire molecular weights and test the stabilities after several freeze-thaw cycles to provide new insight application of fish amylases as enzyme molecular weight markers for substrate zymography.
Materials and methods
Ethics statement
The ethical principles for use and care of animals in science are approved by the Institutional Animal Care and Use Committee of Khon Kaen University (IACUC KKU). All experimental protocols and the care and use of experimental animals complied with animal welfare laws of Thailand, and guidelines and policies approved by ThaiIACUC (permit reference number U1-04,584-2559).
Materials
Starch (from rice) (cat. no. S-7260) was purchased from Sigma-Aldrich, USA. Quick Start Bradford 1 × Dye Reagent (cat.no. 5000205) and gel filtration standard (cat.no. 1511901) were purchased from Bio-Rad, California. Soluble starch (cat.no. SB0904) was from Bio Basic Inc., Canada. 3,5-Dinitrosalicylic acid was purchased from Fluka, USA. Ammonium sulfate was purchased from Ajax Finechem, Univar (USA). Chemical reagents for preparation of buffers were purchased from CARLO ERBA (France), Bio Basic Inc., Sigma-Aldrich, Honeywell Fluka (NJ, USA), and Ajax Finechem. Chemical reagents for gel electrophoresis: acrylamide (cat.no. 17-1302-02) and N,N′-methylenebisacrylamide (cat.no. 17–1304-02) and ammonium persulfate were from GE healthcare (USA), and sodium dodecyl sulfate (cat.no. DB0485) was from Bio Basic Inc.
Sample collection and crude enzyme extraction
Three fish species including Puntioplites proctozystron, Osteochilus hasselti, and Hampala dispar were randomly collected by using nets from Nong Kong Kaew swamp, Chonnabot district, Khon Kaen, Thailand.
The identification of fish species was kindly performed by Assoc. Prof. Apinun Suvarnaraksha, Faculty of Fisheries Technology and Aquatic Resources, Maejo University, Thailand. Fishes with body lengths of 16.64 ± 2.16, 18.42 ± 1.24, and 14.50 ± 1.45 cm for O. hasselti, P. proctozystron, and H. dispar, respectively, were collected. The 110 specimens of O. hasselti, 85 of H. dispar, and 100 of P. proctozystron were sacrificed using 20 mg L−1 MS222. The digestive tracts of fishes were dissected, cleaned with distilled water, and homogenized in the extraction buffer (0.05 M Tris–HCl buffer, pH 8.0) using a blender with the wet weight (g) per buffer volume (mL) of 1:3 ratios. The homogenates were harvested by centrifugation at 24,000 × g, 4 °C for 20 min using the Sorvall™ Legend™ XTR centrifuge (Thermo Scientific). Supernatant, referred to as “crude enzyme extract,” was collected and then stored at − 20 °C until use. All extraction and purification steps were carried out at 4 °C. Protein content was determined according to the Bradford method (Bradford 1976). Standard curve of bovine serum albumin was used to calculate the protein concentration.
Enzyme assay
Amylase assay was carried out by modifying the DNS (3,5-dinitrosalicylic acid) method of Miller (1959). Assays were conducted in a final total volume of 1.0 mL containing 50 µL of crude enzyme extract; 500 µL of 0.5% starch solution in 0.05 M Tris-HCl, pH 8.0; and 450 µL of assay buffer (0.2 M Tris-HCl buffer, pH 8.0). After 15 min of incubation at room temperature (27 °C), the reaction was inactivated, and the sugar product was measured by adding 500 µL of DNS solution and boiling for 10 min. After centrifugation at 16,000 × g for 10 min (Rotina 380R, Hettich) to remove a non-digested starch and denatured enzyme, the supernatant containing amylolytic products was measured by monitoring the absorbance at 540 nm with JASCO V-530 spectrophotometer. The amount of released maltose was calculated from a maltose standard curve. One unit of amylase was defined as the amount of amylase which releases 1 mmol of maltose per min under the assay condition.
Amylase purification
The crude enzyme extracts were precipitated at 80% saturation of ammonium sulfate. The precipitated proteins were collected by centrifugation at 24,000 × g, 4 °C for 20 min and subsequently dissolved in an extraction buffer before dialyzing against 0.02 M Tris-HCl buffer, pH 8.0 containing 0.1 M NaCl and 0.5% glycerol. Protein samples from each fish species were applied to the Q-Sepharose Fast Flow (GE healthcare/Cytiva) resin packed in 1.5 cm × 15 cm column (Bio-Rad). The column was pre-equilibrated with five column volumes of an equilibration buffer (EQ) (0.02 M Tris-HCl buffer, pH 8.0, 0.5% glycerol), and a flow rate was run at 2.5 mL per min. Bound proteins were eluted with a stepwise gradient by using an EQ buffer containing 0.25–1.0 M NaCl. Fractions were collected to measure the protein content by monitoring the absorbance at 280 nm. The active fractions exhibiting high enzymatic activity measured at 540 nm were pooled for further concentration by using Amicon Ultra-15 concentrator (MWCO 10 kDa). Retentate proteins were slowly loaded onto a gel filtration column (1 cm × 100 cm, Bio-Rad) containing Sephacryl S-200 high resolution (GE healthcare/Cytiva). The column was run at a flow rate of 1 mL per 3 min by using 0.05 M Tris-HCl, pH 7.5, 0.15 M NaCl, 0.5% glycerol buffer. Protein content and amylase activity were followed as described above. The active fractions revealing amylase activity were pooled. The purity was analyzed by using SDS-PAGE, and the position of amylase on gel was visualized by substrate zymography.
SDS-PAGE and substrate zymography analysis
SDS-PAGE and zymography were carried out according to the method as previously described (Champasri and Champasri 2017; Champasri et al. 2021) with minor modifications. The protein samples were separated on 10% separating gel and 4% stacking gel in a discontinuous buffer system using a Bio-Rad Tetra cells electrophoresis apparatus. Samples were mixed with 1 × SDS sample buffer (62.5 mM Tris-HCl buffer, pH 6.8 containing 0.02% bromophenol blue, 25% glycerol, 2% SDS) before loading onto a stacking gel. After electrophoresis, proteins on the polyacrylamide gel were detected by staining gel in staining dye solution (0.25% Coomassie brilliant blue R-250, 40% ethanol, 10% glacial acetic acid) and washing with destaining solution (30% ethanol, 10% glacial acetic acid) until the blue bands of proteins were observed against a clear background. A second gel referred to as a zymogram was set up to detect amylase activity and locate the position of the certain enzyme on the gel. The gel was immersed in 0.05 M Tris-HCl buffer, pH 8.0 containing 2% Triton X-100 for 30 min at room temperature with gentle shaking to remove SDS. The gel was subsequently washed for 5–10 min in the same buffer without Triton X-100 to ensure proper folding of the enzyme. To allow the diffusion of substrate into gel, gel was soaked in 2% starch solution (dissolved in 0.05 M Tris-HCl buffer, pH 8.0) at 4 °C for 30 min before moving to incubate at 45 °C with gentle shaking for 1 h to facilitate the catalytic activity of the enzyme. Finally, the gel was stained with iodine solution (5.7% KI, 0.6% I2) for 5 min, and the excess iodine was washed off with distilled water. The clear band against a dark brown background was visualized, and the position of amylolytic enzyme was observed after destaining with distilled water.
Determination of enzyme molecular weights
Determination of molecular weight of the purified amylases was determined by using gel filtration chromatography. Blue dextran with molecular weight of 2000 kDa was firstly applied onto a Sephacryl S-200 high-resolution column (1 cm × 100 cm, Bio-Rad) to determine the void volume (V0) of a column. The gel filtration standard (Bio-Rad) with molecular weights ranging from 1.35 to 670 kDa was dissolved with deionized water and gently mixed well before loading onto the column. Purification was followed with the same protocol as described above for gel filtration in the purification section. The absorbance at 280 nm was followed to monitor the elution of protein. The standard curve of protein molecular weight standard was generated by plotting the Kav values with the logarithm of the known molecular weights of four standard proteins. The Kav was calculated by (Ve − V0/(Vc − V0), where Kav is the distribution coefficient of the protein, Ve is the elution volume of the standard protein or the purified enzyme, V0 is the column void volume, and Vc is the total volume of the column. To determine the molecular weight of the purified amylases, the Kav values of the purified enzymes were calculated from the equation, and the resulting data were subsequently compared with the calibration curve.
Effects of pH and temperature on amylase activity
The effects of pH on amylase activity were evaluated by using 0.2 M buffers consisting sodium acetate (pH 5.0), sodium phosphate (pH 6.0 and 7.0), Tris-HCl (pH 8.0 and 9.0), and glycine-NaOH (pH 10.0 and 11.0). The effects of temperature on amylase activity were determined by incubating the reaction mixtures at 27, 35, 40, 50, 60, and 70 °C. The enzyme activities were conducted in duplicate, and the maximum activity was defined as 100% relative activity.
Effects of temperature on amylase stability
Thermal stabilities were examined by determining enzyme activity after pre-incubation of the purified enzyme in 0.2 M Tris-HCl buffer, pH 8.0 upon incubation for 1, 2, 3, and 24 h. The stabilities were tested at both room temperature (27 °C) and optimal temperatures (40 °C and 50 °C). The pre-incubated enzyme-buffer mixtures were aliquoted to measure the enzymatic activity by adding 0.5% starch according to the enzyme assay protocol. The remaining activities were calculated as the percentage of relative activity, at which 100% relative activity was defined as the activity of enzymes without pre-incubation.
Effects of metal ions, detergent, and chelating agents on amylase activity
Three concentrations (2, 5, and 10 mM) of metal ions (NaCl, CaCl2, MgCl2, and MnCl2), detergent (SDS), and chelating agents (EDTA) were added to the purified enzymes and pre-incubated at room temperature. After incubation for 30 min, the enzyme-ion or enzyme-additive reagent mixture was aliquoted to detect the enzyme activity by adding substrate and buffer. The catalytic activities were allowed at the standard condition as previously described in the enzyme assay section. The enzyme activity without any metal ions, detergent, or chelating agent was defined as 100% relative activity.
Calcium determination by atomic absorption spectrometry
The calcium content in the purified amylase fractions was analyzed by atomic absorption spectrometry (an Instrumentation Laboratory PerkinElmer PinAAcle 900 T atomic absorption spectrophotometer). One hundred microliter of purified enzyme samples was added with 0.5 M HNO3 to the final volume of 1 mL prior to incubation at room temperature for 1 h. After centrifugation at 14,000 × g for 10 min (Rotina 380R, Hettich), clear supernatants were taken to measure the calcium content with an atomic absorption spectrometer at the wavelength of 422.67 nm. The calcium concentration in the purified enzymes was calculated by using calcium standard curve, which was freshly prepared by diluting 1000 ppm stock calcium solution with deionized water to 2–8 ppm. Each analysis was carried out in triplicate, and the average or mean value was presented.
Amylase identification by LC/MS–MS
The partially purified amylases were initially separated by SDS-PAGE. Zymography was also performed in parallel to verify the position of amylase on gel. After electrophoresis, the expected bands of amylases were manually excised and washed twice with deionized water. The bands were subsequently digested with trypsin. The tryptic fragments of the purified peptides were automatically analyzed by liquid chromatography–tandem mass spectrometry (LC/MS–MS) at the Biomolecular Analysis Service Unit (Faculty of Medicine, Khon Kaen University, Thailand). The MS/MS ion results were compared with Swiss-Prot database by Mascot database search online program of Matrix Science (http://www.matrixscience.com). The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (Perez-Riverol et al. 2022) partner repository. The datasets generated and analyzed during the current study are available in PRIDE database (PRIDE—Proteomics Identification Database (ebi.ac.uk) via ProteomeXchange (www.proteomexchange.org) repository with accession number or identifier PXD033432 and PXD033433.
Stability of enzyme molecular weight markers
According to our objective to provide the application of the purified amylases as the enzyme molecular weight markers for zymography, the purified enzymes from different three fish species were mixed together before adding 1 × sample buffer (62.5 mM Tris-HCl, pH 8.0, 1 mM NaN3, 33% glycerol, 2% SDS, 0.01% bromophenol blue) and kept at − 20 °C. The components of sample buffer were prepared based on those of buffer supplied with the commercial unstained protein molecular weight markers. Five microliter of enzyme mixtures stored at − 20 °C was thawed and aliquoted to test their stability every 2 weeks by using SDS-PAGE and substrate zymography. The sharp bands of non-digested enzymes were expected to see. Total 12 weeks of storage at − 20 °C were investigated.
Results
Purification of amylases
Crude enzymes extracted from three fish species were partially purified with 80% saturated ammonium sulfate followed by ion-exchange and size exclusion chromatography. The purification yield and fold were calculated, and the enzyme purity was analyzed by using SDS-PAGE and zymography. After ammonium sulfate precipitation, the fractions containing amylase were dialyzed and loaded onto the Q-Sepharose Fast Flow column. The chromatograms of purification are shown in Fig. 1. Both O. hasselti and H. dispar showed two peaks of amylase activities named as O1, O2, H1, and H2, respectively. Both activity peaks were eluted with 0.25 M and 0.50 M NaCl, respectively. In addition, the amylase activity was also detected in the unbound fraction (UB) of H. dispar. P. proctozystron displayed three peaks of amylase including P1, P2, and P3, which were eluted with 0.25 M, 0.5 M, and 1.0 M NaCl. The certain activity peaks from three fish species were separately pooled for further purification with gel filtration chromatography. Before applying to Sephacryl S-200 HR column, the enzyme samples were concentrated in order to reduce the sample volume. Figure 2 shows the size exclusion chromatograms. Almost all enzyme fractions revealed a single peak of amylase activity except P3 (Fig. 2H), whose activity was exhibited into two peaks. As summarized in Table 1, specific activities were varied from 0.21 to 97.55 U mg−1 protein, whereas the purification folds were presented from 0.18 to 61.48. Among these enzymes, the purified UB and H1 displayed the highest values of both parameters.
Molecular weights of amylases
The molecular weights of the purified amylases were determined by using the calibration standard curve of gel filtration chromatography. The volumes of effluent (Ve) or the elution volume of the purified amylases (Fig. 2) were converted to Kav and compared with the calibration curve. The resulting equation (y = − 0.2916x + 1.7535) with R2 = 0.993 (supplementary information, Fig. S1) was used to calculate the molecular weight (MW) of the purified amylases. The O1 and O2 of O. hasselti had a molecular weight of 49 and 42 kDa (Fig. 2A, B), while UB, H1, and H2 from H. dispar showed the molecular weight about 64, 32, and 25 kDa (Fig. 2C, D, and E), respectively. For P. proctozystron, the molecular weight of P2 amylase was approximately 32 kDa (Fig. 2G), and those of P3, which exhibited two peaks of amylase activities, were 147 and 12 kDa, respectively (Fig. 2H). In case of P1, the enzyme displayed the broad peak activity of the gel filtration column as shown in Fig. 2F; thus, it was not possible to locate the actual Ve.
SDS-PAGE and zymographic results
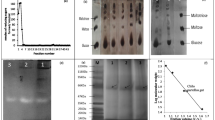
To analyze the purity and verify the presence of an amylase on the purification fractions, SDS-PAGE and zymography were performed every step of purification. Although the SDS gel staining with Coomassie brilliant blue R-250 (CBB) contained a few bands of impurities, zymograms clearly displayed the single isoform of O1, O2, UB, H1, and H2 amylases with different molecular weights (Fig. 3A, B). In the case of P. proctozystron, all three fractions of P1, P2, and P3 showed co-elution of three to six isoforms, which were not able to distinguish by using Sephacryl S-200 column due to the close molecular weights of them (Fig. 3C). Among the purified amylases, the O2 enzyme showed the highest purity as observed on CBB gel.
SDS gels stained with Coomassie brilliant blue R-250 (left panel) and zymogram gels (right panel) showing the purification results of amylases from O. hasselti (A), H. dispar (B), and P. proctozystron (C) (lane M, unstained MW protein markers (Thermo Scientific); lane C, crude enzyme extract; lane A, ammonium sulfate precipitation; lanes O1, O2, UB, H1, H2, P1, P2, and P3, the purified amylases after size exclusion chromatography). Both CBB gels and zymograms were loaded with the similar pattern of enzyme samples. For CBB gel (proteinograms), 25 µg of total protein of all three fish species were loaded in lanes C and A; 10 and 5 µg in O1 and O2; 8 µg in UB, H1, and H2; 20 µg in P1; and 15 µg in P2 and P3. For zymograms, lanes C, O1, O2 contained 200, 125, and 50 mU of amylases; lanes C, A, UB, H1, and H2 contained 75, 70, 65, 42, and 33 mU of the enzymes; lanes C, A, P1, P2, and P3 contained 160, 155, 100, 75, and 60 mU of enzymes. The amounts of total protein loading and enzymatic activity (unit) were loaded and optimized to make clarify in observation on CBB gel upon CBB staining/destaining and could also exhibit the sharp clear band with high intensity on zymograms. Gels were cropped from the beginning of separating gels on top to the bottom edge of the gels. The dye fronts as seen in blue lines were still observed above the bottom edge of CBB gels. More detail of figures was described in supplementary information
Effects of pH and temperature on purified amylase activities
As shown in Fig. 4, the amylases from different species showed different responses on pH and temperatures. Moreover, the different isoforms of the same species displayed the similar pattern of the responses. Interestingly, both O1 and O2 isoforms of O. hasselti showed a broad pH range from 7.0 to 10.0 with nearly 100% of enzymatic activity, and both isoforms revealed the same optimum pH at 8.0 (Fig. 4A). The similar result was also observed in P. proctozystron. Three isoforms exhibited the similar shape of graph at the neutral and alkaline condition but displayed the different response at acidic condition. The P1 and P3 showed the same optimum pH at 8.0, whereas P2 exhibited the optimum pH at 7.0. More than 85% of enzymatic activity was observed at pH 7.0 to 9.0 (Fig. 4E). The result indicated that amylases from both O. hasselti and P. proctozystron had a broad pH range. In contrast, all three isoforms of H. dispar showed a narrow peak of enzyme activity. The enzymes worked properly at pH 7.0. Afterward, the activity slightly dropped at both alkaline and acidic conditions. At pH 6.0 to 9.0, the activity reduced to nearly 60%. However, more than 40% of enzymatic activity was observed in all three isoforms at acidic condition particularly pH 5.0 (Fig. 4C), while only some isoforms of O. hasselti (O2) and P. proctozystron (P2 and P3) were detected. Six isoforms of amylases from both H. dispar and P. proctozystron displayed the same optimum temperatures at 50 °C (Fig. 4D, F) similar to O1 from O. hasselti, whereas O2 revealed the optimum temperature at 40 °C (Fig. 4B). Activities of almost enzymes were significantly declined after temperature above 50 °C except P3 whose activity remained more than 50%.
Effects of pH and temperatures on amylase activities. Amylases from O. hasselti (A, B), H. dispar (C, D) and P. proctozystron (E, F). Reactions were performed in duplicates, and mean values were presented. The maximum activity was defined as 100% of relative activity. (–o–), O1; (–o–), O2; (–◊–), UB; (–o–), H1; (–∆–), H2; (–●–), P1; (–■–), P2 and (–▲–), P3
Thermal stability of enzymes
The stabilities of the purified enzymes were determined at both room temperature (27 °C) and the optimum temperature (50 °C). Figure 5 shows the residual activities of all purified amylases after incubation of enzymes at both temperatures for 1–3 and 24 h. Interestingly, the activities of the amylases from O. hasselti and H. dispar increased up to nearly 150% of initial activity especially O2 and H1 after incubation at room temperature for 1–2 h. Afterward, the activities reduced to around 100% upon incubation for 3 and 24 h suggesting that all isoforms of both species were highly stable (Fig. 5A, C). Moreover, high stability was also observed in P. proctozystron. The relative activities of all three isoforms were constant at 100% until 3 h of incubation, and their activities were then reduced to 93% for P1 and P3 and 69% for P2 after 24 h (Fig. 5E). In contrast, the residual activities of all amylases detected at optimal temperature were lower than those detected at room temperature. The activities of H. dispar amylases continuously decreased with less than 20% of remaining activity after 24 of incubation (Fig. 5D), while the enzymes from O. hasselti were stable at the first 2 h of incubation with the remaining activities about 100% and rapidly decreased after 3 to 24 h (Fig. 5B). However, the amylases from P. proctozystron showed high stability with residual activity more than 80% after incubation for 1–3 h before suddenly declining to less than 20% at 24 h (Fig. 5F).
Effects of temperatures on the stabilities of amylases detected at room temperature (27 °C) (A, C, E) and at optimum temperatures (at 40 °C for O2 and at 50 °C for the rest) (B, D, F). The enzymatic activities of amylases from O. hasselti (A, B), H. dispar (C, D), and P. proctozystron (E, F) were measured after pre-incubation at room temperature and optimum temperatures for 1, 2, 3, and 24 h. The enzymatic activity without pre-incubation (0 h) was defined as 100% of relative activity. Reactions were performed in duplicates, and mean values were presented
Effects of various metal ions, detergent, and chelating agents on amylase activities
In order to determine the effects of metal ions, detergent, and chelating agents on enzyme activity, the purified amylases were pre-incubated in the presence of these reagents, and their effects on enzyme activity were then investigated. Not only different species but also different isoforms showed the different responses in the presence of different ions. Na+ ion significantly activated activities of O1, O2, H2, P1, and P3 similar to Ca2+ ion, which increased the activities of O1 and O2 at 2 mM, UB, P1, and P3 especially UB amylase, whose relative activity increased up to 120% as shown in Fig. 6. Mg2+ and Mn2+ slightly inhibited the activities of all enzymes. In addition, SDS and EDTA dramatically decreased all enzyme activities. The presence of EDTA significantly influenced the O1 activity with the remaining relative activity about 5%, while O2 activity totally disappeared. Interestingly, the remaining of more than 50% of P1 and P3 activities observed in the presence of 2 and 5 mM of SDS indicated the resistance of P1 and P3 to SDS, the protein denaturant. The activation of the enzyme activities in the presence of Ca2+ and the inhibition of the activities in the presence of EDTA, the divalent chelating agent, suggested that the enzymes might be a metalloenzyme and were classified as the Ca2+-dependent manner.
Quantitative analysis of calcium ion
To verify the presence of Ca2+ ion in the purified amylases, the enzyme samples were subjected to atomic absorption spectrophotometry (AAS). Detection was defined as triplicates, and mean values were presented. The level of metal ion was calculated by comparing it with the calibration curve. The equation of the calibration graph of Ca2+ was y = 0.0604x + 0.005 with a correlation coefficient of 0.9925. The results showed that Ca2+ contents were found in the amylases O1 and O2 as 0.05 and 1.35 ppm per mg protein, respectively. The amylases UB, H1, and H2 displayed significant Ca2+ amounts of 13.53, 2.83, and 0.15 ppm per mg protein, and the amylases P1, P2, and P3 revealed 0.02, 0.15, and 0.08 ppm per mg protein, respectively.
Selection of amylases for use as enzyme molecular weight markers for substrate zymography
After purification, the purity and the approximate molecular weights of resulting purified amylases were initially verified and tested by using SDS-PAGE and zymography. Movement pattern of the purified amylases on zymogram gel showed that amylase P1, P2, and P3 revealed three to six isoforms, which some are close in molecular weights that may not be suitable for use as standard molecular weight enzymes. Comparison with amylases from H. dispar, all three fractions showed clear bands of each single isoform with different molecular weights. However, H1 and H2 amylases were similar in sizes as displayed on zymogram gel. The smaller one (H2) was selected due to the sharpness of the clear band. For the amylase O1, the enzyme exhibited the clear band and higher amylolytic activity than O2; thus, the O1 amylase was chosen. Ultimately, the H2, O1, and UB amylases revealed different molecular weights were selected and applied to use as enzyme molecular weight markers. The molecular weights of UB, O1, and H2 amylases analyzed from gel filtration chromatography were 64, 49, and 25 kDa, respectively, as shown in Fig. 7.
SDS gel staining with Coomassie brilliant blue R-250 (left) and zymogram (right) of amylases selected to apply use as the enzyme molecular weight markers for detection and estimation of molecular weights of amylases on zymographic gel. Lane M, unstained protein molecular weight markers (kDa) (Thermo Scientific). Lanes H2, O1, and UB, the selected purified amylases; lane P, pooled fraction of the purified H2, O1, and UB amylases. Black arrows indicated the positions of three amylases. The 8 µg of total protein from each fraction was loaded on CBB gel with 33, 120, and 65 mU of H2, O1, and UB in zymogram, respectively. Pooled fraction (P) contained 24 µg total protein with 218 mU of enzyme mixture. Both CBB gel and zymogram were a full-length gel cropped from the beginning of separating layer on top to the bottom edge of gels. The dye front as seen in a blue line was still observed above the bottom edge of CBB gels. More detail of figures was described in supplementary information
Amylase identification
LC/MS–MS was carried out to confirm the authentic band of the partially purified enzymes. If the protein Mascot scores are greater than 37 indicating that protein is significantly matched with the protein databases with p < 0.05 and a summary result of selected purified amylases was presented in Table 2. The MS analysis confirmed that the protein band of O1 pointed as a black arrow in Fig. 7 matched to the pancreatic alpha-amylase of Struthio camelus (Oosthuizen et al. 1994) and alpha-amylase 2B of Homo sapiens (Yokouchi et al. 1990) with a Mascot score of 89. The selected band of UB amylase matched to pancreatic alpha-amylase, alpha-amylase 1, and alpha-amylase 2B of Rattus norvegicus (MacDonald et al. 1980), Mus musculus (Linnenbrink et al. 2020), Struthio camelus, and Homo sapiens with a Mascot score of 108, whereas the H2 amylase from H. dispar revealed no protein matching in database suggesting that there is no previous report of amylase sequence of H. dispar. The LC–MS/MS analysis was repeated for H2, but the same result was obtained with no protein matching detection. Finally, the results confirmed that the protein bands of O1 and UB were verified as amylase enzymes containing conserved sequence with other organisms.
Stability of enzyme molecular weight markers
As mentioned before, the purified amylases with different molecular weights selected from three fish species were applied to use as enzyme standard markers for zymography. Therefore, UB, O1, and H2 amylases with molecular weights of 64, 49, and 25 kDa, respectively, were selected. Figure 8 shows the SDS gel and zymogram displaying the protein bands and clear bands of amylases. During storage at − 20 °C for 12 weeks, amylase mixture (UB, O1, and H2) dissolved in sample buffers were thawed and aliquoted to analyze with electrophoresis every 2 weeks. After five freeze-thawed cycles, the intensity of blue bands and the amount of protein bands observed on CBB gel were similar to those before storage. No protein degradation was observed. Furthermore, the intensity and sharpness of clear bands on zymographic gel were also similar to the sample before the freezing and thawing process. This result indicated that the purified amylase mixtures were highly stable and were able to be used as enzyme molecular weight markers for both SDS-PAGE and zymography.
SDS gels staining with Coomassie brilliant blue R-250 (left panel) and zymogram (right panel) of the amylase molecular weight markers after storage at − 20 °C. The enzyme markers containing 24 µg total protein with 218 mU enzymes mixture were freeze-thawed and aliquoted to run check on SDS-PAGE upon storage for 2–12 weeks. Lane 1, purified amylase markers before storage at − 20 °C; lanes 2–6, purified enzyme markers after storage at − 20 °C for 2, 4, 6, 8, and 12 weeks, respectively. Gels were separately run every 2 weeks of freeze-thawed processes. In the same week, both CBB gel and zymogram were performed in different gels with the same enzyme mixture samples. Both gels were tested in parallel to monitor the stability of amylases. More detail of figures was described in supplementary information
Discussion
Characterization of amylases has been reported in many organisms including bacteria (Shofiyah et al. 2020; Dash et al. 2015), fungi (Karim et al. 2018; Abdulaal 2018), animals (MacDonald et al. 1980; Hagenbüchle et al. 1980; Oosthuizen et al. 1994; Linnenbrink et al. 2020), and plants (Stanley et al. 2005; Rogers 1985; Bertoft et al. 1984). Different sources of enzymes have revealed both difference and similarity of enzyme properties such as the optimum pH, optimum temperature, pH and thermal stabilities, effects of ions, chemicals, and detergents on enzymatic activity. Different features of enzymes are useful for different and various applications, i.e., therapeutic uses, food and beverage industries, detergent, and biofuel. Many enzymes have been used in industrial processes such as protease, lipase, xylanase, and cellulase. Amylases are also one of the main enzymes used in important industries such as conversion of starch to glucose in food and beverage industries and stain removal in detergent processing. However, application of amylase, especially fish amylases, as enzyme molecular weight markers for zymography has not been reported.
Amylase activities have been characterized by using crude enzymes or purified enzymes. The extraction and purification of native proteins often result in low yields. Therefore, high total protein as well as enzyme content of specimens is required to gain more and enough enzymes for purification. In contrast, microbial enzymes and recombinant enzymes exhibit high amounts of total protein upon extraction and high yield of purification due to the rapid growth of microorganisms and the overexpression of recombinant proteins. For certain reasons, we got a low yield after purification that might be due to low enzyme content in digestive tracts of fishes or not enough digestive tracts, which led to low protein content in extracted samples. In addition, the presence of several isoforms of amylases in fish is sometimes difficult to isolate or may get lost during purification that leads to the low yield of purification of each isoform. Most amylases from microorganisms have been reported in single isoform (Shofiyah et al. 2020; Abdulaal 2018), while amylases in shrimp (Coccia et al. 2011) and fishes (Fernández et al. 2001; Bai et al. 2012; Ji et al. 2012) have been found in several isoforms with different molecular weights. The enzymes have been characterized (Al-Tameemi et al. 2010; Champasri and Champasri 2017; Fernández et al. 2001; Ji et al. 2012; Champasri et al. 2021; Bai et al. 2012). However, the purification of fish amylase has been rarely presented, and the application as enzyme molecular weight markers for zymographic gel has not been found yet.
The recovery yields of all amylases in this study were between 0.17 and 0.68%, which were lower than those reported in bacteria such as Dociostaurus maroccanus (Rafiei et al. 2016) and Corynebacterium alkanolyticum ATH3 (Banerjee et al. 2016), whose recovery yields are 10.78% and 17.55%, respectively. However, the purification yields obtained in this study were higher than those reported in Escherichia coli (Hassan et al. 2018) (0.093%). Besides the low amount of native proteins in digestive tracts of fish, the presence of several isoenzymes of fish digestive enzymes was another reason to get low yield of purification of each isoform. The characterizations of amylase have been previously reported with optimum pH at 8.0 in seven cyprinid fishes (Champasri and Champasri 2017), thick-lipped gray mullet (Pujante et al. 2017), and Italian locust (Darvishzadeh and Bandani 2012). The optimum temperatures of amylases have been found at 40 °C in the Italian locust (Darvishzadeh and Bandani 2012) and 45 to 55 °C in the cyprinid fishes (Champasri and Champasri 2017). The same optimum pH may be due to the enzymes sharing the conserved amino acids and exposing the similar shape at the active site upon ionization.
Our previous studies have shown that different freshwater fish species exhibit different amylase activities and different isoenzyme patterns. High enzymatic activity also displays the strong clear bands on zymograms (Champasri and Champasri 2017; Champasri et al. 2021). In this study, the isoenzymes of amylase with different molecular weights were purified. The result showed that amylase from H. dispar (carnivore) was separated into two isoforms, while those from O. hasselti (herbivore) and P. proctozystron (omnivore) were separated into three isoforms. The presence of several numbers and different sizes of isoenzymes and high activity in a wide pH and temperature ranges particularly P. proctozystron and O. hasselti amylases might due to the ability to digest different kinds of food, different feeding behavior and biochemical composition of food (Fernández et al. 2001; Al-Tameemi et al. 2010), or adaptation for survival from pH, temperature, water environmental, or climate changes. Two, three, and four isoforms of crude amylases have been reported in paddlefish, bighead carp, and hybrid sturgeon with the molecular weights of 156.3 and 129.9, 111.2–74.2, and 156.3–116.2 kDa, respectively (Ji et al. 2012). Two isoforms were also detected in all wild fish (Pagrus pagrus, Pagellus erythrinus, P. bogaraveo, Boops boops, and Diplodus annularis (Fernández et al. 2001). The estimated molecular weights were around 135 and 175 kDa compared to those of Oreochromis niloticus reported by Champasri et al. (2021). Noticeably, the molecular sizes of freshwater fish amylases were smaller than those reported in marine fishes. The examples of certain species were Ompok bimaculatus, Kryptopterus geminus, Hemibagrus spilopterus, and Puntius gonionotus. The molecular weights displayed in the range of 35–95 kDa (Champasri et al. 2021). The molecular weights of the purified amylases reported here were between 12 and 147 kDa, in the range of those produced from Monascus sanguineus (Tallapragada et al. 2017) and Aspergillus flavus NSH9 (Karim et al. 2018) whose molecular weights have been reported as 56 and 54 kDa, respectively. The limitation of our study is the low number of enzyme candidates with a wide range of molecular weights. Only three amylases (UB, O1, H2) with 64, 49, and 25 kDa were selected. Other isoforms were not chosen due to the impurity, the blurred clear bands, and the close molecular weights of the selected one. However, our results showed the high stability of the purified enzymes after storage in sample buffers at − 20 °C and several freeze-thawed cycles. Moreover, all amylases showed high stabilities especially at room temperature (27 °C). Almost enzymes except P2 displayed the remaining activities nearly 100% upon incubation for 24 h, which were higher than those reported in Bacillus subtilis BI19 (Dash et al. 2015), miswak amylases (Mohamed et al. 2014) (A1, A4a, A4b, A5a, and A5b), and sparid fishes (Fernández et al. 2001) (Boops boops, Pagellus erythrinus, and Pagellus bogaraveo) with the residue activities less than 40% after 1 h of incubation at 50 °C.
α-Amylases have been reported as Ca2+-dependent or Ca2+-independent enzymes. The Ca2+-dependent amylases contain Ca2+ in the enzyme structures. The presence of Ca2+ ion helps to maintain enzyme structure in the correct conformation (Normurodova et al. 2007; Yin et al. 2017; Bush et al. 1989; Larson et al. 1994; Buisson et al. 1987), prevent the thermal inactivation (Goyal et al. 2005; Khajeh et al. 2001), and influence the thermostability of the enzymes (Liao et al. 2019). However, some α-amylases are Ca2+ independence (Hmidet et al. 2008; Asoodeh et al. 2010; Sharma and Satyanarayana 2010), and the activities are inhibited by Ca2+ ion (Tanaka and Hoshino 2003; Mehta and Satyanarayana 2013). From our result, the activities of almost purified amylases were accelerated by Ca2+ ion, which correspond to amylases reported in barley malt (Bertoft et al. 1984), Pichia pastoris (Sun et al. 2018), wild Farfantepenaeus subtilis, Litopenaeus schmitti, Litopenaeus vannamei (Castro et al. 2012), and thick-lipped gray mullet fish (Pujante et al. 2017). Furthermore, the presence of Ca2+ ions in the purified enzymes was confirmed by an atomic absorption spectrophotometer. The strong inhibition of the enzymatic activities by EDTA (the divalent cation chelating agent) suggests that our purified amylases are Ca2+ dependent. Our results suggested that the purified amylases from digestive tracts of fishes were metalloenzymes which contained calcium ions in their structure. Satisfyingly, the enzyme molecular weight markers showed high stability with sharp bands on zymogram, and no protein degradation was observed on CBB gels. Moreover, the investigation of enzyme stability at − 20 °C in the presence of sample buffer and after several freeze-thawing cycles of fish amylases was firstly reported here. This study provides new insight into the application of the purified stable amylase as the enzyme molecular weight markers for zymography.
Conclusions
After purification with ammonium sulfate precipitation, ion-exchange, and size exclusion chromatography, the amylase enzymes were separated into two isoforms of O. hasselti (O1, O2) and three isoforms of both H. dispar (UB, H1, H2) and P. proctozystron (P1, P2, P3) with different folds and recovery yields of purification. All isoenzymes were characterized and determined their molecular weights. The optimum pH values displayed at 7.0 and 8.0, while the optimum temperatures revealed at 40 and 50 °C. The different shapes or patterns of graphs upon response to different pH and temperatures were observed in different isoenzymes. Studies of the influences of metal ions, SDS, and EDTA showed that almost all isoenzyme activities were activated by NaCl and CaCl2, whereas EDTA and SDS strongly inhibited all enzymatic activities. Verification with an atomic absorption spectrophotometry exhibited the presence of Ca2+ ions in the range of 0.02–13.53 ppm per mg protein indicating that amylases are Ca2+ dependent. Molecular weight analysis revealed 12 to 147 kDa of amylases. In order to investigate the application of the purified fish amylase as enzyme molecular weight markers for zymography, the UB, O1, and H2 amylases with appropriate molecular masses of 64, 49, and 25 kDa were selected. Validation with LC–MS/MS showed that O1 matched the pancreatic alpha-amylase of Struthio camelus and alpha-amylase 2B of Homo sapiens with a Mascot score of 89. UB amylase matched to pancreatic alpha-amylase, alpha-amylase 1, and alpha-amylase 2B of Rattus norvegicus, Mus musculus, Struthio camelus, and Homo sapiens with a Mascot score of 108, whereas the H2 amylase revealed no protein matching in database. The three selected amylases revealed high stability in the presence of a sample buffer after five cycles of freeze-thawing process upon storage at − 20 °C for 12 weeks. No protein degradation was observed on polyacrylamide gel, and the enzymes still displayed sharp and clear bands on zymograms.
Data availability
All data included in this study are available upon request by contact with the corresponding author. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (Perez-Riverol et al. 2022) partner repository. The datasets generated and analyzed during the current study are available in PRIDE database (PRIDE—Proteomics Identification Database (ebi.ac.uk) via ProteomeXchange (www.proteomexchange.org) repository with accession number or identifier PXD033432 and PXD033433.
References
Abdulaal WH (2018) Purification and characterization of α-amylase from Trichoderma pseudokoningii. BMC Biochem 19:4. https://doi.org/10.1186/s12858-018-0094-8
Al-Tameemi R, Aldubaikul A, Salman NA (2010) Comparative study of α-amylase activity in three Cyprinid species of different feeding habits from Southern Iraq. Turkish J Fish Aquat Sci 10(3):411–414. https://doi.org/10.4194/trjfas.2010.0315
Asatsuma S, Sawada C, Itoh K, Okito M, Kitajima A, Mitsui T (2005) Involvement of α-amylase I-1 in starch degradation in rice chloroplasts. Plant Cell Physiol 46(6):858–869. https://doi.org/10.1093/pcp/pci091
Asoodeh A, Chamani J, Lagzian M (2010) A novel thermostable, acidophilic alpha-amylase from a new thermophilic “Bacillus sp. Ferdowsicous” isolated from Ferdows hot mineral spring in Iran: purification and biochemical characterization. Int J Biol Macromol 46(3):289–297. https://doi.org/10.1016/j.ijbiomac.2010.01.013
Bai Y, Huang H, Meng K, Shi P, Yang P, Luo H, Luo C, Feng Y, Zhang W, Yao B (2012) Identification of an acidic α-amylase from Alicyclobacillus sp. A4 and assessment of its application in the starch industry. Food Chem 131(4):1473–1478. https://doi.org/10.1016/j.foodchem.2011.10.036
Banerjee G, Mukherjee S, Bhattacharya S, Ray AK (2016) Purification and characterization of extracellular protease and amylase produced by the bacterial strain, Corynebacterium alkanolyticum ATH3 isolated from fish gut. Arab J Sci Eng 41:9–16. https://doi.org/10.1007/s13369-015-1809-4
Bertoft E, Andtfolk C, Kulp SE (1984) Effect of pH and temperature, and calcium ions on barley malt α-amylase isoenzymes. J Inst Brew 90(5):298–302. https://doi.org/10.1002/j.2050-0416.1984.tb04278.x
Bradford MM (1976) A rapid and sensitive method for the quantitation microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254. https://doi.org/10.1016/0003-2697(76)90527-3
Buisson G, Duee E, Haser R, Payan F (1987) Three-dimensional structure of porcine pancreatic alpha-amylase at 2.9 Å resolution. Role of calcium in structure and activity. EMBO J 6(13):3909–3916. https://doi.org/10.1002/j.1460-2075.1987.tb02731.x
Bush DS, Sticher L, van Huystee R, Wagner D, Jones RL (1989) The calcium requirement for stability and enzymatic activity of two isoforms of barley aleurone alpha-amylase. J Biol Chem 264(32):19392–19398. https://doi.org/10.1016/S0021-9258(19)47314-6
Cano-Ramírez C, Santiago-Hernández A, Rivera-Orduña FN, Pineda-Mendoza RM, Zúñiga G, Hidalgo-Lara ME (2017) One-step zymogram method for the simultaneous detection of cellulase/xylanase activity and molecular weight estimation of the enzyme. Electrophoresis 38(3–4):447–451. https://doi.org/10.1002/elps.201600347
Castro PF, Freitas ACV, Santana WM, Costa HMS, Carvalho LB, Bezerra RS (2012) Comparative study of amylase from the midgut gland of three species of penaeid shrimp. J Crust Biol 32(4):607–613. https://doi.org/10.1163/193724012X634206
Champasri C, Champasri T (2017) Biochemical characterization, activity comparison and isoenzyme analysis of amylase and alkaline proteases in seven Cyprinid fishes. J Fish Aquat Sci 12(6):264–272. https://doi.org/10.3923/jfas.2017.264.272
Champasri C, Champasri T, Woranam K (2015) Purification, biochemical characterization of a Macrotermes gilvus cellulase and zymogram analysis. Asian J Biochem 10(5):190–204. https://doi.org/10.3923/ajb.2015.190.204
Champasri C, Phetlum S, Pornchoo C (2021) Diverse activities and biochemical properties of amylase and proteases from six freshwater fish species. Sci Rep 11:5727. https://doi.org/10.1038/s41598-021-85258-7
Coccia E, Varricchio E, Paolucci M (2011) Digestive enzymes in the crayfish Cherax albidus: polymorphism and partial characterization. Int J Zool. https://doi.org/10.1155/2011/310371
Darvishzadeh A, Bandani AR (2012) Identification and characterization of α-amylase in the Italian locust, Calliptamus italicus (Linnaeus, 1758) (Orthoptera: Acrididae). Mun Ent Zool 7:1028–1034
Dash BK, Rahman MM, Sarker PK (2015) Molecular identification of a newly isolated Bacillus subtilis BI19 and optimization of production conditions for enhanced production of extracellular amylase. Biomed Res Int. https://doi.org/10.1155/2015/859805
Fernández I, Moyano FJ, Díaz M, Martínez T (2001) Characterization of α-amylase activity in five species of Mediterranean sparid fishes (sparidae, teleostei). J Exp Mar Bio Ecol 262(1):1–12. https://doi.org/10.1016/S0022-0981(01)00228-3
Gioda CR, Pretto A, de Souza Freitas C, Leitemperger J, Loro VL, Lazzari R, Lissner LA, Baldisserotto B, Salbego J (2017) Different feeding habits influence the activity of digestive enzymes in freshwater fish. Cienc Rural 47(3):1-7. https://doi.org/10.1590/0103-8478cr20160113
Goyal N, Gupta JK, Soni SK (2005) A novel raw starch digesting thermostable α-amylase from Bacillus sp. I-3 and its use in the direct hydrolysis of raw potato starch. Enzyme Microb Technol 37(7):723–734. https://doi.org/10.1016/j.enzmictec.2005.04.017
Hagenbüchle O, Bovey R, Young RA (1980) Tissue-specific expression of mouse-alpha-amylase genes: nucleotide sequence of isoenzyme mRNAs from pancreas and salivary gland. Cell 21(1):179–187. https://doi.org/10.1016/0092-8674(80)90125-7
Hassan BA, Alsalami NY, Jebor MA (2018) Amylase production, purification and characterization from Escherichia coli. J Pharm Sci Res 10(7):1691–1696
Hmidet N, Bayoudh A, Berrin JG, Kanoun S, Juge N, Nasri M (2008) Purification and biochemical characterization of a novel α-amylase from Bacillus licheniformis NH1: Cloning, nucleotide sequence and expression of amyN gene in Escherichia coli. Process Biochem 43(5):499–510. https://doi.org/10.1016/j.procbio.2008.01.017
Ji H, Sun HT, Xiong DM (2012) Studies on activity, distribution, and zymogram of protease, α-amylase, and lipase in the paddlefish Polyodon spathula. Fish Physiol Biochem 38:603–613. https://doi.org/10.1007/s10695-011-9541-9
Jones GT (2014) Chapter seven - matrix metalloproteinases in biologic samples. Adv Clin Chem 65:199–219. https://doi.org/10.1016/B978-0-12-800141-7.00007-3
Kaplan F, Guy CL (2004) β-Amylase induction and the protective role of maltose during temperature shock. Plant Physiol 135(3):1674–1684. https://doi.org/10.1104/pp.104.040808
Karim KMR, Husaini A, Sing NN, Sinang FM, Roslan HA, Hussain H (2018) Purification of an alpha amylase from Aspergillus flavus NSH9 and molecular characterization of its nucleotide gene sequence. 3 Biotech 8(204):1–14. https://doi.org/10.1007/s13205-018-1225-z
Khajeh K, Ranjbar B, Naderi-Manesh H, Ebrahim Habibi A, Nemat-Gorgani M (2001) Chemical modification of bacterial alpha-amylases: changes in tertiary structures and the effect of additional calcium. Biochim Biophys Acta 1548(2):229–237. https://doi.org/10.1016/s0167-4838(01)00236-9
Larson SB, Greenwood A, Cascio D, Day J, McPherson A (1994) Refined molecular structure of pig pancreatic α-amylase at 2.1 Å resolution. J Mol Biol. 235(5):1560–1584. https://doi.org/10.1006/jmbi.1994.1107
L-hui S, Qin T, Liu Y, Zhao H, Xia X, Lei X (2018) Cloning, expression, and characterization of a porcine pancreatic α-amylase in Pichia pastoris. Anim Nutr 4(2):234–240. https://doi.org/10.1016/j.aninu.2017.11.004
Linnenbrink M, Ullrich KK, McConnell E, Tautz D (2020) The amylase gene cluster in house mice (Mus musculus) was subject to repeated introgression including the rescue of a pseudogene. BMC Evol Biol 20:56. https://doi.org/10.1186/s12862-020-01624-5
Liao SM, Liang G, Zhu J, Lu B, Peng LX, Wang QY, Wei YT, Zhou GP, Huang RB (2019) Influence of calcium ions on the thermal characteristics of α-amylase from Thermophilic anoxybacillus sp. GXS-BL Protein Pept Lett 6(2):148–157. https://doi.org/10.2174/0929866526666190116162958
MacDonald RJ, Crerar MM, Swain WF, Pictet RL, Thomas G, Rutter WJ (1980) Structure of a family of rat amylase genes. Nature 287(5778):117–122. https://doi.org/10.1038/287117a0
Mehta D, Satyanarayana T (2013) Biochemical and molecular characterization of recombinant acidic and thermostable raw-starch hydrolysing α-amylase from an extreme thermophile Geobacillus thermoleovorans. J Mol Catal B Enzym 85–86:229–238. https://doi.org/10.1016/j.molcatb.2012.08.017
Mohamed SA, Almulaiky YQ, Ahmed YM, Al-Bar OAM, Ibrahim IH (2014) Purification and characterization of α-amylase from miswak Salvadora persica. BMC Complement Altern Med 14(1):1–10. https://doi.org/10.1186/1472-6882-14-119
Miller GL (1959) Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal Chem 31(3):426–428. https://doi.org/10.1021/ac60147a030
Normurodova KT, Nurmatov SK, Alimova BK, Pulatova OM, Akhmedova ZR, Makhsumkhanov AA (2007) Isolation and characteristics of highly active α-amylase from Bacillus subtilis-150. Chem Nat Compd 43(4):454–457. https://doi.org/10.1007/s10600-007-0159-1
Oosthuizen V, Naude RJ, Oelofsen W, Muramoto K, Kamiya H (1994) Ostrich pancreatic alpha-amylase: kinetic properties, amino terminal sequence and subsite structure. Int J Biochem 26(10–11):1313–1321. https://doi.org/10.1016/0020-711X(94)90101-5
Perez-Riverol Y, Bai J, Bandla C, Hewapathirana S, García-Seisdedos D, Kamatchinathan S, Kundu D, Prakash A, Frericks-Zipper A, Eisenacher M, Walzer M, Wang S, Brazma A, Vizcaíno JA (2022) The PRIDE database resources in 2022: a hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res 50(D1):D543–D552. https://doi.org/10.1093/nar/gkab1038
Pujante IM, Díaz-López M, Mancera JM, Moyano FJ (2017) Characterization of digestive enzymes protease and alpha-amylase activities in the thick-lipped grey mullet (Chelon labrosus, Risso 1827). Aquac Res 48(2):367–376. https://doi.org/10.1111/are.13038
Rafiei B, Ghadamyari M, Imani S, Hosseininaveh V, Ahadiyat A (2016) Purification and characterization of α -amylase in Moroccan locust, Dociostaurus maroccanus Thunberg (Orthoptera: Acrididae) and its inhibition by inhibitors from Phaseolus vulgaris L. Toxin Rev 35(3–4):90–97. https://doi.org/10.1080/15569543.2016.1207190
Rogers JC (1985) Conserved amino acid sequence domain in alpha amylase from plant, mammal and bacteria. Biochem Biophys Res Commun 128:470–476. https://doi.org/10.1016/0006-291X(85)91702-4
Saini R, Saini HS, Dahiya A (2017) Amylases: characteristics and industrial applications. J Pharmacogn Phytochem 6(4):1865–1871
Sharma A, Satyanarayana T (2010) High maltose-forming, Ca2+-independent and acid stable α-amylase from a novel acidophilic bacterium. Bacillus Acidicola Biotechnol Lett 32(10):1503–1507. https://doi.org/10.1007/s10529-010-0322-9
Shofiyah SS, Yuliani D, Widya N, Sarian FD, Puspasari F, Radjasa OK, Ihsanawati I, Natalia D (2020) Isolation, expression, and characterization of raw starch degrading α-amylase from a marine lake Bacillus megaterium NL3. Heliyon 6(12):e05796. https://doi.org/10.1007/s10529-010-0322-9
Sodhi HK, Sharma K, Gupta JK, Soni SK (2005) Production of a thermostable α-amylase from Bacillus sp. PS-7 by solid state fermentation and its synergistic use in the hydrolysis of malt starch for alcohol production. Process Biochem 40(2):525–534. https://doi.org/10.1016/j.procbio.2003.10.008
Stanley D, Farnden KJF, MacRae EA (2005) Plant α-amylases: functions and roles in carbohydrate metabolism. Biologia 16(6):65–71 (Bratisl)
Svensson B (1994) Protein engineering in the α-amylase family: catalytic mechanism, substrate specificity, and stability. Plant Mol Biol 25:141–157. https://doi.org/10.1007/BF00023233
Tallapragada P, Dikshit R, Jadhav A, Sarah U (2017) Partial purification and characterization of amylase enzyme under solid state fermentation from Monascus sanguineus. J Genet Eng Biotechnol 15(1):95–101. https://doi.org/10.1016/j.jgeb.2017.02.003
Tanaka A, Hoshino E (2003) Secondary calcium-binding parameter of Bacillus amyloliquefaciens α-amylase obtained from inhibition kinetics. J Biosci Bioeng 96(3):262–267. https://doi.org/10.1016/s1389-1723(03)80191-3
Van de Louw A, Jean D, Frisdal E, Cerf C, d’Ortho MP, Baker AH, Lafuma C, Duvaldestin P, Harf A, Delclaux C (2002) Neutrophil proteinases in hydrochloric acid- and endotoxin-induced acute lung injury: evaluation of interstitial protease activity by in situ zymography. Lab Invest 82(2):133–145. https://doi.org/10.1038/labinvest.3780406
Vandooren J, Geurts N, Martens E, Van den Steen PE, Opdenakker G (2013) Zymography methods for visualizing hydrolytic enzymes. Nat Methods 10:211–220. https://doi.org/10.1038/nmeth.2371
Wong JEMM, Blaise M (2020) Report of false positives when using zymography to assess peptidoglycan hydrolytic activity of an endopeptidase with multiple LysM domains. Biochimie 177:25–29. https://doi.org/10.1016/j.biochi.2020.07.014
Wu X, Wang Y, Tong B, Chen X, Chen J (2018) Purification and biochemical characterization of a thermostable and acid-stable alpha-amylase from Bacillus licheniformis B4–423. Int J Biol Macromol 109:329–337. https://doi.org/10.1016/j.ijbiomac.2017.12.004
Xian L, Wang F, Luo X, Feng YL, Feng JX (2015) Purification and characterization of a highly efficient calcium-independent α-amylase from Talaromyces pinophilus 1–95. PLoS ONE 10(3):e0121531. https://doi.org/10.1371/journal.pone.0121531
Yin H, Yang Z, Nie X, Li S, Sun X, Gao C, Wang Z, Zhou G, Xu P, Yang C (2017) Functional and cooperative stabilization of a two-metal (Ca, Zn) center in α-amylase derived from Flavobacteriaceae species. Sci Rep 7(1):1–8. https://doi.org/10.1038/s41598-017-18085-4
Yokouchi H, Horii A, Emi M, Tomita N, Doi S, Ogawa M, Mori T, Matsubara K (1990) Cloning and characterization of a third type of human alpha-amylase gene, AMY2B. Gene 90(2):281–286. https://doi.org/10.1016/0378-1119(90)90191-s
Funding
This research was supported by the research capability enhancement program through graduate student scholarship, Faculty of Science, Khon Kaen University.
Author information
Authors and Affiliations
Contributions
Suthathip Phetlum performed the experiments and wrote the first draft of the manuscript. Chamaiporn Champasri acquired the funding, prepared materials and methods, wrote the final draft, revised and edited the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethical approval
Experiments performed under the ethical principles for use and care of animals in science are approved by the Institutional Animal Care and Use Committee of Khon Kaen University (IACUC KKU). All experimental protocols and the care and use of experimental animals complied with animal welfare laws of Thailand, and guidelines and policies approved by ThaiIACUC (permit reference number U1-04,584-d2559). Fish collection and digestive tract preparation were done to minimize fish suffering.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Phetlum, S., Champasri, C. Purification and characterization of amylases from three freshwater fish species providing new insight application as enzyme molecular markers for zymography. Fish Physiol Biochem 49, 1257–1276 (2023). https://doi.org/10.1007/s10695-023-01255-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10695-023-01255-9





 ,
,
 ,
,
 , amylase activity detected at 540 nm; –-, NaCl gradient)
, amylase activity detected at 540 nm; –-, NaCl gradient)
 ,
,
 ,
,
 , amylase activity detected at 540 nm)
, amylase activity detected at 540 nm)




