Abstract
Accumulating evidence suggests that the growth hormone (GH)/insulin-like growth factor (IGF) system participates in fish reproduction. To understand the physiological functions of the GH/IGF system, the mRNA expression profiles of all known members of the GH/IGF system, including hepatic and ovarian gh, GH receptor (ghr), IGFs (igf-i, igf-ii), IGF-I receptor (igf-ir) and IGF binding protein (igfbp1, igfbp2), pituitary gh, and hepatic vitellogenin (vtg) were investigated during ovarian development in turbot Scophthalmus maximus. Results showed that ghr, igf-i, igf-ii, igf-ir, and igfbp2 were expressed in the liver and ovary, whereas igfbp1 and gh were undetected. The hepatosomatic index (HSI) and gonadosomatic index (GSI) gradually increased and peaked during the late vitellogenesis (Latvtg) and migratory nucleus (Mig-nucl) stages, respectively. The mRNA expression profiles of ovarian ghr, igf-ii, hepatic igf-ir, vtg, and pituitary gh were similar to the HSI; ovarian igf-i and igf-ir expression was close to the GSI. However, the hepatic mRNA levels of ghr, igf-i, and igf-ii peaked at the early vitellogenesis (Evtg) stage, and then drastically declined during ovarian development. The mRNA expression of hepatic igfbp2 decreased and reached the lowest at the atresia (Atre) stage, whereas that of ovarian igfbp2 increased and peaked at Latvtg stage. Furthermore, significant correlations between pituitary gh, ovarian ghr, igf-i, and igf-ii, and hepatic ghr, igf-i, igf-ir, and igf-ii were observed, respectively. These results suggest that GH/IGF members appear to play distinct roles in the regulation of ovarian development in turbot and will be valuable for fish reproduction and broodstock management of aqua-cultured fish species.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Growth and reproduction are two closely related major physiological processes that occur during the life cycle of vertebrates. The growth hormone (GH)/insulin-like growth factor (IGF) system is involved in the regulation of growth and reproduction in both mammals and fish (Kiapekou et al. 2005; Spicer and Aad 2007; Reinecke 2010; Ahumada-Solorzano et al. 2012, 2016; Zhou et al. 2016). The GH/IGF system includes GH, GH receptors (GHRs), GH-binding proteins (GHBPs), insulin-like growth factors (IGFs), IGF receptors (IGFRs), and IGF-binding protein (IGFBPs). The complexity of the fish GH/IGF system is attributed to a past genome duplication event in their lineage that has resulted in multiple subtypes of GHBPs, GHRs, IGFRs, and IGFBPs (Duan 2002; Kamangar et al. 2006; Reindl and Sheridan 2012). The classical perspective on the GH/IGF system holds that the pituitary gland produces GH and secretes it into the blood; circulating GH stimulates the synthesis and secretion of hepatic and other extrahepatic tissue IGF-I, and IGF-I, in turn, stimulates cell proliferation and differentiation in various target tissues by binding to IGF-I receptors (Wood et al. 2005; Laviola et al. 2007). GH/IGF system members, including GH, IGFs, and their receptors, are expressed in the ovaries of humans, rats, and fish (Abir et al. 2008; Zhao et al. 2002; Reinecke 2010). Baker et al. (1996) found that IGF-I gene knockout in mice results in reproductive incompetence. GH-insufficient states disrupt ovarian function, causing problems in sexual maturation, and the reproductive ability of the female (Spiliotis 2003). It has been identified that GH directly stimulates steroidogenesis in spotted seatrout (Singh and Thomas 1993) and promotes the final maturation of postvitellogenic oocytes in the catfish (Sarang and Lal 2005). In addition, gh mRNA has been detected in full-grown zebrafish oocytes (Zhou et al. 2016). IGF-I and IGF-II stimulate oocyte maturation in numerous fish species including zebrafish (Nelson and Van Der Kraak 2010), rainbow trout (Bobe et al. 2004), and sea bream (Kagawa et al. 1994). Wang et al. (2008) were the first to discover another subtype of IGF, namely, IGF-3, which is only expressed in the gonad of tilapia. In zebrafish, the gonad-specific IGF-3 acts as an important mediator of the action of luteinizing hormone in oocyte maturation (Li et al. 2015). IGFBPs are synthesized in various types of tissue and play pivotal roles in the stabilization of IGFs, as well as regulate the availability of IGFs to target tissues. In mammals, IGFBPs participate in the regulation of follicular growth, maturation, and the selection of dominant follicles (Mazerbourg et al. 2003; Mazerbourg and Monget 2018). Zhou et al. (2016) reported that an intrafollicular network that involves the GH/IGF mini-axis exists in the zebrafish ovary. Therefore, accumulating evidence suggests that the GH/IGF system is involved in regulating ovarian development via complex mechanisms and plays important roles in the reproductive cycles of mammals and fishes.
Turbot (Scophthalmus maximus) is a commercially valuable flatfish species that is widely cultured in Europe and China. The production has been maintained at 50,000–60,000 t during the past decade, which account for approximately 80% of the world’s total output of aqua-cultured turbot (Lei et al. 2012). Some members of the turbot GH/IGF system have been cloned, and their physiological functions have been characterized (Berwert et al. 1995; Eliès et al. 1999; Duval et al. 2002). Wen et al. (2015) isolated IGF-II and analyzed the expression patterns of IGFs during embryogenesis and early larval development. They reported that the highest and lowest mRNA expression levels of igf-ii and igf-i were observed during the gastrula stage of embryonic development. Meng et al. (2016) found that IGF-I and IGF-II have different functions during the metamorphic development of turbot. In fish broodstock management, exogenous hormones can be applied to induce reproductive maturation and promote high-quality egg production (Mylonas et al. 2010). In farmed fish, the GH/IGF axis is involved in regulating gonadal growth and development and is correlated with puberty control (Taranger et al. 2010). However, the physiological function of the GH/IGF system in the regulation of oocyte development during the turbot reproductive cycle has yet to be completely elucidated. Thus, the present study aimed to investigate the hepatic and ovarian mRNA expression patterns of all known GH/IGF system members (igf-i, igf-ii, igf-ir, gh, ghr, igfbp1, and igfbp2), pituitary gh, and hepatic vtg during the reproductive cycle of turbot. The finding of this work will be valuable for fish reproduction and broodstock management.
Materials and methods
Animals and tissue sampling
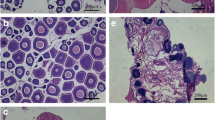
Four-year-old sexually mature female turbots weighting 3500–4000 g were obtained from Tianyuan Aquaculture Co., Ltd. of Yantai Economic Development Zone, Yantai, China. The fish (30 females) were kept in round tanks (30 m3 in volume) supplied with recirculating water at a rate of 25 L/min and exposed to a constant photoperiod (16-h light/8-h dark). Water salinity and oxygen were ranged from 20 to 25 g/L and 5 to 9 mg/L, respectively. Temperature was maintained between 12 and 13 °C. Fish were fed with a diet of frozen sardines, squid, and shrimp. The experiment period commenced in mid-October and ended in mid-December. All fish experimental procedures were conducted and approved according to the guidelines established by the Institutional Animal Care and Use Committee at Yellow Sea Fisheries Research Institute, Chinese Academy of Fishery Science. The fish were anesthetized with 100 mg/L tricaine methanesulfonate (MS-222, Sigma, St. Louis, MO). Body, liver, and ovary weights were recorded to calculate the hepatosomatic index [HSI = (liver weight / body weight)] and gonadosomatic index [GSI = (gonad weight / body weight)]. Hepatic, ovarian, and pituitary tissues were collected from each fish and stored in liquid nitrogen for RNA extraction. The ovaries were placed in Bouin’s solution for hematoxylin and eosin (HE) staining to identify the oocyte developmental stages. The stages of ovarian development were classified based on our previous study (Jia et al. 2014): briefly, pre-vitellogenesis stages (Prevtg, appearance of multiple nucleoli at the periphery of the nucleus), early vitellogenesis stages (Evtg, oocytes contain small vitellogenic yolk granules/globules), late vitellogenesis stages (Latvtg, oocytes contain many vitellogenic granules), migratory nucleus stage (Mig-nucl, oocytes became larger and the nucleus migrated toward the periphery of the cell), and atresia stage (Atre, oocytes shrunk or collapse).
RNA isolation, reverse transcription, and polymerase chain reaction
Total RNA was extracted from the collected samples with TRIzol reagent (Gibco-BRL, USA) and then quantified with NanoDrop 2000 (Thermo Fisher Scientific, Rockford, IL, USA). The extracted total RNA was treated for 30 min with DNase I (Qiagen) at 37 °C to prevent genomic DNA contamination. Subsequently, 1 μg of total RNA from each sample was reverse-transcribed with a Thermo Fisher One-Step RT-PCR Kit in accordance with the manufacturer’s instructions. To identify the distribution of igf-i, igf-ii, gh, igfbp1, igfbp2, igf-ir, and ghr in the liver and ovary, 2 μL of the reverse transcription products was used for amplification by polymerase chain reaction (PCR). The PCR cycling condition was as follows: denaturation at 94 °C for 30 s, annealing at 56 °C for 30 s, and extension at 72 °C for 1 min for 32 cycles. The primer sequences and PCR product lengths are listed in Table 1. The amplified products were verified by 1.2% agarose gel electrophoresis, and individual DNA fragments were collected with TIANamp Marine Animal DNA kit (Tiangen Biotech, China) in accordance with the manufacturer’s protocol. Subsequently, the individual DNA fragments were sequenced on an ABI 3730XL instrument. Sequence reads were assembled and evaluated using Geneious 8 software. The NCBI BLAST function (https://blast.ncbi.nlm.nih.gov/Blast.cgi) was used to identify turbot nucleotide.
Real-time quantitative PCR
To understand the GH/IGF network and its functions during turbot ovarian development, the mRNA expression levels of hepatic and ovarian ghr, igf-i, igf-ii, igfbp2, and igf-ir, pituitary gh, and hepatic vtg at different stages of ovarian development were quantified via quantitative real-time quantitative polymerase chain reaction (qRT-PCR). The reaction was performed on an ABI StepOnePluse Sequence Detection System (Applied Biosystems, USA) in accordance with the manufacturer’s instructions. The SYBR Premix Ex Taq™ kit (Takara Bio., China) was used for amplification, and the reaction mixture contained 10 μL of SYBR® Premix Ex Taq™, 0.8 μL of each primer (10 μM), 0.4 μL of ROX dye (50×), 2 μL of cDNA sample (25 ng/μL), and 6 μL of sterile distilled water. Initial denaturation was conducted at 95 °C for 10 s, followed by 40 cycles at 95 °C for 5 s and at 60 °C for 30 s. The qRT-PCR efficiency (E) was established on the basis of the slopes of standard curves that were generated by using a 10-fold dilution series of purified PCR fragments (1:10,000 dilution) as a template. E was calculated using the formula E (%) = (10–1/slope − 1) × 100, and the E values of 90% to 110% were considered acceptable. The melt curve for each amplicon of all aforementioned genes exhibited a distinct peak (Supplemental data, S1). For normalization, four commonly used housekeeping genes [elongation factor-1α (ef1α), cathepsin d (ctsd), glyceraldehyde-3-phosphate-dehydrogenase (gapdh), and beta-actin (β-actin)] were evaluated to compare the CT values of a subset of samples. Evaluation revealed that ctsd is the most constantly expressed housekeeping gene in this study. Thus, ctsd was selected as the housekeeping gene, and the relative abundance of the mRNA was normalized to ctsd by using the 2−∆∆CT method (Livak and Schmittgen 2001). The qRT-PCR primers are listed in Table 1. All samples were amplified in triplicate.
Statistical analysis
All data were expressed as the means ± standard error of the mean. One-way analysis of variance was conducted using SPSS16.0 software (SPSS Inc., USA). Differences were considered significant when P < 0.05, and Duncan’s test was conducted when necessary for multiple comparisons. Levels of correlation were determined by calculating the Person correlation coefficient (r). Statistical significance was considered at P < 0.05.
Results
GH/IGF system-related gene expression in turbot liver and ovary was analyzed by PCR. igf-i, igf-ii, igfbp2, igf-ir, and ghr were detected in the hepatic and ovarian tissues, whereas gh and igfbp1 were undetected (Fig. 1). HSI remained unchanged during the Prevtg and Evtg stages. However, HSI significantly increased to the peak level at the Latvtg stage and then began to decrease until the Atre stage (Fig. 2a, P < 0.05). GSI significantly increased from the Prevtg to the Mig-nucl stage, with the highest values observed at the Mig-nucl stage, and then decreased significantly at the Atre stage (Fig. 2b, P < 0.05).
Reproductive index of female turbot during ovarian development. a Hepatosmatic index (HSI); b gonadosomatic index (GSI). Values represent the mean ± SEM (n = 6). Bars with different superscripts differ (P < 0.05). Prevtg previtellogenesis, Evtg early vitellogenesis, Latvtg late vitellogenesis, Mig-nucle migratory nucleus, Atre atresia
Given that gh and igfbp1 were undetected in the liver and ovary, we only examined the expression profiles of igf-i, igf-ii, igfbp2, igf-ir, and ghr at different stages of ovarian developmental via qRT-PCR. The hepatic mRNA levels of ghr, igf-i, and igf-ii gradually increased during the Prevtg to Evtg stages, with the highest values observed at Evtg stage, and then significantly decreased at the Latvtg stage (Fig. 3a–c; P < 0.05). During the Latvtg to the Atre stages, hepatic igf-ii mRNA remained unchanged (Fig. 3b; P ˃ 0.05), and ghr and igf-ii mRNA significantly decreased and increased (Fig. 3a, c; P < 0.05), respectively. However, the hepatic mRNA levels of ghr, igf-i, and igf-ii from the Latvtg to the Atre stages were significantly lower than Prevtg and Evtg stages (Fig. 3a–c; P < 0.05). Similarly, the hepatic mRNA levels of igf-ir gradually increased from the Prevtg to the Latvtg stages, peaking at the Latvtg stage, and then significantly decreased from the Mig-nucle to the Atre stages (Fig. 3e; P < 0.05). By contrast, hepatic igfbp2 mRNA levels significantly decreased during ovarian development and reach the lowest value at the Atre stage (Fig. 3d; P < 0.05). The results for ovarian igf-i, igf-ii, and igf-ir mRNA were similar to those for hepatic igf-ir, whereas ovarian igf-i and igf-ir peak at the Mig-nucle stage (Fig. 3e, g, h, j; P < 0.05). In addition, the greatest increase in ovarian ghr and igf-ir was observed at the Latvtg and Mig-nucle stages, respectively, which were > 20-fold compared with that at the Prevtg stage (Fig. 3f, j; P < 0.05). Hepatic vtg and pituitary gh mRNA showed similar results to ovarian ghr (Fig. 4). Statistically significant correlations were observed between HSI and hepatic vtg, between pituitary gh and ovarian igf-i and ghr, between hepatic ghr and hepatic igf-i and igf-ii, between hepatic igf-i and hepatic igf-ii and ovarian igf-ii, between hepatic igf-ii and ovarian igf-ii, and between hepatic igf-ir and hepatic vtg and ovarian ghr and igf-i (Table 2, P < 0.05). Strong but statistically nonsignificant correlations (Table 2, P ˃ 0.05) were also observed between GSI and hepatic igf-i and igf-ii and ovarian igf-ii and igf-ir; between HSI and pituitary gh, hepatic igf-ir, ad ovarian igf-i and igfbp2; between pituitary gh and hepatic igf-ir and vtg, and ovarian igfi-r; between hepatic ghr and ovarian igf-ii; between hepatic igf-i and hepatic vtg; between hepatic igf-ir and ovarian igf-ir, hepatic vtg, and ovarian igf-i; between ovarian ghr and ovarian igf-i and igf-ir; and between ovarian igf-i and ovarian igf-ir, whereas these factors showed no significantly correlation.
The expression of GH/IGF system genes’ (ghr, igf-i, igf-ii, igfbp2, igf-ir) mRNAs in liver and ovary during ovarian development of turbot. Values represent the mean ± SEM (n = 6). Bars with different superscripts differ (P < 0.05). Prevtg previtellogenesis, Evtg early vitellogenesis, Latvtg late vitellogenesis, Mig-nucle migratory nucleus, Atre atresia
The expression of pituitary gh and hepatic vtg mRNAs during ovarian development of turbot. Values represent the mean ± SEM (n = 6). Bars with different superscripts differ (P < 0.05). Prevtg previtellogenesis, Evtg early vitellogenesis, Latvtg late vitellogenesis, Mig-nucle migratory nucleus, Atre atresia
Discussion
The GH/IGF system plays important roles in coordinating the growth and reproduction of vertebrates, including fish. The liver stores and provides massive amount of energy for oocyte growth and maturation during ovarian development. HSI reflects hepatic energy content and physiological status. Turbot oocytes increase in number and size from the Prevtg to Evtg stages (Jia et al. 2014). This finding suggests that hepatic energy accumulation is just beginning and hepatic vitellogenesis not occurring. These phenomena may account for the HSI remained unchanged between the Prevtg and Evtg stages in the present study. Xue et al. (2018) found similar results in the ovarian development of turbot. In the present study, we detected for the first time all known isoforms of igfs (igf-i, igf-ii), igf-ir, gh, ghr, and igfbps (igfbp1, igfbp2) in the liver and ovary, and quantified their mRNA expression during turbot ovarian development. The GH/IGF system members, GHR, IGFs, IGFRs, and IGFBPs, are widely expressed in the tissues of fish and other vertebrates, and the liver is the primary site for the localization of GHR and IGFs (Reindl and Sheridan 2012). In the current study, igf-i, igf-ii, igf-ir, ghr, and igfbp2 were expressed in turbot liver and ovary, whereas gh and igfbp1 were undetected. GH is a pluripotent hormone that is produced primarily by the pituitary gland and distributed in extrapituitary tissues (Dai et al. 2015). Yang et al. (1999) found that high levels of GH mRNA were primarily detected in the pituitary gland, brain, gill, and heart and low levels of GH mRNA were also detected in the kidney, liver, and ovary in adult rainbow trout. In zebrafish, igfbp1 is expressed in the liver and absent in extrahepatic tissues, whereas igfbp2 was detected in extrahepatic tissues (Duan et al. 1999). Kamangar et al. (2006) found that igfbp2 is expressed in the liver and ovary and hypothesized that this gene participates in the modulation of follicular steroid production by IGFs during follicular competence acquisition and oocyte maturation. Meanwhile, gh, ghrs, igfs, and igf-irs have all been detected in the zebrafish ovary (Zhou et al. 2016). We speculate that species difference and other uncertain factors account for the absence of igfbp1 and gh in turbot liver and ovary. These results also suggest that igfs (igf-i, igf-ii), igf-ir, ghr, and igfbp2 may participate in the regulation of turbot ovarian development.
The development and maturation of ovarian oocytes are key physiological processes in female reproduction. The GSI and the HSI are good indicators of gonadal development and liver energy content, respectively (Dahle et al. 2003). In the present study, HSI significantly increased at Latvtg and then gradually decreased from Mig-nucle to Atre stages. GSI also showed similar results, with the highest value observed at the Mig-nucle stages. These results indicated that hepatic vitellogenesis occurred and gradually transferred to the ovary during the Latvtg to Mig-nucle stages. The decrease in HSI at the Atre stage may be caused by the reduction in hepatic metabolic activity. The turbot pituitary gh mRNAs showed similar results as HSI, and their levels peaked at the Lavtg stage. The pituitary gland secrets GH and is transferred to blood bound to high-affinity GH binding protein (GHBP), which has been shown to correspond to the extracellular part of hepatic GHR (Baumann 1991; Baumann and Mercado 1993; Fisker et al. 1998). Increasing concentrations of GHBP may modulate the binding of GH to its receptor and exert a negative effect on receptor recycling and synthesis (Amit et al. 1992; Gonzalez et al. 2007). Meanwhile, the circulating concentrations of human GHBP peak in early adulthood (Schilbach and Bidlingmaier 2015). Turbot plasma GHBP concentrations were significantly higher at the Latvtg stage than Evtg stage during ovarian development (Supplemental data, S2) and may be attributed to hepatic ghr peaking earlier than pituitary gh during turbot ovarian development. However, the precise mechanism of endocrine regulation remains unknown and requires further investigation. In current study, significantly correlations and strong correlations were observed between pituitary gh and ovarian ghr and igf-i and between pituitary gh and hepatic igf-ir and vtg, respectively. Zhou et al. (2016) state that GH may be involved in regulating the fast-secondary growth phase starting from Prevtg and plays a local role in the early follicle stage of zebrafish. In rat, the GH protein is present in oocytes and follicle cells of the pre-antral follicles (Zhao et al. 2002). These results suggest that GH is involved in the regulation of oocyte growth during turbot ovarian development. In addition, IGF and IGFBP members also play important roles during final oocyte maturation in teleost. Weber and Sullivan (2005) found that IGF-I induces oocyte maturational competence but not meiotic resumption in white bass. IGF-II could directly induce maturational competence in southern flounder by upregulating the mRNA and protein expression levels of mPRα (Picha et al. 2012). IGFBPs are a family of secreted proteins that specifically bind to IGF-I and IGF-II that prolong the half-lives of IGFs and buffer the potential hypoglycemic effects of high concentrations of IGFs in the circulation. In addition, biological actions of some IGFBPs are IGF-independent. It has been identified that IGFBP 2, 3, 4, 5, and 6 manifested different expression patterns and involve in regulating oocyte maturation during reproductive cycle in rainbow trout (Kamangar et al. 2006). To identify the possible mechanisms involving the GH/IGF system during turbot ovarian development, we investigated the mRNA expression patterns of igf-i, igf-ii, igf-ir, ghr, and igfbp2 in the liver and ovary at different stages of ovarian development. Interestingly, the mRNA expression levels of ovarian ghr, igf-ii, and igfbp2 and hepatic igf-ir gradually increased from the Prevtg to the Latvtg stages and peaked at the Latvtg stage. Ovarian igf-i and igf-ir showed similar expression patterns, but peaked at the Mig-nucl stage. However, the mRNA levels of hepatic igf-i, igf-ii, and ghr peaked at the Evtg stage and then dramatically declined during reproductive cycle. Previous research identified the existence of an intraovarian network in zebrafish follicles that involves GH/IGF members (Zhou et al. 2016). Meanwhile, the hepatic mRNA levels of vtg gradually increased from the Prevtg stage to the Latvtg stage during reproductive cycle in turbot. Significantly correlations were observed among HSI, ovarian ghr, igf-i, and igf-ii, and hepatic vtg, ghr, igf-i, igf-ir, and igf-ii in the present study. Therefore, these results suggest that the GH/IGF system is involved in regulating turbot ovarian development and affects oocyte growth and maturation by complex endocrine, paracrine, and autocrine manner. However, the detailed mechanisms remain unknown and await thorough investigation.
In conclusion, all known members of the GH/IGF system present distinct expression profiles during turbot ovarian development. The results indicate that GH/IGF members are involved in the regulation of oocyte growth, maturation, and ovulation during turbot ovarian development. These findings extend our knowledge of the expression patterns of the GH/IGF system during the reproductive cycle in turbot and will be valuable for fish reproduction and broodstock management. However, the direct evidence and detailed mechanism to evaluate the GH/IGF member roles regulate turbot ovarian development need to be further studied. Fish possess multiple isoforms of GH/IGF ligands, receptors, and binding proteins and that these isoforms exhibit distinct expression patterns and functions. Therefore, other isoforms of the members of the GH/IGF system should be identified and investigated through whole-genome sequencing. The roles of these isoforms must be determined to obtain information regarding the molecular basis of the roles of GH/IGF in the turbot life cycle.
References
Abir R, Garor R, Felz C, Nitke S, Krissi H, Fisch B (2008) Growth hormone and its receptor in human ovaries from fetuses and adults. Fertil Steril 90:1333–1339
Ahumada-Solorzano SM, Carranza ME, Pedernera E, Rodriguez-Mendez AJ, Luna M, Aramburo C (2012) Local expression and distribution of growth hormone and growth hormone receptor in the chicken ovary: effects of GH on steroidogenesis in cultured follicular granulosa cells. Gen Comp Endocrinol 175:297–310
Ahumada-Solórzano SM, Martínez-Moreno CG, Carranza M, Ávila-Mendoza J, Luna-Acosta JL, Harvey S, Luna M, Arámburo C (2016) Autocrine/paracrine proliferative effect of ovarian GH and IGF-I in chicken granulosa cell cultures. Gen Comp Endocrinol 234:47–56
Amit T, Ish-Shalom S, Glaser B, Youdim MB, Hochberg Z (1992) Growth-hormone-binding protein in patients with acromegaly. Horm Res 37(6):205–211
Baker J, Hardy MP, Zhou J, Bondy C, Lupu F, Bellve AR, Efstratiadis A (1996) Effects of an Igf1 gene null mutation on mouse reproduction. Mol Endocrinol 10:903–918
Baumann G (1991) Growth hormone-binding proteins: biochemical characterization and assays. Acta Endocrinol 124:21–26
Baumann G, Mercado M (1993) Growth hormone-binding proteins in plasma. Nutrition 9:546–553
Berwert L, Segnert H, Reinecke M (1995) Ontogeny of IGF-1 and the classical islet hormones in the turbot, Scophthalmus maximus. Peptides 16:113–122
Bobe J, Nguyen T, Jalabert B (2004) Targeted gene expression profiling in the rainbow trout (Oncorhynchus mykiss) ovary during maturational competence acquisition and oocyte maturation. Biol Reprod 71:73–82
Dahle R, Taranger GL, Karlsen O, Kjesbu OS, Norberg B (2003) Gonadal development and associated changes in liver size and sexual steroids during the reproductive cycle of captive male and female Atlantic cod (Gadus morhua L.). Comp Biochem Physiol A 136:641–653
Dai XY, Zhang W, Zhuo ZJ, He JY, Yin Z (2015) Neuroendocrine regulation of somatic growth in fishes. Sci China Life Sci 58(2):137–147
Duan C (2002) Specifying the cellular responses to IGF signals: roles of IGF-binding proteins. J Endocrinol 175:41–54
Duan C, Ding J, Li Q, Tsai W, Pozios K (1999) Insulin-like growth factor binding protein 2 is a growth inhibitory protein conserved in zebrafish. Proc Natl Acad Sci U S A 96:15274–15279
Duval H, Rousseau K, Eliès G, Le Bail PY, Dufour S, Boeuf G, Boujard D (2002) Cloning, characterization, and comparative activity of turbot IGF-I and IGF-II. Gen Comp Endocrinol 126:269–278
Eliès G, Duval H, Bonnec G, Wolff J, Boeuf G, Boujard D (1999) Insulin and insulin-like growth factor-1 receptors in an evoluted fish, the turbot: cDNA cloning and mRNA expression. Mol Cell Endocrinol 158:173–185
Fisker S, Ebdrup L, Ørskov H (1998) Influence of growth hormone binding protein on growth hormone estimation in different immunoassays. Scand J Clin Lab Invest 58:373–381
Gonzalez L, Curto LM, Miquet JG, Bartke A, Turyn D, Sotelo AI (2007) Differential regulation of membrane associated-growth hormone binding protein (MA-GHBP) and growth hormone receptor (GHR) expression by growth hormone (GH) in mouse liver. Growth Hormon IGF Res 17:104–112
Jia Y, Meng Z, Niu H, Hu P, Lei J (2014) Molecular cloning, characterization, and expression analysis of luteinizing hormone receptor gene in turbot (Scophthalmus maximus). Fish Physiol Biochem 40:1639–1650
Kagawa H, Kobayashi M, Hasegawa Y, Aida K (1994) Insulin and insulin-like growth factors I and II induce final maturation of oocytes of red seabream, Pagrus major, in vitro. Gen Comp Endocrinol 95:293–300
Kamangar BB, Gabillard JC, Bobe J (2006) Insulin-like growth factor-binding protein (IGFBP)-1, −2, −3, −4, −5, and −6 and IGFBP-related protein 1 during rainbow trout post vitellogenesis and oocyte maturation: molecular characterization, expression profiles, and hormonal regulation. Endocrinology 147(5):2399–2410
Kiapekou E, Loutradis D, Drakakis P, Zapanti E, Mastorakos G, Antsaklis A (2005) Effects of GH and IGF-I on the in vitro maturation of mouse oocytes. Hormones 4:155–160
Laviola L, Natalicchio A, Giorgino F (2007) The IGF-I signaling pathway. Curr Pharm Des 13(7):663–669
Lei J, Liu X, Guan C (2012) Turbot culture in China for two decades: achievements and prospect. Prog Fish Sci 33:123–130 in Chinese; with English abstract
Li J, Chu L, Sun X, Liu Y, Cheng CH (2015) IGFs mediate the action of luteinizing hormone on oocyte maturation in zebrafish. Mol Endocrinol 29(3):373–383
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real time quantitative PCR and the 2-∆∆CT method. Methods 25:402–408
Mazerbourg S, Monget P (2018) Insulin-like growth factor binding proteins and IGFBP proteases: a dynamic system regulating the ovarian folliculogenesis. Front Endocrinol 9:134
Mazerbourg S, Bondy CA, Zhou J, Monget P (2003) The insulin-like growth factor system: a key determinant role in the growth and selection of ovarian follicles? A comparative species study. Reprod Domest Anim 38:247–258
Meng Z, Hu P, Lei JL (2016) Expression of insulin-like growth factors at mRNA levels during the metamorphic development of turbot (Scophthalmus maximus). Gen Comp Endocrinol 235:11–17
Mylonas CC, Fostier A, Zanuy S (2010) Broodstock management and hormonal manipulations of fish reproduction. Gen Comp Endocrinol 165:516–534
Nelson SN, Van Der Kraak G (2010) Characterization and regulation of the insulin-like growth factor (IGF) system in the zebrafish (Danio rerio) ovary. Gen Comp Endocrinol 168:111–120
Picha ME, Shi B, Thomas P (2012) Dual role of IGF-II in oocyte maturation in southern flounder Paralichthys lethostigma: up-regulation of mPRα and resumption of meiosis. Gen Comp Endocrinol 177:220–230
Reindl KM, Sheridan MA (2012) Peripheral regulation of the growth hormone-insulin-like growth factor system in fish and other vertebrates. Com Biochem Physiol A 163:231–245
Reinecke M (2010) Insulin-like growth factors and fish reproduction. Biol Reprod 82:656–661
Sarang MK, Lal B (2005) Effect of piscine GH/IGF-I on final oocyte maturation in vitro in Heteropneustes fossilis. Fish Physiol Biochem 31:231–233
Schilbach K, Bidlingmaier M (2015) Growth hormone binding protein-physiological and analytical aspects. Best Pract Res Clin Endocrinol 29(5):671–683
Singh H, Thomas P (1993) Mechanism of stimulatory action of growth hormone on ovarian steroidogenesis in spotted seatrout, Cynoscion nebulosus. Gen Comp Endocrinol 89:341–353
Spicer LJ, Aad PY (2007) Insulin-like growth factor (IGF) 2 stimulates steroidogenesis and mitosis of bovine granulosa cells through the IGF1 receptor: role of follicle-stimulating hormone and IGF2 receptor. Biol Reprod 77:18–27
Spiliotis BE (2003) Growth hormone insufficiency and its impact on ovarian function. Ann N Y Acad Sci 997:77–84
Taranger GL, Carrillo M, Schulz RW, Fontaine P, Zanuy S, Felip A, Weltzien FA, Dufour S, Karlsen Ø, Norberg B, Andersson E, Hansen T (2010) Control of puberty in farmed fish. Gen Comp Endocrinol 165:483–515
Wang DS, Jiao B, Hu C, Huang X, Liu Z, Cheng CH (2008) Discovery of a gonad specific IGF subtype in teleost. Biochem Biophys Res Commun 367:336–341
Weber GM, Sullivan CV (2005) Insulin like growth factor-I induces oocyte maturational competence but not meiotic resumption in white bass (Morone chrysops) follicles in vitro: evidence for rapid evolution of insulin-like growth factor action. Biol Reprod 72:1177–1186
Wen HS, Qi Q, Hu J, Si YF, He F, Li J (2015) Expression analysis of the insulin like growth factors I and II during embryonic and early larval development of turbot (Scophthalmus maximus). J Ocean Univ China 14:309–316
Wood AW, Duan C, Bern HA (2005) Insulin-like growth factor signaling in fish. Int Rev Cytol 243:215–285
Xue R, Wang X, Xu S, Liu Y, Feng C, Zhao C, Liu Q, Li J (2018) Expression profile and localization of vitellogenin mRNA and protein during ovarian development in turbot (Scophthalmus maximus). Comp Biochem Physiol B 226:53–56
Yang BY, Greene M, Chen TT (1999) Early embryonic expression of the growth hormone family protein genes in the developing rainbow trout, Oncorhynchus mykiss. Mol Reprod Dev 53:127–134
Zhao J, Taverne MA, van der Weijden GC, Bevers MM, van den Hurk R (2002) Immunohistochemical localisation of growth hormone (GH), GH receptor (GHR), insulin-like growth factor I (IGF-I) and type I IGF-I receptor, and gene expression of GH and GHR in rat pre-antral follicles. Zygote 10:85–94
Zhou R, Yu SMY, Ge W (2016) Expression and functional characterization of intrafollicular GH-IGF system in the zebrafish ovary. Gen Comp Endocrinol 232:32–42
Acknowledgments
This study was supported by National Natural Science Foundation of China (31302205), Shandong Major Science and Technology Innovation Projects (2018YFJH0703), and China Agriculture Research System (CARS-49-G24).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Additional information
Publisher’s Note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOC 1209 kb)
Rights and permissions
About this article
Cite this article
Jia, Y., Jing, Q., Gao, Y. et al. Involvement and expression of growth hormone/insulin-like growth factor member mRNAs in the ovarian development of turbot (Scophthalmus maximus). Fish Physiol Biochem 45, 955–964 (2019). https://doi.org/10.1007/s10695-018-0604-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10695-018-0604-z