Abstract
Perilipin family is the main structural proteins of lipid droplet (LD) that is intracellular neutral lipid store ponds, and regulates LD assembly and formation, and is crucial for lipid metabolism. Here three paralogs of perilipin family were characterized from grass carp and their complete coding sequences (CDS) were obtained, including perilipin1, perilipin2, and perilipin3, coding peptides of 492, 454, and 419 amino acids, respectively. The alignment of the homology of grass carp perilipin deduced amino acid sequences with other teleost species showed that the homology with mammalian was less than 55%. PAT (perilipin) domain in mammalian was also predicted in grass carp perilipin 1–3 proteins. Genomic organization analysis revealed that grass carp perilipin1 contained 6 coding exons, while both perilipin2 and perilipin3 consisted of 7 coding exons. The mRNA encoding three paralogs were expressed in a wide range of tissues; perilipin1–3 were primarily expressed in adipose tissue and liver; besides, perilipin3 was also highly expressed in the heart. In vitro, 200 μM DHA increased the proportion of smaller lipid droplets effectively in fully differentiated adipocytes of grass carp. The mRNA expression of perilipin1, perilipin2, and perilipin3 was significantly increased in the adipocytes treated with DHA (P < 0.05, P < 0.01). The same responses of different paralogs in the adipocytes during DHA treatment suggest that they might play synergistic roles in the formation of LDs.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Disturbances in lipid metabolism and imbalance between energy intake and energy expenditure spark a range of metabolic syndrome, such as systemic insulin resistance, inflammation, obesity, and diabetes (Ozcan et al. 2004; Esser et al. 2014; Cotillard et al. 2014). Adipose tissue, having the highlighted plasticity, facilitates energy storage and influences aspects of metabolic disease risk, and is also participated in the regulation of both glucose and energy homeostasis (Meshkani and Adeli 2009; Wozniak et al. 2009; Hajer et al. 2008; Flachs et al. 2013). Adipocytes are cells specialized for storage of neutral lipids in lipid droplets, mainly triacylglycerol (TAG). Lipid droplets (LDs) can provide a rapidly mobilized lipid source for many important biological processes and regulate cellular energy balance (Murphy et al. 2009). Therefore, understanding the function and structure of lipid droplets in adipocytes is essential to the study of lipid metabolism.
LDs are omnipresent that can be found in almost all eukaryotic cell (yeast, C. elegans, drosophila, zebrafish, algae, plants, insects, vertebrates, and vertebrates), and their characteristic features and sizes considerably vary among cells (Zweytick et al. 2000; Arrese et al. 2014). Structurally, lipid droplet contains three parts: a neutral lipid core surrounded with a phospholipid monolayer, which is embedded with multiple proteins involved in the processes of lipid metabolism (Bozza et al. 2009). Of which, the most abundant proteins in lipid droplets are the perilipin family proteins. Perilipins, an ancient gene family, identified specific LD marker proteins from seminal in 1999 firstly, and play a critical role in neutral lipid metabolism (Greenberg et al. 1991). Perilipin1 is highly expressed in adipose tissue, which is involved in adipocyte lipolysis processes. During basal conditions in adipocytes, perilipin1 protects the LDs from lipases, including hormone-sensitive lipase (HSL) and adipocytes triglyceride lipase (ATGL). However, in stimulation conditions, perilipin1 is multiphosphorylated by cAMP-dependent protein kinase A, thereby stimulating the activity of TAG lipase to promoting lipolysis (Moore et al. 2005). Furthermore, perilipin1-knockout mice display reduced adipose tissue mass about 30% when compared with wild-type mice, and exhibit elevated basal lipolysis due to loss of the protective function of perilipin (Tansey et al. 2001). Additionally, Perilipin1 promotes the formation of unilocular lipid droplet by activating fat-specific protein 27 (Fsp27) in adipocytes (Sun et al. 2013). Perilipin2, adipophilin, is ubiquitously expressed which plays an essential role in LD formation and maintains lipid homeostasis, and is considered to be involved in both LD biogenesis and the regulation of lipolysis (Brasaemle et al. 1997; Fukushima et al. 2005; Listenberger et al. 2007). Perilipin2-deficient mice increased physical activity and reduced energy intake arrests high-fat diet (HFD)–induced adipose tissue inflammatory foci and liver steatosis, and its basal lipolysis was dramatically upregulated during OA treatment by increasing the expression of both HSL and phosphorylated HSL (McManaman et al. 2013; Xu et al. 2019). In macrophages, perilipin2 deficiency decreased the content of cellular lipids and the size and number of LD (Larigauderie et al. 2006). Unlike perilipin1/2, perilipin3 functions in the biogenesis and degradation of LD, which has apolipoprotein-like properties and reorganizes liposomes into small lipid discs. Remarkably, it remains stable in the cytoplasm when perilipin3 do not associate with LDs (Hocsak et al. 2010; Bulankina et al. 2009). It was found in HeLa cells that interfering perilipin3 led to a cellular growth rest, decreased the integration of TAG into lipid droplet, and blocked their maturation (Bulankina et al. 2009; Ganley et al. 2004). And in myotubes from leans, perilipin3 knockdown showed sharp alleviations in lipid oxidation (Covington et al. 2015). Furthermore, the mitochondrial membrane integrity can be protected through combining with perilipin3, as well as prevent cell death induced by oxidative stress (Hocsak et al. 2010).
Docosahexaenoic acid (DHA), one number of n-3 polyunsaturated fatty acids (PUFAs), has been reported to improve obesity-associated metabolic disorders and modulated lipid metabolisms in several ways (Martínez-Fernández et al. 2015). Increasing evidence suggested that n-3 PUFAs have an effect on LD biogenesis and the number of LDs per cell was rest with both concentration and type of fatty acid (Lecchi et al. 2013). In the liver of Fa/fa Zucker rats, DHA was more effective than eicosapentaenoic acid (EPA) for adding the percentage of smaller lipid droplets (Hong et al. 2019). Meanwhile, DHA and EPA reduced the number of lipid droplets by 56% and 42% respectively in C3H10 T1/2 cells (late phase of differentiation) (Warnke et al. 2011). The study in caprine monocytes showed that both DHA and EPA upregulated the gene expression of perilipin2 and perilipin3 and increased accumulation of lipid droplets in intracellular; however, DHA had fewer effects compared with EPA (Lecchi et al. 2013). Furthermore, human adipocytes treated by DHA significantly increased the release of glycerol and the mRNA of perilipin decreased about 40% compared with the control group (Wang et al. 2010). Currently, most researches of lipid droplets mainly focus mammalian. However, especially in teleostean the exact molecular mechanism of n-3 PUFAs affecting the formation of LD is still obscure.
Grass carp (Ctenopharyngodon idella), in terms of freshwater fish, is the largest scale of breeding in China and accumulates excess fat in the liver and adipose tissue during farming. So it is considered a good model for the study of lipid metabolism (Shi et al. 2017). Fortunately, the draft genome of grass carp has been released, which is a useful tool for identifying genomic structure of genes involved in lipid metabolism (Wang et al. 2015). Herein, three paralogs of perilipin were cloned from grass carp. To investigate its role in the effect of LD formation influenced by DHA in grass carp adipocytes (the late phase of differentiation), the mRNA expression of three paralogs in the DHA-induced adipocytes was also evaluated. The results will provide a theoretical basis for extending our acknowledgment of the physiological function of perilipins in fish adipocytes.
Materials and methods
Grass carp culture and cDNA samples
Experimental grass carps, weighing 40–50 g, were obtained from a local fish rearing farm in Ankang, Shaanxi, P. R. China. Before the experiment, animals were adapted to laboratory conditions for at least 14 days in 300-L square tanks (approximately 25 fish per tank) with living water supply at about 28 °C, ~ 6.0 mg/L of dissolved oxygen, under a light 12 h: dark 12 h photoperiod. Grass carps were fed commercial pellet diet (crude protein: 35%; crude lipid: 7%) 3 times/day.
For cDNA cloning and analysis of tissue distribution, grass carp was anesthetized by immersion in MS-222 (100 mg/L) (Sigma, St. Louis, MO, USA). Tissues including the heart, liver, intestinal tract, muscle, spleen, adipose tissue, and kidney were rapidly dissected, frozen in liquid nitrogen, and stored at − 80 °C until experiments. All procedures were performed in accordance with the Guide for Care and Use of Laboratory Animals and approved by the Northwest A&F University Institutional Animal Care and Use Committee.
Identification and cloning of perilipin family
Sequences of grass carp perilipin1, perilipin2, and perilipin3 were searched by gene annotation in our transcriptomic database (accession number: SRP044769). We used the SeqMan program of DNAstar software to assemble the ESTs (expressed sequence tags) into a consensus sequence including the complete open reading frame (ORF) in silico (Burland 2000). PCR specific primers were designed to amplify and verify the whole length of perilipin1, perilipin2, and perilipin3 cDNAs (Table 1).
For perilipin1–3 cloning, using TRIzol Reagent (TaKaRa, Dalian, China), total RNA was extracted separately from grass carp liver, muscle, and adipose tissue according to the instructions of the manufacturer. The integrity, concentration, and purity of total RNA were measured by 1.5% agarose gel electrophoresis and a spectrophotometer at 260/280 nm and 260/230 respectively. Five micrograms of total RNA, 1 μg of Oligo (dT)18 primer, and 1 μg Random Hexamer primer were used to synthesize first-strand cDNA by reverse transcription using RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific, USA). The resulting product was used as template for PCR amplification. PCR amplification was made with 0.5 μL of cDNA, 0.5 μM forward primer and reverse primer each, 4.75 μL of ddH2O, and 6.25 μL of 2 × TSINGKE® MASTER MIX in a total volume of 12.5 μL. PCR was performed 30 cycles at 94 °C for 30 s, 58 °C for 30 s, and 72 °C for 2 min, with an additional initial 4-min denaturation at 94 °C and a 10-min final extension at 72 °C. 1.5% agarose gel electrophoresis was used to detect PCR products, and the bands of the expected size were purified by PCR Purification Kit (Tiangen, China).
Sequence characterization
The protein sequence was predicted by ORF Finder (https://www.ncbi.nlm.nih.gov/orffinder/). The theoretical molecular mass and isoelectric point (pI) were analyzed by ProtParam (http://br.expasy.org/tools/protparam.html). And SignalP 4.0 (http://www.cbs.dtu.dk/services/SignalP/), NetOGlyc 3.1 (http://www.cbs.dtu.dk/services/NetOGlyc/), NetNGlyc 1.0 (http://www.cbs.dtu.dk/services/NetNGlyc/), and NetPhos 2.0 (http://www.cbs.dtu.dk/services/NetPhos/) were used to verify whether signal peptides, O-glycosylation and N-glycosylation sites, or phosphorylation sites existed in grass carp perilipin1, perilipin2, and perilipin3 protein sequences, respectively. The protein domains of grass carp perilipin family were predicted using the conserved domain profile (http://smart.emblheidelberg.de/).
Gene organization
Gene structure (exons/introns) of grass carp perilipin1, perilipin2, and perilipin3 was characterized by aligning the cloned cDNA with the genome (http://www.ncgr.ac.cn/grasscarp/), and their gene structures were compared with the ENSEMBL gene predictions (http//www.ensembl.org) in human (Homo sapiens), mouse (Mus musculus), and other representative teleosts, including zebrafish (Danio rerio), Atlantic salmon (Salmo salar), fugu (Takifugu rubripes), spotted gar (Lepisosteus oculatus), and tilapia (Oreochromis niloticus).
Phylogenetic analysis of perilipin family
Sequence alignments and percentage of amino acid conservation were assessed with the Clustal-W multiple alignment algorithm by DNAMAN8.0. The phylogenetic tree was constructed with MEGA 7.0 (Kumar et al. 2016) by the neighbor-joining (NJ) method based on the JTT + G model (Jones et al. 1992). The confidence of each node was assessed by 1000 bootstrap replicates. We provided detail information of species in attachment. The deduced amino acid sequences of perilipin 1–3 from grass carp and other organisms were selected to construct the phylogenetic tree.
Cell culture and treatment
The method of culturing grass carp pre-adipocytes as described in Liu et al. (2015) was slightly modified. Briefly, the adipose tissues were isolated from the abdominal and then washed four times with phosphate-buffered saline (PBS, pH 7.4), followed by a 30-min incubating in 2% ABV (Albumin Bovine V) with 0.1% Type I collagenase (Sigma, USA) at room temperature. Before centrifugation (590 g, 10 min), filtration is required through a 200-μm nylon filter. The cell pellet was incubated in erythrocyte lysing buffer for 6 min, and terminated the reaction immediately using DMEM complete medium and washed twice with fresh medium. Afterwards, cells were resuspended in growth medium (GM, containing Dulbecco’s modified Eagle’s medium (DMEM), 10% FBS, and 100 U/mL streptomycin, and 100 U/mL penicillin) and were seeded in plates which are precoated with gelatin at a density of approximately 20 g tissue/25 cm2. The cells were cultured at 28 °C with 5% CO2 until reaching 85–90% confluence. At confluence, pre-adipocytes were incubated in adipogenic medium (AM) containing GM supplemented with 10 μg/mL insulin, 10 nmol/L triiodothyronine, 1 μmol/L dexamethasone, and 0.5 mmol/L 3-isobutyl-1-methylxanthine (IBMX). The medium was changed every 2 days. In late phase of differentiation (about differentiation day 6), cells were cultured for additional 48 h in AM supplemented with DHA (0, 100, 200 μmol/L) and extracted RNA after 48 h. Three independent experiments were conducted in each control and treatment group.
Quantitative real-time PCR
The extracted and purified total RNA was treated with 4 × gDNA wiper Mix to prevent genomic DNA amplification. One microgram of total RNA was used for reverse transcription with First Strand cDNA Synthesis Kit (Vazyme Biotech Co., Ltd., Nanjing, China). Quantitative real-time PCR was performed using a CFX96TM Real-Time PCR Detection System (Bio-Rad, USA). The amplification was performed in a final volume of 20 μL containing 1 μL of the diluted cDNA (8-fold), 0.6 μL of each primer (0.5 μM), 7.8 μL of sterilized double-distilled water, and 10 μL of 2 × SYBR®Premix Ex TaqTMII (Vazyme Biotech Co., Ltd., Nanjing, China). The quantitative real-time PCR contained an initial activation step at 95 °C for 30 s, followed by 40 cycles of 95 °C for 15 s and 60 °C for 15 s. The comparative Ct method (2−ΔΔCt), described in the literatures (Pfaffl 2001), was used to calculate the gene expression values.
Statistical analysis
Statistical analyses were performed with SPSS 13.0 software (SPSS, Chicago, IL, USA). Data are expressed as mean ± SEM and were analyzed using one-way analysis of variance (ANOVA). Differences were considered to be significant at P < 0.05, P < 0.01.
Results
Identification of perilipin family in grass carp
In the present study, the complete CDS sequences of the three perilipin paralogs, named as perilipin1 (GenBank accession no. KY684179), perilipin2 (GenBank accession no. MN650220), and perilipin3 (GenBank accession no. MN650221) were obtained for grass carp. The open reading frame of perilipin1, perilipin2, and perilipin3 from grass carp was 1266 bp, 1455 bp, and 1598 bp long, and encoded 492, 454, and 419 deduced amino acids respectively. The molecular weight of the three paralogs is 53,564.87 Da, 50,381.17 Da, and 45,230.24 Da. Furthermore, the theoretical isoelectric point was 8.90, 5.58, and 5.14 respectively. We provided a detailed description of basic characteristics about the grass carp perilipin1, perilipin2, and perilipin3 in Table 2. The results from multiple alignments of the perilipin paralogs using DNAMAN 8.0 are shown in supplementary Fig. 1a–c. For perilipin1: Atlantic salmon (Salmo salar), 42.51%; coho salmon (Oncorhynchus kisutch), 44.84%; common carp German mirror (Cyprinus carpio), 77.24%; horned golden-line barbell (Sinocyclocheilus rhinocerous), 70.12%; human (Homo sapiens), 37.65%; mouse (Mus musculus), 38.98%; zebrafish (Danio rerio), 69.28% (supplementary Fig. 1a). For perilipin2: Atlantic salmon (Salmo salar), 54.27%; Burton’s mouthbrooder (Haplochromis burtoni), 56.05%; coelacanth (Latimeria chalumnae), 50.79%; coho salmon (Oncorhynchus kisutch), 56.07%; common carp German mirror (Cyprinus carpio), 72.19%; eastern happy (Astatotilapia calliptera), 55.80%; horned golden-line barbell (Sinocyclocheilus rhinocerous), 77%; human (Homo sapiens), 50.85%; mouse (Mus musculus), 55%; spotted gar (Lepisosteus oculatus), 53.76%; Nile tilapia (Oreochromis niloticus), 56.23%; zebrafish (Danio rerio), 71.03% (supplementary Fig. 1b). For perilipin3: Atlantic salmon (Salmo salar), 67.59%; Burton’s mouthbrooder (Haplochromis burtoni), 65.72%; common carp German mirror (Cyprinus carpio), 89.74%; eastern happy (Astatotilapia calliptera), 65.96%; fugu (Takifugu rubripes), 65.48%; human (Homo sapiens), 42.83%; Japanese medaka (Oryzias latipes), 58.20%; mouse (Mus musculus), 42.06%; Nile tilapia (Oreochromis niloticus), 66.43%; zebrafish (Danio rerio), 87.11% (supplementary Fig. 1c). Furthermore, the visualization of perilipin1–3 protein domains in grass carp was predicted (supplementary Fig. 1d). The results showed that three paralogs all have perilipin (PAT) domain, position at 11–360, 247–411, 18–400 respectively.
Gene structure and phylogenetic analysis
To assess the gene structural similarities and differences between the three perilipin paralogs of grass carp, the sequences of perilipin1, perilipin2, and perilipin3 were aligned. As shown in Fig. 1, grass carp perilipin1 gene consisted of 6 exons, which lost 2 exons compared with mammalian and lost 1 exon compared with zebrafish. Grass carp perilipin2 gene is composed of 7 exons, which is highly similar among mammals. However, the number of exons for perilipin2 among teleosts was varied. Grass carp perilipin3 gene contained 7 coding exons which were highly conserved among mammalian and representative teleosts (except spotted gar).
Phylogenetic trees for the proteins identified as homologous to perilipins are showed in Fig. 2a–c. The perilipin1 and perilipin3 amino acid sequences of teleost fish species clustered into the same subgroup (except coelacanth perilipin1), showed a close phylogenetic relationship with the grass carp, zebrafish, common carp German mirror, and horned golden-line barbell, while mammalian, chicken, and tropical clawed frog clustered into the other subgroup. Similarly, the perilipin2 amino acid sequences of grass carp, zebrafish, common carp German mirror, and horned golden-line barbell were the most closely related, while mammalian, chicken, tropical clawed frog, and coelacanth were far from the grass carp sequence.
Tissue distribution of perilipin1–3 paralogs in grass carp
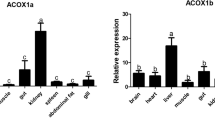
The expression profile of three perilipin paralogs in grass carp tissue, including the heart, liver, intestinal tract, muscle, spleen, adipose tissue, and kidney, was studied at the mRNA levels (Fig. 3). Both perilipin1 and perilipin2 paralog transcripts were abundant in adipose tissue and liver. The highest mRNA expression levels of perilipin3 were observed in the heart, liver, and adipose tissue; moderate levels in the kidney following in intestinal tract and muscle; and the lowest levels in the spleen (P < 0.05). Although the abundance of each mRNA is distinct among tissues, they were broadly distributed.
Perilipin 1–3 might be involved in DHA-induced lipid drop formation
To investigated the effect of LD formation influenced by DHA in grass carp adipocytes. The cells were treated with DHA at various concentrations (0, 100, 200 μmol/L) for 48 h. The results suggested that DHA increased the proportion of smaller lipid droplets effectively in grass carp adipocytes of the late phase of differentiation (Fig. 4). Meanwhile, DHA significantly increased the mRNA expressions of perilipin1, perilipin2, and perilipin3 respectively compared with the control and in a dose-dependent manner (Fig. 5a, P < 0.05, P < 0.01). Furthermore, we detected the mRNA expressions of peroxisome proliferator activated receptor γ (PPARγ); the results are similar with perilipin1–3 (Fig. 5b, P < 0.05, P < 0.01).
Quantitative analysis of the relative mRNA expression levels of perilipin family (a) and PPARγ (b) expression in grass carp adipocyte that treating with different concentrations of DHA. Data (mean ± SEM, n = 4) were expressed relative to expression of β-actin gene (#p < 0.05, ##p < 0.01 versus model; *P < 0.05, **P < 0.01 versus control)
Discussion
Lipid droplets (LDs), dynamic cellular organelles, are deemed to originate in the endoplasmic reticulum (ER) (Martin and Parton 2006), which serve crucial storerooms of lipids. These lipids furnish energy and serve as substrates for membrane synthesis, ATP production, and gene regulation, making LDs vital metabolic hubs (Wilfling et al. 2014). Perilipins, the main structural proteins of LDs (Bickel et al. 2009), adjust biogenesis and degradation of LD tightly and are involved in maintaining LD structural integrity. Although lots of studies exploring the function of perilipin protein have been conducted in mammalian systems, concentrating on lipid metabolism, studies on non-mammalian vertebrates have been performed relatively little (Granneman et al. 2017).
In the present study, three paralogs of perilipin were cloned and characterized from grass carp, and evaluated their roles in lipid drop formation induced by DHA. Currently, there are five known paralogs of perilipin family which is also called “PAT” family proteins (perilipin1, perilipin2/ADRP, perilipin3/Tip47, perilipin4/S3–12, perilipin5/OXPAT) identified in mammalian (Kimmel et al. 2010). The region of perilipin family in mammalian approximately 100 amino acids at the amino terminus is highly conserved; that sequence is known as the PAT (renamed the perilipin) domain (Londos et al. 1995; Miura et al. 2002). Similarly, we also predicted the PAT (perilipin) domain in this study (supplementary Fig. 1d). Hence, we can surmise that perilipins of grass carp may have the similar function as that of mammalian perilipins. PAT domain has been speculated to be related to the lipid droplet binding properties of the protein, but previous studies have shown that this domain does not perform the function of protein and lipid droplet binding (Liu and Xu 2006).
In order to better understand the physiological functions of genes, their tissue expression pattern was detected. Perilipin1 only expressed in mammary gland which contains vast adipocytes, and in visceral adipose tissue of rat. Similar to the study in C57BL/6J mouse, perilipin1 mRNA detected in adipose and adrenal, no expression or little was detected in other tissues (Nishiu et al. 1998; Yamaguchi et al. 2006). Nevertheless, perilipin1 mRNA of grass carp was highly expressed in liver and adipose, which are active tissues for lipid metabolism, but in other tissues little expressed (Fig. 3a). Perilipin2 mRNA was expressed in a variety of tissues of mouse and especially abundant in adipose tissue (Jiang and Serrero 1992). Inconsistently, perilipin2 mRNA was highest observed in adrenal of C57BL/6J mouse, and then in the liver (Yamaguchi et al. 2006). In our current study, Perilipin2 mRNA of grass carp was expressed in all tissues and especially abundant in adipose tissue and liver (Fig. 3b), consistent with mentioned above reports. Some studies have shown that Perilipin3 is expressed ubiquitously (Subramanian et al. 2004; Ducharme and Bickel 2008; Paul et al. 2008). In grass carp the result was detected similarly with mentioned; Perilipin3 mRNA expression also detected in all tissues, but the mRNA expression level is relatively low (Fig. 3c). To clarify, perilipin2 is abundantly expressed in the early stage differentiation of 3T3-L1 adipocytes; however with the process of differentiation, although its mRNA expression is increased, the protein expression is rapidly reduced (Ducharme and Bickel 2008; Brasaemle et al. 1997). At the same time the expression of perilipin1 protein gradually increases, which is the phenomenon of the conversion of the protein on the surface of the LDs from perilipin2 to perilipin1 during the differentiation process, so there is a large amount of perilipin1 in the surface of the LDs in the late differentiation and mature adipocytes without the presence of perilipin2 (Ducharme and Bickel 2008; Brasaemle et al. 1997). Remarkably, the perilipin1–3 of grass carp all expressed in adipose tissue and liver, the main organs that store and mobilize LDs in fish. The function of perilipin family in regulating lipid metabolism is entwined with its tissue expression pattern.
DHA (docosahexaenoic acid, C22:6) has been shown to effect proliferation, differentiation, lipolysis, lipogenesis, and apoptosis of adipocytes through several pathways (Wang et al. 2010; Kim et al. 2006). Lipid droplet acts as the hubs of lipid metabolism, its size, number, and membrane components influenced by n-3PUFA (Lecchi et al. 2013; Hong et al. 2019; Warnke et al. 2011). In our study, similar to the study in fat Zucker rats (Hong et al. 2019), 200 μM DHA increased the proportion of smaller lipid droplets effectively in grass carp adipocytes of the late phase of differentiation (Fig. 4). The mRNA expression of perilipin1, perilipin2, and perilipin3 significantly increased, induced by DHA through a dose-dependent manner (Fig. 5a). Perilipin family takes part in LD assembly and biogenesis (Bickel et al. 2009), the function of perilipin1 is mainly involved in adipocyte lipolysis, and perilipin2 covers many aspects, for example, lipid-droplet formation, lipolysis, and lipid accumulation (Fukushima et al. 2005). When cellular lipid influx adds, perilipin3 takes part in the formation of nascent lipid-droplet (Wolins et al. 2006). Because the change in mRNA expression levels of perilipin1–3 in grass carp adipocytes is related to the adding availability of DHA, we hypothesized that DHA may affect the formation of LDs by regulating the mRNA expression of perilipins.
Peroxisome proliferator-activated nuclear receptors (PPARs), vital nuclear receptors, distributing predominantly in the adipocytes and liver, regulate lipid metabolism, energy homeostasis, and the expression of LD-associated proteins (Yamaguchi et al. 2006). On the one hand, activation of PPARγ has been shown to increased perilipin expression in fully differentiated 3T3-L1 adipocytes (Tamori et al. 2002) and added the number of little adipocytes containing small LDs in white adipose tissues of fat Zucker rats, presumably via activating peroxisome proliferator-activated nuclear receptor–γ (PPARγ) (Okuno et al. 1998). On the other hand, some studies have demonstrated that PPARγ can be triggered by DHA and EPA (Li et al. 2005). Consistently with our previous study found that DHA increased the mRNA and protein expression levels of PPARγ in adipocytes of grass carp (Jin et al. 2018). We then tried to establish whether the activation of PPARγ by DHA led to perilipin1–3 mRNA expression in grass carp. Our present study showed that DHA treatments increased the mRNA expression of PPARγ (Fig. 5b) to potentially upregulate the mRNA expression of perilipin1–3.
In conclusion, the present study identified for the first time that three paralogs of perilipin were cloned and characterized from grass carp. Their high expression in liver and adipose tissues which are the principal bank store of LDs supports their roles in LD formation and lipid metabolism of fish. Presumably via activating PPARγ, DHA intervention not only increases the proportion of smaller lipid droplets effectively in fully differentiated grass carp adipocytes but also upregulates the mRNA expression levels of perilipin1–3, indicating that during DHA treatment perilipin family might play synergistic roles in the formation of LDs. This study is just an underlying exploration of the functions of perilipin gene in grass carp. Notably, more accurate molecular mechanism exploration is needed to determine specific ways of grass carp perilipin1–3 participating in LD formation and lipid metabolism induced by n-3 PUFAs.
References
Arrese EL, Saudale FZ, Soulages JL (2014) Lipid droplets as signaling platforms linking metabolic and cellular functions. Lipid Insights 7:7–16
Bickel PE, Tansey JT, Welte MA (2009) PAT proteins, an ancient family of lipid droplet proteins that regulate cellular lipid stores. Biochim Biophys Acta 1791(6):419–440
Bozza PT, Magalhães KG, Weller PF (2009) Leukocyte lipid bodies - biogenesis and functions in inflammation. Biochim Biophys Acta 1791(6):540–551
Brasaemle DL, Barber T, Wolins NE, Serrero G, Blanchette-Mackie EJ, Londos C (1997) Adipose differentiation-related protein is an ubiquitously expressed lipid storage droplet-associated protein. J Lipid Res 38(11):2249–2263
Bulankina AV, Deggerich A, Wenzel D, Mutenda K, Wittmann JG, Rudolph MG, Burger KN, Höning S (2009) TIP47 functions in the biogenesis of lipid droplets. J Cell Biol 185(4):641–655
Burland TG (2000) DNASTAR’s Lasergene sequence analysis software. Methods Mol Biol 132:71–91
Cotillard A, Poitou C, Torcivia A, Bouillot JL, Dietrich A, Klöting N, Grégoire C, Lolmede K, Blüher M, Clément K (2014) Adipocyte size threshold matters: link with risk of type 2 diabetes and improved insulin resistance after gastric bypass. J Clin Endocrinol Metab 99(8):E1466–E1470
Covington JD, Noland RC, Hebert RC, Masinter BS, Smith SR, Rustan AC, Ravussin E, Bajpeyi S (2015) Perilipin 3 differentially regulates skeletal muscle lipid oxidation in active, sedentary, and type 2 diabetic males. J Clin Endocrinol Metab 100(10):3683–3692
Ducharme NA, Bickel PE (2008) Lipid droplets in lipogenesis and lipolysis. Endocrinology 149(3):942–949
Esser N, Legrand-Poels S, Piette J, Scheen AJ, Paquot N (2014) Inflammation as a link between obesity, metabolic syndrome and type 2 diabetes. Diabetes Res Clin Pract 105(2):141–150
Flachs P, Rossmeisl M, Kuda O, Kopecky J (2013) Stimulation of mitochondrial oxidative capacity in white fat independent of UCP1: a key to lean phenotype. Biochim Biophys Acta 1831(5):986–1003
Fukushima M, Enjoji M, Kohjima M, Sugimoto R, Ohta S, Kotoh K, Kuniyoshi M, Kobayashi K, Imamura M, Inoguchi T, Nakamuta M, Nawata H (2005) Adipose differentiation related protein induces lipid accumulation and lipid droplet formation in hepatic stellate cells. In vitro cellular & developmental biology. Animal 41(10):321–324
Ganley IG, Carroll K, Bittova L, Pfeffer S (2004) Rab9 GTPase regulates late endosome size and requires effector interaction for its stability. Mol Biol Cell 15(12):5420–5430
Granneman JG, Kimler VA, Zhang H, Ye X, Luo X, Postlethwait JH, Thummel R (2017) Lipid droplet biology and evolution illuminated by the characterization of a novel perilipin in teleost fish. eLife 6:e21771
Greenberg AS, Egan JJ, Wek SA, Garty NB, Blanchette-Mackie EJ, Londos C (1991) Perilipin, a major hormonally regulated adipocyte-specific phosphoprotein associated with the periphery of lipid storage droplets. J Biol Chem 266(17):11341–11346
Hajer GR, van Haeften TW, Visseren FL (2008) Adipose tissue dysfunction in obesity, diabetes, and vascular diseases. Eur Heart J 29(24):2959–2971
Hocsak E, Racz B, Szabo A, Mester L, Rapolti E, Pozsgai E, Javor S, Bellyei S, Gallyas F Jr, Sumegi B, Szigeti A (2010) TIP47 protects mitochondrial membrane integrity and inhibits oxidative-stress-induced cell death. FEBS Lett 584(13):2953–2960
Hong L, Zahradka P, Cordero-Monroy L, Wright B, Taylor CG (2019) Dietary docosahexaenoic acid (DHA) and eicosapentaenoic acid (EPA) operate by different mechanisms to modulate hepatic steatosis and hyperinsulemia in fa/fa Zucker rats. Nutrients 11(4):917
Jiang HP, Serrero G (1992) Isolation and characterization of a full-length cDNA coding for an adipose differentiation-related protein. Proc Natl Acad Sci U S A 89(17):7856–7860
Jin A, Shi XC, Liu Y, Sun J, Ji H (2018) Docosahexaenoic acid induces PPARγ-dependent preadipocytes apoptosis in grass carp Ctenopharyngodon idella. Gen Comp Endocrinol 266:211–219
Jones DT, Taylor WR, Thornton JM (1992) The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci 8:275–282
Kim HK, Della-Fera M, Lin J, Baile CA (2006) Docosahexaenoic acid inhibits adipocyte differentiation and induces apoptosis in 3T3-L1 preadipocytes. J Nutr 136(12):2965–2969
Kimmel AR, Brasaemle DL, McAndrews-Hill M, Sztalryd C, Londos C (2010) Adoption of PERILIPIN as a unifying nomenclature for the mammalian PAT-family of intracellular lipid storage droplet proteins. J Lipid Res 51(3):468–471
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33(7):1870–1874
Larigauderie G, Cuaz-Pérolin C, Younes AB, Furman C, Lasselin C, Copin C, Jaye M, Fruchart JC, Rouis M (2006) Adipophilin increases triglyceride storage in human macrophages by stimulation of biosynthesis and inhibition of beta-oxidation. FEBS J 273(15):3498–3510
Lecchi C, Invernizzi G, Agazzi A, Modina S, Sartorelli P, Savoini G, Ceciliani F (2013) Effects of EPA and DHA on lipid droplet accumulation and mRNA abundance of PAT proteins in caprine monocytes. Res Vet Sci 94(2):246–251
Li H, Ruan XZ, Powis SH, Fernando R, Mon WY, Wheeler DC, Moorhead JF, Varghese Z (2005) EPA and DHA reduce LPS-induced inflammation responses in HK-2 cells: evidence for a PPAR-gamma-dependent mechanism. Kidney Int 67(3):867–874
Listenberger LL, Ostermeyer-Fay AG, Goldberg EB, Brown WJ, Brown DA (2007) Adipocyte differentiation-related protein reduces the lipid droplet association of adipose triglyceride lipase and slows triacylglycerol turnover. J Lipid Res 48(12):2751–2761
Liu MF, Xu GH (2006) Function of pat family proteins in the lipid metabolism. Sheng LI Ke Xue Jin Zhan 37(2):103–107
Liu P, Ji H, Li C, Chen LQ, Du ZY (2015) Morphology, mitochondrial development and adipogenic-related genes expression during adipocytes differentiation in grass carp(ctenopharyngodon idellus). Sci Bull (14):21–31
Londos C, Brasaemle DL, Gruia-Gray J, Servetnick DA, Schultz CJ, Levin DM, Kimmel AR (1995) Perilipin: unique proteins associated with intracellular neutral lipid droplets in adipocytes and steroidogenic cells. Biochem Soc Trans 23(3):611–615
Martin S, Parton RG (2006) Lipid droplets: a unified view of a dynamic organelle. Nature reviews. Mol Cell Biol 7(5):373–378
Martínez-Fernández L, Laiglesia LM, Huerta AE, Martínez JA, Moreno-Aliaga MJ (2015) Omega-3 fatty acids and adipose tissue function in obesity and metabolic syndrome. Prostaglandins Other Lipid Mediat 121(Pt A):24–41
McManaman JL, Bales ES, Orlicky DJ, Jackman M, MacLean PS, Cain S, Crunk AE, Mansur A, Graham CE, Bowman TA, Greenberg AS (2013) Perilipin-2-null mice are protected against diet-induced obesity, adipose inflammation, and fatty liver disease. J Lipid Res 54(5):1346–1359
Meshkani R, Adeli K (2009) Hepatic insulin resistance, metabolic syndrome and cardiovascular disease. Clin Biochem 42(13–14):1331–1346
Miura S, Gan JW, Brzostowski J, Parisi MJ, Schultz CJ, Londos C, Oliver B, Kimmel AR (2002) Functional conservation for lipid storage droplet association among Perilipin, ADRP, and TIP47 (PAT)-related proteins in mammals, Drosophila, and Dictyostelium. J Biol Chem 277(35):32253–32257
Moore HP, Silver RB, Mottillo EP, Bernlohr DA, Granneman JG (2005) Perilipin targets a novel pool of lipid droplets for lipolytic attack by hormone-sensitive lipase. J Biol Chem 280(52):43109–43120
Murphy S, Martin S, Parton RG (2009) Lipid droplet-organelle interactions; sharing the fats. Biochim Biophys Acta 1791(6):441–447
Nishiu J, Tanaka T, Nakamura Y (1998) Isolation and chromosomal mapping of the human homolog of perilipin (PLIN), a rat adipose tissue-specific gene, by differential display method. Genomics 48(2):254–257
Okuno A, Tamemoto H, Tobe K, Ueki K, Mori Y, Iwamoto K, Umesono K, Akanuma Y, Fujiwara T, Horikoshi H, Yazaki Y, Kadowaki T (1998) Troglitazone increases the number of small adipocytes without the change of white adipose tissue mass in obese Zucker rats. J Clin Invest 101(6):1354–1361
Ozcan U, Cao Q, Yilmaz E, Lee AH, Iwakoshi NN, Ozdelen E, Tuncman G, Görgün C, Glimcher LH, Hotamisligil GS (2004) Endoplasmic reticulum stress links obesity, insulin action, and type 2 diabetes. Science (New York, NY) 306(5695):457–461
Paul A, Chan L, Bickel PE (2008) The PAT family of lipid droplet proteins in heart and vascular cells. Curr Hypertens Rep 10(6):461–466
Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucl Acids Res 29(9):e45
Shi XC, Sun J, Yang Z, Li XX, Ji H, Li Y, Chang ZG, Du ZY, Chen LQ (2017) Molecular characterization and nutritional regulation of carnitine palmitoyltransferase (CPT) family in grass carp (Ctenopharyngodon idellus). Comp Biochem Physiol B Biochem Mol Biol 203:11–19
Subramanian V, Garcia A, Sekowski A, Brasaemle DL (2004) Hydrophobic sequences target and anchor perilipin A to lipid droplets. J Lipid Res 45(11):1983–1991
Sun Z, Gong J, Wu H, Xu W, Wu L, Xu D, Gao J, Wu JW, Yang H, Yang M, Li P (2013) Perilipin1 promotes unilocular lipid droplet formation through the activation of Fsp27 in adipocytes. Nat Commun 4:1594
Tamori Y, Masugi J, Nishino N, Kasuga M (2002) Role of peroxisome proliferator-activated receptor-gamma in maintenance of the characteristics of mature 3T3-L1 adipocytes. Diabetes 51(7):2045–2055
Tansey JT, Sztalryd C, Gruia-Gray J, Roush DL, Zee JV, Gavrilova O, Reitman ML, Deng CX, Li C, Kimmel AR, Londos C (2001) Perilipin ablation results in a lean mouse with aberrant adipocyte lipolysis, enhanced leptin production, and resistance to diet-induced obesity. Proc Natl Acad Sci U S A 98(11):6494–6499
Wang YC, Kuo WH, Chen CY, Lin HY, Wu HT, Liu BH, Chen CH, Mersmann HJ, Chang KJ, Ding ST (2010) Docosahexaenoic acid regulates serum amyloid A protein to promote lipolysis through down regulation of perilipin. J Nutr Biochem 21(4):317–324
Wang Y, Lu Y, Zhang Y, Ning Z, Li Y, Zhao Q, Lu H, Huang R, Xia X, Feng Q, Liang X, Liu K, Zhang L, Lu T, Huang T, Fan D, Weng Q, Zhu C, Lu Y, Li W, Wen Z, Zhou C, Tian Q, Kang X, Shi M, Zhang W, Jang S, du F, He S, Liao L, Li Y, Gui B, He H, Ning Z, Yang C, He L, Luo L, Yang R, Luo Q, Liu X, Li S, Huang W, Xiao L, Lin H, Han B, Zhu Z (2015) The draft genome of the grass carp (Ctenopharyngodon idellus) provides insights into its evolution and vegetarian adaptation. Nat Genet 47(6):625–631
Warnke I, Goralczyk R, Fuhrer E, Schwager J (2011) Dietary constituents reduce lipid accumulation in murine C3H10 T1/2 adipocytes: a novel fluorescent method to quantify fat droplets. Nutr Metab 8(1):30
Wilfling F, Haas JT, Walther TC, Farese RV Jr (2014) Lipid droplet biogenesis. Curr Opin Cell Biol 29:39–45
Wolins NE, Brasaemle DL, Bickel PE (2006) A proposed model of fat packaging by exchangeable lipid droplet proteins. FEBS Lett 580(23):5484–5491
Wozniak SE, Gee LL, Wachtel MS, Frezza EE (2009) Adipose tissue: the new endocrine organ? A review article. Dig Dis Sci 54(9):1847–1856
Xu SM, Zou F, Diao ZQ, Zhang SY, Deng YQ, Zhu XT, Cui LJ, Yu JH, Zhang ZG et al (2019) Perilipin 2 and lipid droplets provide reciprocal stabilization. Biophys Rep 3:145–160
Yamaguchi T, Matsushita S, Motojima K, Hirose F, Osumi T (2006) MLDP, a novel PAT family protein localized to lipid droplets and enriched in the heart, is regulated by peroxisome proliferator-activated receptor alpha. J Biol Chem 281(20):14232–14240
Zweytick D, Athenstaedt K, Daum G (2000) Intracellular lipid particles of eukaryotic cells. Biochim Biophys Acta 1469(2):101–120
Funding
This work was financially supported by the National Nature Science Foundation of China (NSFC, Grant Number: 31772863) and China Postdoctoral Science Foundation Funded Project (2019M660266).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
All procedures were performed in accordance with the Guide for Care and Use of Laboratory Animals and approved by the Northwest A&F University Institutional Animal Care and Use Committee.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOCX 1275 kb)
Rights and permissions
About this article
Cite this article
Huang, X., Sun, J., Bian, C. et al. Perilipin 1–3 in grass carp Ctenopharyngodon idella: molecular characterization, gene structure, tissue distribution, and mRNA expression in DHA-induced lipid droplet formation in adipocytes. Fish Physiol Biochem 46, 2311–2322 (2020). https://doi.org/10.1007/s10695-020-00857-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10695-020-00857-x