Abstract
Polyspermy is the most commonly observed cause of embryonic abnormalities in fertilization, often resulting in death. In sterlet (Acipenser ruthenus), however, polyspermic embryos have high survival (similar to a control group) and morphological development is similar to monospermic larvae. Ploidy of these individuals is n/2n mosaic (whereas the normal state for A. ruthenus is a functional diploid). This study was undertaken to test whether sturgeon eggs can be fertilized by several spermatozoa from different species to produce viable offspring from three interspecific parents: A. ruthenus (2n), A. gueldenstaedtii (4n), and A. baerii (4n). Four trials were performed: (1) and (2) A. baerii eggs were fertilized with a mixture of A. ruthenus and A. gueldenstaedtii sperm; (3) A. gueldenstaedtii eggs were fertilized with a mixture of A. baerii and A. ruthenus sperm; and (4) A. gueldenstaedtii eggs were fertilized with a mixture of A. gueldenstaedtii and A. ruthenus sperm. Fertilized embryos with abnormal cleavage (3, 5, 6, 7, 9, and 10 cells) were collected and kept separately until 14 days post-fertilization. Ploidy level of 25 larvae (hatched from abnormal cleaved embryos) was evaluated by flow cytometry. Forty-four percent of observed hybrids had a mosaic 2n/3n ploidy. Five larva were processed further with microsatellite analysis and demonstrated that three specimens were heterospecific polyspermic larvae, containing alleles from three parents, and two specimens were conspesific polyspermic larvae from two parents. This astonishing phenomenon was emphasized by the fact that it was generated without any significant intervention.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Sturgeons (Acipenseridae) are an ancient family (Bemis and Grande 1997) with unique biological characteristics that can be challenging for research (Carmona et al. 2009). Sturgeon eggs have multiple micropyles, and the number varies among females of different species and between eggs of individual females (Dettlaff et al. 1993): Acipenser ruthenus eggs contain five to 13 micropyles (Zalenskii 1878); up to 52 micropyles in A. gueldenstaedtii eggs (Dettlaff and Vassetzky 1991); and A. baerii eggs have two to 16 micropyles (Debus et al. 2002).
Ploidy in sturgeon fishes also varies and can be categorized into three classes depending on chromosome number: functional diploids (2n) have ~ 120 chromosomes (Huso huso, A. sturio, A. ruthenus); functional tetraploids (4n) have ~ 240 chromosomes (A. baerii, A. gueldenstaedtii, A. naccarii, A. mikadoi); and functional hexaploids (6n) have ~ 360 chromosomes (A. brevirostrum) (Ludwig et al. 2001; Fontana and Colombo 1974; Fontana 1994; Kim et al. 2005; Birstein and Vasilev 1987; Vasil’ev 2010). However, sturgeons hybridize more easily than other vertebrates (Birstein et al. 1997) between species with the same and/or different ploidy level (Billard and Lecointre 2001) and demonstrate interspecific and intergeneric hybridization in the wild and under artificial conditions (Havelka et al. 2011).
Multiple micropyles, which are typical for sturgeon eggs, and fertilization with high concentrated sperm suspensions are associated with the occurrence of polyspermy (Dettlaff et al. 1981). It is believed that polyspermy is the most commonly observed reason for embryonic abnormalities in fertilization, often resulting in death (Boveri 1901; Wang et al. 2003). However, recent research by Iegorova et al. (2018) demonstrates that sturgeon polyspermic embryos are unique in their ability to survive and to develop into morphologically normal fry after abnormal cleavage as 3, 5, 6, 7, 9, and 10 blastomeres at the 2- to 4-cell stage. It was described that ploidy level of A. ruthenus larvae from abnormally divided embryos (abnormally divided embryos = AD embryos) was n/2n, and A. gueldenstaedtii x A. ruthenus hybrids had 2n/3n mosaic ploidy. That was the reason of fusion of one sperm pronucleus with the egg pronucleus generating a zygote, and the accessory sperm pronucleus or pronuclei developed separately from the zygote (Iegorova et al. 2018).
Based on these characteristics of sturgeon, we tested the possibility of producing a polyspermic interspecific hybrid by using gametes from three parents.
Broodfish and gamete collection
Two A. baerii females, two A. gueldenstaedtii females, four A. ruthenus males, three A. gueldenstaedtii males, and one A. baerii male were held in 4 m3 tanks at 15 °C in the hatchery of the Research Institute of Fish Culture and Hydrobiology in Vodnany, Czech Republic. To induce spermiation, sturgeon males were injected with acetone-dried homogenized carp pituitary extract (CPE) at a dose of 4 mg/kg body weight (BW). Using a catheter, sperm was collected from the urogenital papilla at 42 h post-injection, transferred to a 250 ml cell-culture container, and stored at 4 °C until fertilization. Sperm concentration was estimated using a Burker cell hemocytometer (Meopta, Czech Republic) at 20× magnification on a Nikon ECLIPSE Ci-S phase contrast microscope (Nikon, Japan). Ovulation in sturgeon females was induced with two doses of CPE by intramuscular injection: the first was given 36 h before stripping (0.5 mg/kg BW) and the second 24 h before stripping (4.5 mg/kg BW) (Dettlaff et al. 1981). Ovulated eggs were collected by microsurgical incision of oviducts as described by Podushka (1999).
Fertilization and identification of AD embryos
Four separate fertilizations were completed: (1) Eggs from one A. baerii (4n) female were fertilized with a mixture of sperm from one A. ruthenus (2n) and one A. gueldenstaedtii (4n) male; (2) eggs from one A. baerii (4n) female were fertilized with a mixture of sperm from one A. ruthenus (2n) and one A. gueldenstaedtii (4n) male; (3) eggs from one A. gueldenstaedtii (4n) female were fertilized with a mixture of sperm from one A. baerii (4n) and one A. ruthenus (2n) male; and (4) eggs from one A. gueldenstaedtii (4n) female were fertilized with a mixture of sperm from one A. gueldenstaedtii (4n) and one A. ruthenus (2n) male. Approximately 450 eggs (10 g) were fertilized for each group, using 2.3312 109 spz × 1 ml. To remove the stickiness of the outer layer, fertilized eggs were treated with 0.1% tannic acid solution (three times in 10 min). Embryos with 3, 5, 6, 7, 9, and 10 blastomeres at the 2- to 4-cell stage were considered as AD embryos. AD embryos and normally dividing embryos were separated and counted.
Survival and ploidy analysis
Embryos were incubated in dechlorinated water at 15 ± 1 °C. After 14 days, survival was noted and ploidy level of 25 larvae, developed from ADs, were randomly chosen from all four groups in total and evaluated by flow cytometry (Paa Partec CCA I; Partec GmbH, Münster, Germany) using 4′, 6-diamidino-2-phenylindole (CyStain DNA 2step kit; Partec GmbH) according to manufacturer’s instructions. Normally developed hybrids were used as controls, and their expected ploidy was 3n and 4n.
Microsatellite genotyping
To prove an existence of several interspecific spermatozoa in the egg, five randomly chosen AD larvae were processed for microsatellite genotyping: two larvae from A. baerii x A. ruthenus x A. gueldenstaedtii: AD1 and AD2; two larvae from A. gueldenstaedtii x A. baerii x A. ruthenus fertilizations: AD3 and AD4; and one larva from A. gueldenstaedtii x A. gueldenstaedtii x A. ruthenus group: AD5. Genomic DNA was extracted using a DNA extraction kit (GenElute Mammalian Genomic DNA Miniprep Kit; Sigma-Aldrich®) according to manufacturer’s instructions. Presence of multiple spermatozoa in AD larvae was estimated by parentage-like assignment using seven informative microsatellite markers, i.e., AfuG_135 (Welsh et al. 2003), Aox_27, Aox_45 (King et al. 2001), Spl_101, Spl_107, Spl_163, and Spl_173 (McQuown et al. 2000). Amplification was carried out according to the protocol described by Havelka et al. (2013). Microsatellite fragment analysis was performed on an Applied Biosystems SeqStudio Genetic Analyzer using a GeneScan LIZ 600 size standard (Applied Biosystems), and genotypes were scored in GENEIOUS 8.1.9, using a Microsatellite Plugin 1.4.4. The complexity of the duplicated sturgeon genome and the nature of current microsatellite genotyping make it impossible to reliably determine allele dosage behind a specific peak. Hence, peak patterns were treated as dominant data and interpreted as “allele phenotypes” (Rodzen et al. 2004). In several cases, we were able to estimate a real genotype at a fully heterozygote loci. Alleles that were not mutually shared by males, and by males and female were identified private alleles and tracked in allele phenotypes of AD larvae.
Results
Survival rate of three species hybrids
AD embryos were detected in all groups. The percentage of abnormally divided embryos ranged from 5 to 10% in each group. Hatching was observed at 10 dpf (days post-fertilization). Survival of abnormally cleaved embryos was up to 49% at 4 dpf; 15–42% at 7 dpf; and 15–27% of AD survived to 14 dpf. Survival of normally cleaved embryos was 77–90% at 4 dpf; 76–88% at 7 dpf; and 76–81% by 14 dpf (Table 1, Fig. 1).
Demonstration of 22-day post-fertilization larvae, (dorsal and ventral view) from the combination: A. baerii eggs x mix of sperm from A. ruthenus and A. gueldenstaedtii. a, b Produced three species larvae. Scale bar, 2 mm. c, d A. gueldenstaedtii larva. Scale bar, 2 mm. e, f A. baerii larva. Scale bar, 2 mm. g, h A. ruthenus larva. Scale bar, 1 mm
Ploidy level of obtained AD embryos
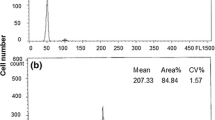
From 25 analyzed larvae, 88% of AD embryos were mosaic: 44% had 2n/3n ploidy (three-species progenies) (Fig. 2a), 40% had 2n/4n ploidy (polyspermic A. baerii x A. gueldenstaedtii larvae), and 4% had 2n/5n ploidy (unexpected ploidy). The remaining 12% of AD embryos consisted of 4% of diploids, 4% of triploids, and 4% of tetraploids (Table 1). Controls contained triploids and tetraploids, as it was expected (Fig. 2b).
Microsatellite genotyping
Fertilization A. baerii x A. ruthenus x A. gueldenstaedtii. Allele phenotypes of sample AD1 showed the presence of private alleles of both A. gueldenstaedtii male and A. ruthenus male at five informative loci AfuG_135, Aox_27, Aox_45, Spl_101, and Spl_107. At the most informative locus Aox_45, the specimen had five alleles clearly showing that the specimen originated by five genomes, two from diploid oocyte (with genotype 164/167) of tetraploid A. baerii female, two from diploid spermatozoon (with genotype 134/158) of tetraploid A. gueldenstaedtii male, and one from haploid spermatozoon (with genotype 155) of diploid A. ruthenus male. It clearly shows that specimen AD1 originated from heterospecific polyspermy (Table 2).
Allele phenotypes of the second specimen in this group (AD2) had only private alleles transmitted from A. gueldenstaedtii male with no private alleles from A. ruthenus male. At two informative loci AfuG_135 and Aox_27, the specimen presented three phenotypic alleles transmitted from A. gueldenstaedtii male (Table 2). It shows that the specimen originated from conspecific polyspermy.
Fertilization A. gueldenstaedtii x A. baerii x A. ruthenus. Allele phenotypes of specimen AD3 showed the presence of private alleles of both A. baerii male and A. ruthenus male at four informative loci, i.e., Aox_45, Spl_101, Spl_163, and Spl_173. On contrary, allele phenotypes of the second specimen in this group had only private alleles derived from A. ruthenus male with no private alleles from A. baerii male. At the most informative locus Spl_173, the specimen AD3 inherited phenotypic allele 254 from diploid oocyte (genotype 254/254) of tetraploid A. gueldenstaedtii female, alleles 266 and 270 from diploid spermatozoon of tetraploid A. baerii male, and one allele 236 from haploid spermatozoon of diploid A. ruthenus male (Table 3). Specimen AD4 inherited two alleles 254 and 258 from diploid oocyte of tetraploid A. gueldenstaedtii female and two alleles 236 and 266 from two haploid spermatozoa of diploid A. ruthenus male (Table 3). It shows that the specimen AD3 originated from heterospecific polyspermy while the specimen AD4 from conspecific polyspermy.
Fertilization A. gueldenstaedtii x A. gueldenstaedtii x A. ruthenus. Allele phenotypes of the specimen AD5 showed the presence of private alleles of both A. gueldenstaedtii male and A. ruthenus male at three informative loci, i.e., AfuG_135, Aox_45, and Spl 163. The most informative was locus Aox_45. At this locus, the specimen AD5 had five alleles clearly showing that the specimen originated from five genomes, i.e., two from diploid oocyte (with genotype 140/158) of tetraploid A. gueldenstaedtii female, two from diploid spermatozoon (with genotype 134/176) of tetraploid A. gueldenstaedtii male, and one from haploid spermatozoon (with genotype 146) of diploid A. ruthenus male (Table 4). It clearly showed that specimen AD5 originated from heterospecific polyspermy. Other loci were not fully informative for the estimation of polyspermy; however, no locus was contradictory to the conclusion.
Discussion
This study is a first report about three interspecific parent fertilization in the animal kingdom. Here, we demonstrate sturgeon fertilization characteristics, great plasticity for hybridization between species and survival.
Our previous research described physiological polyspermy in sturgeons. Cytologically analyzed A. ruthenus and A. baerii embryos demonstrated high number of spermatozoa in the cytoplasm right after fertilization and that number of supernumerary spermatozoa is significantly decreasing before first cleavage. At the same time, sometimes, additional spermatozoa were participating in the development in a way “karyogamy with additional plasmogamy.” A sperm nucleus or nuclei destined to form an additional blastomere began developing independently and probably 1 cycle later than a zygote (Iegorova et al. 2018). Our present work demonstrates that no significant intervention was needed to generate interspecific three-parent hybrids, where two species gave rise to a hybrid zygote and the third species contributed supplementary sperm-derived cells.
In Fig. 3, we list all possible variations that could be obtained using gametes from three species (as an example fertilization of A. baerii (4n) eggs with a mixture of sperm from A. ruthenus (2n) and A. gueldenstaedtii (4n) as an example). Monospermic fertilization of A. baerii egg by a single A. ruthenus spermatozoon would produce a normally divided triploid embryo. If A. baerii egg was fertilized by a single A. gueldenstaedtii spermatozoa, the embryo should be a normally divided tetraploid. Due to multiple micropyles, A. baerii egg could be fertilized by several spermatozoa. Polyspermic fertilization of A. baerii egg with several A. ruthenus spermatozoa should result in abnormally divided mosaics with ploidy level 1n/3n. Polyspermic fertilization of A. baerii egg by several A. gueldenstaedtii spermatozoa would produce an abnormally divided embryo with a mosaic ploidy of 2n/4n. Using gametes from three parents, and fertilizing A. baerii egg with A. gueldenstaedtii spermatozoa and A. ruthenus spermatozoa, would produce either 2n/3n or 1n/4n mosaic hybrid. The 2n/3n mosaics would be created if the A. ruthenus sperm pronucleus fused with A. baerii egg pronucleus (3n) and the accessory A. gueldenstaedtii sperm pronucleus developed singly as 2n. However, 1n/4n mosaics would appear if A. gueldenstaedtii sperm pronucleus fused with A. baerii egg pronucleus (4n) and the accessory A. ruthenus sperm pronucleus developed singly as 1n.
In our study, 88% of AD embryos displayed mosaic ploidy: 44% of polyspermic embryos from three species: A. baerii x A. gueldenstaedtii x A. ruthenus, 40% of polyspermic embryos from two species: A. baerii x A. gueldenstaedtii, and 4% of 2n/5n embryos, which could be an unexplained fusion of pronuclei. Triploids and tetraploids could be obtained due to asymmetrical division of blastomeres, as described by Dettlaff et al. (1993), which we could consider as AD embryos. Their ploidy suggests that they were monospermic progeny from two species. Besides mosaics, triploid and tetraploid was detected one diploid fish. Probably it was occurred due to androgenotes: one decondensed sperm nuclei from A. baerii or A. gueldenstaedtii became a male pronuclei and continued development without fusion with female pronucleus and produced diploid larva. Successful amplification of DNA fragments from three sturgeon species proved the heterospecific polyspermy: three larvae contained alleles from three parents.
Survival of AD three parents’ hybrid embryos
During the last few decades, it was believed to be fatal if more than one spermatozoa fused with the oocyte: polyspermic embryos develop abnormally and perish before hatching or can develop into abnormal larvae that subsequently die. However, Iegorova et al. (2018) showed that polyspermic embryos had almost the same survival rate as controls. Our present work demonstrates that three-parent hybrids still have high survival, although the survival rate was lower than the control (27 vs 81% at 14 dpf), which indicated that probably some combinations of interspecific pronuclei can be lethal. Interestingly, we did not find any 1n/4n mosaics (A. baerii x A. gueldenstaedti hybrid with A. ruthenus accessory sperm pronuclei) in our trials, which could represent a lethal combination.
Do AD embryos produce clonal gametes?
Morishima et al. (2004) have shown a diploid/triploid loach (Misgurnus anguillicaudatus) males produce clonal unreduced diploid spermatozoa. When normal diploid female was crossed with mosaic diploid/triploid male, only triploid progeny appeared and exhibited microsatellite genotypes with two alleles identical to the clonal genotype and one allele derived from female. When UV-irradiated eggs were fertilized by diploid spermatozoa from the mosaic male, they gave rise to the occurrence of androgenetic diploids and they exhibited microsatellite genotypes and DNA fingerprints, absolutely identical to those of the natural clones. These results of Morishima et al. (2004) clearly concluded that the diploid-triploid mosaic male generate clonal diploid spermatozoa, with genetically identical genotypes.
In our previous research (Iegorova et al. 2018), we produced mosaic 1n/2n sterlet (A. ruthenus). We found that haploid cells were distributed among all germ layer derivatives, including gonads. Hypothetically, additional blastomeres produced by supernumerary spermatozoa exhibit the paternal genome exclusively. Thus, if these haploid-derived germ cells produce gametes, the gametes must carry only the paternal genome, and all haploid gametes will be clonal. If AD fish can produce clonal gametes, induction of multiple-sperm mosaicism might be a useful tool for production of isogenic strains in sturgeons in a short time.
Conclusion
Our findings indicated that sturgeons present a developmental pattern unique in the animal kingdom. This study clearly refutes statements about fatality of organisms if more than one spermatozoa fertilizes the oocyte, especially when gametes belong to interspecific animals. Here, we demonstrate a great plasticity in sturgeon hybridization, and easy ploidy manipulations, which could be an important strategy in aquaculture for mass production of clonal gametes.
References
Bemis WE, Grande L (1997) An overview of Acipenseriforms. Environ Biol Fish 48:25–71
Billard R, Lecointre G (2001) Biology and conservation of sturgeon and paddlefish. Rev Fish Biol Fish 10:355–392
Birstein VJ, Vasiliev VP (1987) Tetraploid-octaploid relations and karyologycal evolution in the order Acipenseriformes (Pieces). Karyotypes, nucleoli, and nucleolus organized regions in four acipenserid species. Genetica 72:3–12
Birstein VJ, Hanner R, DeSalle R (1997) Phylogeny of the Acipenseriformes: cytogenetic and molecular approaches. Environ Biol Fish 48:127–155
Boveri T (1901) Zellen-Studien: Über die Natur der Centrosomen, vol 28. Fisher Z Med Naturw, Jena, pp 1–220
Carmona R, Domezain A, Garcia-Gallego M, Hernando JA, Rodriguez F, Ruiz-Rejon M (2009) Biology,conservation and sustainable development of sturgeons. Springer Science + Business Media B. V.
Debus L, Winkler M, Billard R (2002) Structure of micropyle surface on oocytes and caviar grains in sturgeons. Int Rev Hydrobiol 87(5–6):685–603
Dettlaff TA, Vassetzky SG (1991) Animal species for developmental studies. Volume 2. Vertebrates. Consultants Bureau - NY and London, pp 22–23
Dettlaff TA, Ginsburg AS, Schmalhausen OI (1981) Development of sturgeon fishes (Razvitie osetrovyh ryb) in Russian. Moscow "Nauka" pp 77–91
Dettlaff TA, Ginsburg AS, Schmalhausen OI (1993) Sturgeon fishes. Developmental Biology and Aquaculture, Berlin; NY: Springer-Verlag, pp 24–28
Fontana F (1994) Chromosomal nucleolar organizer regions in four sturgeon species as markers of karyotype evolution in Acipenseriformes (Pisces). Genome 37(5):888–892
Fontana F, Colombo G (1974) The chromosomes of Italian sturgeons. Experientia 30:739–742
Havelka M, Kašpar V, Hulák M, Flajšhans M (2011) Sturgeon genetics and cytogenetics: a review related to ploidy levels and interspecific hybridization. Folia Zool 60(2):93–103
Havelka M, Hulak M, Bailie DA et al (2013) Extensive genome duplication in sturgeons: new evidence from microsatellite data. J Appl Ichthyol 29:704–708
Iegorova V, Psenicka M, Lebeda I, Rodina M, Saito T (2018) Polyspermy produces viable haploid/diploid mosaics in sturgeon. Biol Reprod. https://doi.org/10.1093/biolre/ioy092
Kim DS, Nam YK, Noh JK, Park CH, Chapman FA (2005) Karyotype of north American shortnose sturgeon Acipenser brevirostrum with the highest chromosome number in Acipenseriformes. Ichthyol 52:94–97
King TL, Lubinski BA, Spidle AP (2001) Microsatellite DNA variation in Atlantic sturgeon Acipenser oxyrinchus: and cross-species amplification in the Acipenseridae. Conserv Genet 2:103–119
Ludwig A, Belfiore NM, Pitra C, Svirsky V, Jenneckens I (2001) Genome duplication events and functional reduction of ploidy levels in sturgeon (Acipenser, Huso and Scaphirhynchus). Genetics 158:1203–1215
McQuown EC, Sloze BL, Sheehan RJ et al (2000) Microsatellite analysis of genetic variation in sturgeon (Acipenseridae): new primer sequences for Scaphirhynchus and Acipenser. Trans Am Fish Soc 129:1380–1388
Morishima K, Oshima K, Horie S, Fujimoto T, Yamaha E, Arai K (2004) Clonal diploid sperm of the diploid-triploid mosaic loach, Misgurnus anguillicaudatus (Teleostei:Cobitidae). J Exp Zool A Comp Exp Biol 301(6):502–511
Podushka SB (1999) New method to obtain sturgeon eggs. J Appl Ichthyol 15(4–5):319
Rodzen JA, Famula TR, May B (2004) Estimation of parentage and relatedness in the polyploid white sturgeon (Acipenser transmontanus) using a dominant marker approach for duplicated microsatellite loci. Aquaculture 232:165–182
Vasil’ev VP (2010) How many times has polyploidization occurred during acipenserid evolution? New data on the karyotypes of sturgeons (Acipenseridae, Actinopterygii) from the Russian Far East. J Ichthyol 10:950–959
Wang WH, Day BN, Wu GM (2003) How does polyspermy happen in mammalian oocytes. Microsc Res Tech 61:335–341
Welsh AB, Blumberg M, May B (2003) Identification of microsatellite loci in lake sturgeon, Acipenser fulvescens, and their variability in green sturgeon, A. medirostris. Mol Ecol Notes 3:47–55
Zalenskii VV (1878) Evolution of Acipenser ruthenus development. In Russian (Istoriya razvitiya sterlyadi (Acipenser ruthenus)). Kazan: Typography of the Imperial University 7(3), pp 18–30, part 1 Embryonic development (Embryonalnoye razvitiye)
Acknowledgments
We thank all members of the Laboratory of Germ Cells, Faculty of Fisheries and Protection of Waters, University of South Bohemia in Ceske Budejovice for their support.
Funding
This study was supported by the Ministry of Education, Youth and Sports of the Czech Republic, projects CENAKVA (CZ.1.05/2.1.00/01.0024), “CENAKVA II” (LO1205 under the NPU I program), the Grant Agency of the University of South Bohemia in Ceske Budejovice (125/2016/Z), by the Czech Science Foundation (17-19714Y), and project Biodiversity (CZ.02.1.01/0.0/16_025/0007370).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
All experimental procedures were performed in accordance with national and institutional guidelines on animal experimentation and care and were approved by the Animal Research Committee of the Faculty of Fisheries and Protection of Waters in Vodnany, Czech Republic.
Rights and permissions
About this article
Cite this article
Iegorova, V., Havelka, M., Psenicka, M. et al. First evidence of viable progeny from three interspecific parents in sturgeon. Fish Physiol Biochem 44, 1541–1550 (2018). https://doi.org/10.1007/s10695-018-0553-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10695-018-0553-6