Abstract
A high colorectal cancer (CRC) incidence is observed in Tunisia, with a relatively high proportion of patients developing CRC before the age of 40. While this suggests a genetic susceptibility, only a few Tunisian Lynch Syndrome families have been described. In this study we aimed to identify the underlying genetic cause in 32 patients with early onset CRC and/or a positive family history. Of twenty-four patients’ tumor or biopsies could be analyzed with immunohistochemical staining to detect loss of expression of one of the MMR proteins. Ten tumors showed loss of expression, of which one tumor was from a patient where a germline pathogenic MSH2 variant was detected previously with Sanger sequencing. Next generation sequencing of the MMR, POLE and POLD1 genes was performed in leukocyte and tumor DNA of the remaining nine patients, as well as in two patients with MMR-proficient tumors, but with severe family history. In six of 11 patients a germline variant was detected in MLH1 (n = 5) or MSH2 (n = 1). Two of six patients were from the same family and both were found to carry a novel in-frame MLH1 deletion, predicted to affect MLH1 function. All MLH1 variant carriers had loss of heterozygosity with retention of the variant in the tumors, while a somatic pathogenic variant was detected in the patient with the germline MSH2 variant.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Lynch syndrome (LS;[MIM] 120435), is an autosomal dominant disease with early onset of colorectal cancer (CRC) and other extracolonic cancers including endometrium-, stomach-, small intestine-, hepatobiliary tract-, urinary tract-, ovarian-, brain- and skin cancer [1]. LS is caused by heterozygous pathogenic germline variants in the DNA mismatch repair (MMR) genes: MLH1 (MIM 120436), MSH2 (MIM 609309), MSH6 (MIM 600678) and PMS2 (MIM 600259) [2, 3]. The majority of causative germline variants are located in MLH1 and MSH2 (50% and 40% respectively), while 10% of variants are located in MSH6 and PMS2 [4]. Additionally, deletions in EPCAM (MIM 185535) have been described to result in hypermethylation of the MSH2 promoter and subsequent silencing of MSH2 [5].
The inactivation of the MMR pathway can also be caused by somatic events, such as MLH1 promoter hypermethylation or bi-allelic somatic mutations in MMR genes [6,7,8]. MLH1 promoter hypermethylation is present in 15% of sporadic CRCs and is more frequent in older patients without family history of CRC [9].
Few studies of CRC in Tunisia are reported and little is known about the underlying genetics [10]. The incidence of CRC in Tunisia in 2009 was about 12.4/100.000 inhabitants [11]. A higher CRC rate has been described in European countries such as Danish population where CRC has an incidence of 160/100.000 [12]. However, since a consistent nationwide screening program is lacking, this number is likely underestimated and is predicted to more than double in 2024 [11]. Furthermore, a relatively high proportion of patients develop tumors before the age of 40 years (14.2%), suggesting a genetic susceptibility [10].
Until now, only few Tunisian LS families were described. A 2011 study sequencing the MMR genes in 31 unrelated Tunisian families suspected of LS based on age of onset or positive family history, found germline MMR variants in 52.6% of male and 8.3% of female patients tested, leaving a large fraction of patients unexplained [10]. Previously, bi-allelic somatic inactivation of the MMR genes and germline variants in other CRC susceptibility genes, as POLE and POLD1, have been described in suspected Lynch Syndrome patients, but this has never been tested in a Tunisian population and prevalence of these variants in Tunisia remains unknown [8, 13]. The aim of the current study was to identify the underlying genetic cause in 24 Tunisian index patients with suspected LS, early-onset CRC and/or a positive family history. To this end, next-generation sequencing of the MMR genes and POLE and POLD1 was performed.
Materials and methods
Cohort description
In this study, 32 patients suspected for Lynch Syndrome from 29 different Tunisian families were investigated. All patients developed colorectal cancer, with an average onset age of 43.2 years (range 19–75 years, Table 1). Leukocyte- and tumor DNA was available for 24 patients (75%) and tumor DNA of these patients was isolated either from biopsies (n = 6) or formalin-fixed paraffin-embedded (FFPE, n = 18) tissue blocks. Of the remaining cases (n = 8) FFPE and biopsies were not available. Patients were recruited between 2009 and 2017 from University Hospital Farhat Hached—Sousse (n= 12), University Hospital Sahloul—Sousse (n= 6), University Hospital Fatouma Bourguiba—Monastir (n= 2), University Hospital Tahar Maamouri—Nabeul (n= 1) and private gastro-intestinal doctors (n = 3).
Available information concerning sex, age of onset, site and size of the tumor and CRC familial history were verified. The majority of patients tested (72%) had a first-degree relative with a Lynch-associated tumor and 26 of 32 patients were from families fulfilling Amsterdam II criteria (Table 1).
Sanger sequencing
First, 18 patients were pre-screened with Sanger Sequencing for variants in MLH1 and MSH2 (Supplementary Table 1). Sequencing was done as previously described [14]. Primers are shown in Supplementary Table 2.
Immunohistochemical staining
Immunohistochemical staining (IHC) of the four MMR proteins (MLH1, MSH2, MSH6 and PMS2) was performed on available FFPE tissue blocks or paraffin-embedded biopsies (n = 24, Supplementary Table 1), using a previously described standard protocol [15].
Next-generation sequencing
Total DNA was isolated from all FFPE-tissue blocks or biopsies [16] that showed expression loss of at least one of the MMR proteins by immunohistochemistry (n = 11) with the tissue preparation system [17]. DNA concentration was measured with the Qubit® 3.0 fluorometer. Of all patients with MMR-deficient tumors, leukocyte (n = 11) and tumoral DNA (n = 10) was sequenced with the Ion Proton System (Life Technologies, Carlsbad, CA, USA) using a custom MMR panel [8]. Libraries were prepared with Ion AmpliSeq™ Library Kit 2.0 according to the manufacturer’s protocol. The Proton sequencer generated unaligned BAM which were mapped against the human reference genome (GRCh37/hg19) using the TMAP 5.0.7 software with default parameters (https://github.com/iontorrent/TS).
All variants were filtered on minor allele frequency (< 0.05), variant frequency (> 10%) and coverage (> 50x). The following Genbank reference sequences were used: NM_000249.3 for MLH1, NM_000251.2 for MSH2, NM_000179.2 for MSH6, NM_000535.5 for PMS2, NM_006231.2 for POLE and NM_001256849.1 for POLD1. In silico analysis of all variants was done using Mutation Taster [17], Polyphen 2 [18] and UniProt [19].
Variants classification was done according to the InSiGHT MMR gene variant classification criteria (https://www.insight-group.org). Variants were classified as: Class 5: pathogenic, Class 4: likely pathogenic, Class 3: Variant of uncertain significance (VUS), Class 2: Likely not pathogenic/little clinical significance, Class 1: Not pathogenic/low clinical significance.
Results
Immunohistochemical staining was performed on sections from all available biopsies (n = 6) and formalin-fixed paraffin-embedded (FFPE) tissue blocks (n = 18). Thirteen tumors/biopsies (54%) showed positive staining of all four MMR proteins, whereas 10 tumors (42%) showed loss of expression of MLH1/PMS2 (n = 6) or MSH2/MSH6 (n = 4). In the final patient staining results were inconclusive due to poor quality of the tumor tissue.
Sanger sequencing was firstly performed on 18 patients, with a pre-screening for some germline exonic variants in MLH1 and MSH2. Two patients were found to carry a germline variant, either a nonsense variant (MSH2 c.1413dupA, patient TUN4) or a variant of uncertain significance (MLH1 c.218T > C, patient TUN13), while the rest tested negative for germline variants.
Tumor DNA was isolated from 9 of 10 MMR-deficient tumors, as well as two MMR-proficient tumors (TUN3 and TUN7) with MMR-proficient tumors, but with severe family history (Supplemental Fig. 1). NGS of the four MMR genes, POLE and POLD1 was performed on tumor- and normal DNA of all 11 patients.
With NGS, a germline variant was detected in the MMR gene that showed expression loss with IHC (Table 2) in six patients. Patient TUN25 and TUN26 are related (Fig. 1), and in both patients a germline in-frame MLH1 deletion was detected. The remaining four detected variants were a nonsense variant (n = 1) and non-synonymous missense variants of uncertain significance (VUS, n = 3). All missense variants showed loss of heterozygosity (LOH) in the tumor, while the patient with the nonsense variant (TUN28), was found to carry a second somatic nonsense variant (MSH2 c.2557G > T) explaining the tumor phenotype. Patient TUN19 and TUN30 were found to carry the same MLH1 VUS (c.218 T > C), however, patients were not related. In the two tested patients with MMR- proficient tumors (TUN3 and TUN7), no germline or somatic MMR variants were detected. Additionally, no POLE or POLD1 exonuclease domain variants were found.
Pedigrees of MMR variant carriers. Pedigrees of patients with germline MMR variants. CRC colorectal cancer, BrC breast cancer, Uter uterine cancer, StC stomach cancer, CHC hepatocarcinoma. Age at diagnosis is mentioned with the type of the cancer; circles represent females; squares represent males; diamonds represent undisclosed gender; cross striped individuals are deceased. Filled symbols represent affected family members (CRC), while the open symbols are unaffected family members black right upper corner presents family member with Uter cancer, black left upper corners presents family member with StC, black right lower corner presents family member with BrC. Index patients are indicated with the arrow
Discussion
The incidence of colorectal cancer (CRC) in Tunisia is increasing and is expected to rise to 39.3/100.000 patients in 2024 [11]. However, even though a high proportion (14.2%) of these CRC patients are under 40, indicating a possible genetic susceptibility, only a few Tunisian Lynch Syndrome (LS) families have been described [10].
In the current study, DNA from 32 patients from 29 families was collected to test for germline or somatic variant in the MMR genes, but also in POLE and POLD1. Pre-screening of leukocyte DNA of 18 patients only detected a pathogenic variant in one patient (TUN4) and a variant of uncertain significance (VUS) in a second patient (TUN13). The pathogenic variant (MSH2 c.1413dupA), is a frameshift variant resulting in a premature stop codon in exon 9, previously described in multiple Lynch Syndrome families. The patient carrying this mutation (TUN4) was a male with a 10 cm sigmoid tumor appearing at the age of 49 years (Supplemental Table 1). The sigmoid tumor of patient TUN13 showed loss of MSH2 and MSH6 staining with immunohistochemistry (IHC). No material was available of other family members.
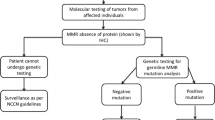
Immunohistochemical staining of MLH1, MSH2, MSH6 and PMS2 in 24 tumors and biopsies showed loss of expression in ten patients (from nine families). Next generation sequencing (NGS) was performed on leukocyte- and tumor DNA for nine of ten patients (except TUN4), with MMR-deficient tumors, as well as two patients TUN3 and TUN7, (Supplemental Fig. 1) with MMR-proficient tumors but with a strong family history of CRC. Patient TUN4 was not screened with NGS, since a pathogenic variant was already detected with Sanger in the gene that showed immunohistochemical loss of expression (Fig. 2).
With NGS, six germline variants were detected in the gene that showed immunohistochemical expression loss. One variant was detected before with Sanger (MLH1 c.218T > C). A MLH1 c.1918C > A variant was found in two obviously unrelated patients (TUN19 and TUN30) who were not pre-screened with Sanger Sequencing. While both variants are still classified as class 3 (variant of uncertain significance) by the LOVD database according to InSiGHT classifications, previous studies have shown partial loss of MMR for both variants [20]. In silico analysis of the MLH1 c.1918C > A predicted a damaging effect, through affecting the interaction with the Exonuclease I (EXO1) protein, and a previous study shows the variant results in only 53% MMR activity [20]. The MLH1 c.218T > C variant is also predicted to affect function, and a previous study shows 34–66% loss of MMR function [21]. The patients’ affected sister (CRC22, Fig. 1), was tested for the variant and was also found to be a carrier, supporting possible pathogenicity of the variant.
Patients TUN25 and TUN26 (two sisters) were found to carry the same novel MLH1 c.1940_1951delTGCCCCCTTTGG in-frame deletion. Both showed loss of heterozygosity with retention of the variant in the tumor. In silico analysis predicts loss of the MLH1-EXO1 interaction. Known missense variants within this region (c.1942C > T, c.1943C > T) have been described to be pathogenic and have been shown to result in altered protein localization [22, 23]. Patient TUN28 was found to carry a germline MSH2 c.1255C > T nonsense variant, with an additional somatic MSH2 c.2557G > T nonsense variant in the tumor. This is a known pathogenic variant, previously described in a Dutch population [24].Unexpectedly, the patients affect sister (CRC34), did not carry this variant.
In three of 10 patients (TUN8, TUN11 and TUN15) with MMR-deficient tumors no germline or somatic MMR, POLE or POLD1 variant was detected. Interestingly, the average age of onset of these three patients was 26,6 years (19–37 years), and two of three were from Amsterdam II positive families. This strongly suggests that there is an underlying genetic cause in these families, which has currently not been detected.
In this study, two assays were performed on somatic tumor cells sections or DNA to perform IHC and NGS. They represent robust and complemented techniques for a direct suspicion and confirmation of the right diagnosis. Together they allow a major gain in time and cost.
A recent study, published by Hampel H et al. in 2018, showed that NGS sequencing of tumor DNA can replace all current standard tests, including those used universally for tumor screening in LS [25].
In 13 index patients no loss of expression was detected in any of the MMR genes. While Lynch Syndrome is unlikely in these patients, an underlying genetic cause cannot be excluded. Four of 13 families fulfilled Amsterdam II criteria, and could therefore be classified as familial colorectal type X (FCCTX) families, for which currently no one underlying gene defect is found [26,27,28]. Previous studies found variants in SEMA4A, ACVRLK3 and SETD6 that could possibly explain a small fraction of these patients, but the majority of FCCTX patients remain unexplained [29]. Furthermore, germline variants in the exonuclease domain of POLE and POLD1 have been described to cause polymerase associated polyposis syndrome, in which patients predispose to early onset colorectal cancer, often with polyps [30,31,32]. While these tumors are often microsatellite stable, secondary MMR variants resulting in MMR loss also have been described in PPAP (Polymerase Proofreading-Associated Polyposis) patients, and the phenotype remains broad [8, 13]. For this reason in this study, three families with MMR- proficient tumors, but with a severe family history (Supplemental Table 1) were tested for variants in these genes, but no genetic variant was detected. Implementation of a large gene panel or whole exome sequencing could possibly find the genetic cause in these patients [33].
In summary, with a custom NGS panel and Sanger sequencing we were able to detect a germline MMR variant in seven of ten patients with MMR-deficient tumors. In 13 patients no MMR-deficiency was found in the tumor and the underlying genetic cause remains unknown.
References
Lynch HT, de la Chapelle A (2003) Hereditary colorectal cancer. N Engl J Med 348:919–932
Leach FS, Nicolaides NC, Papadopoulos N et al (1993) Mutations of a mutS homolog in hereditary nonpolyposis colorectal cancer. Cell 75:1215–1225
Nicolaides NC, Papadopoulos N, Liu B et al (1994) Mutations of two PMS homologues in hereditary nonpolyposis colon cancer. Nature 371:75–80
Peltomäki P (2001) Deficient DNA mismatch repair. Hum Mol Genet 7:735–740
Ligtenberg MJ, Kuiper RP, Chan TL et al (2009) Heritable somatic methylation and inactivation of MSH2 in families with Lynch syndrome due to deletion of the 3’ exons of TACSTD1. Nat Genet 41:112–117
Mensenkamp AR, Vogelaar IP, van Zelst-Stams WA et al (2014) Somatic mutations in MLH1 and MSH2 are a frequent cause of mismatch-repair deficiency in Lynch syndrome-like tumors. Gastroenterology 146(643–646):e8
Geurts-Giele WR, Leenen CH, Dubbink HJ et al (2014) Somatic aberrations of mismatch repair genes as a cause of microsatellite-unstable cancers. J Pathol 234:548–559
Jansen AM, Van Wezel T, Van den Akker BE et al (2016) Combined mismatch repair and POLE/POLD1 defects explain unresolved suspected Lynch Syndrome cancers. Eur J Hum Genet 24:1089–1092
Herman JG, Umar A, Polyak K et al (1998) Incidence and functional consequences of hMLH1 promoter hypermethylation in colorectal carcinoma. Proc Natl Acad Sci USA 95:6870–6875
Moussa SA, Moussa A, Kourda N et al (2011) Lynch syndrome in Tunisia: first description of clinical features and germline mutations. Int J Colorectal Dis 26:455–467
Khiari H, Ben Ayoub HW, Ben Khadhra H et al (2017) Colorectal cancer incidence trend and projections in Tunisia (1994–2024). Asian Pac J Cancer Prev 18(10):2733–2739
Bjerrum A, Andersen O, Fischer A et al (2016) Colorectal cancer mortality 10 years after a single round of guaiac faecal occult blood test (gFOBT) screening: experiences from a Danish screening cohort. BMJ Open Gastroenterol 3(1):e000120
Elsayed FA, Kets CM, Ruano D et al (2015) Germline variants in POLE are associated with early onset mismatch repair deficient colorectal cancer. Eur J Hum Genet 23:1080–1084
Mili A, Ben Charfeddine I, Amara A et al (2012) A c.3216_3217delGA mutation in AGL gene in Tunisian patients with a glycogen storage disease type III: evidence of a founder effect. Clin Genet 82(6):534–539
De Jong AE, van Puijenbroek M, Hendriks Y et al (2004) Microsatellite instability, immunohistochemistry, and additional PMS2 staining in suspected hereditary nonpolyposis colorectal cancer. Clin Cancer Res 10:972–980
van Eijk R, Stevens L, Morreau H et al (2013) Assessment of a fully automated high-throughput DNA extraction method from formalin-fixed, paraffin-embedded tissue for KRAS, and BRAF somatic mutation analysis. Exp Mol Pathol 94(1):121–125
Schwarz JM, Cooper DN, Schuelke M et al (2014) MutationTaster2: mutation prediction for the deep-sequencing age. Nat Methods 11(4):361–362
Adzhubei IA, Schmidt S, Peshkin L et al (2010) A method and server for predicting damaging missense mutations. Nat Methods 7(4):248–249
The UniProt Consortium (2017) UniProt: the universal protein knowledgebase. Nucleic Acids Res 45:D158–D169
Takahashi M, Shimodaira H, Andreutti-Zaugg C et al (2007) Functional analysis of human MLH1 variants using yeast and in vitro mismatch repair assays. Cancer Res 67(10):4595–4604
Ellison AR, Lofing J, Bitter GA (2004) Human MutL homolog (MLH1) function in DNA mismatch repair: a prospective screen for missense mutations in the ATPase domain. Nucleic Acids Res 32(18):5321–5338
Raevaara TE, Gerdes AM, Lönnqvist KE et al (2004) HNPCC mutation MLH1 P648S makes the functional protein unstable, and homozygosity predisposes to mild neurofibromatosis type 1. Genes Chromosom Cancer 40:261–265
Drost M, Je Zonneveld, van Dijk L et al (2010) A cell-free assay for the functional analysis of variants of the mismatch repair protein MLH1. Hum Mutat 31:247–253
Overbeek L, Kets CM, Hebeda KM et al (2007) Patients with an unexplained microsatellite instable tumour have a low risk of familial cancer. Br J Cancer 96:1605–1612
Hampel H, Pearlman R, Beightol M et al (2018) Assessment of tumor sequencing as a replacement for lynch syndrome screening and current molecular tests for patients with colorectal cancer. JAMA Oncol 4(6):806–813
Lindor NM, Rabe K, Petersen GM et al (2005) Lower cancer incidence in Amsterdam-I criteria families without mismatch repair deficiency - familial colorectal cancer type X. Jama-J Am Med Assoc 293:1979–1985
Balmana J, Castells A, Cervantes A et al (2010) Familial colorectal cancer risk: ESMO clinical practice guidelines. Ann Oncol 5:v78–v81
Shiovitz S, Copeland WK, Passarelli MN et al (2014) Characterisation of familial colorectal cancer Type X, Lynch syndrome, and non-familial colorectal cancer. Br J Cancer 111:598–602
Schulz E, Klampfl P, Holzapfel S et al (2014) Germline variants in the SEMA4A gene predispose to familial colorectal cancer type X. Nat Commun 5:5191
Briggs S, Tomlinson I (2013) Germline and somatic polymerase epsilon and delta mutations define a new class of hypermutated colorectal and endometrial cancers. J Pathol 230:148–153
Palles C, Cazier JB, Howarth KM et al (2013) Germline mutations affecting the proofreading domains of POLE and POLD1 predispose to colorectal adenomas and carcinomas. Nat Genet 45:136–144
Heitzer E, Tomlinson I (2014) Replicative DNA polymerase mutations in cancer. Curr Opin Genet Dev 24:107–113
Stoffel EM, Koeppe E, Everett J et al (2017) Germline genetic features of young individuals with colorectal cancer. Gastroenterology 154(4):897–905
Acknowledgements
We thank all team members of Human Cytogenetic, Molecular Genetics and Biology of Reproduction laboratory, Farhat HACHED Hospital Sousse-Tunisia, and the Molecular Diagnostics of the Pathology department of the Leiden University Medical Center. Also, we thank all patients and their families for their contribution and great help.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
10689_2019_130_MOESM1_ESM.tif
Supplementary material 1—Presentation of suspected Lynch Syndrome patients tested by NGS. Pedigrees showing candidates with detected variants by MMR Panel and the segregation of the CRC in their families. CRC: Colorectal Carcinoma, BrC: Breast Cancer, Uter: Uterine Cancer, StC: Stomach Cancer, CHC: Hepatocarcinoma, BrnC:Brain Cancer ,LeukC: Leukemia Cancer, LunC:Lung Cancer.Age at diagnosis is mentioned with the type of the cancer; circles represent females; squares represent males; diamonds represent undisclosed gender; cross striped individuals are deceased. Black blocks present patients and family members with CRC, black right upper corner presents family member with Uter cancer, black left upper corners presents family member with StC , black right lower corner presents family member with BrC,black left lower corner presents family member with BrnC, left Black half presents family member with LeukC, right black half presents family member with LunC (TIFF 117 kb)
Rights and permissions
About this article
Cite this article
Ben Sghaier, R., Jansen, A.M.L., Bdioui, A. et al. Targeted next generation sequencing screening of Lynch syndrome in Tunisian population. Familial Cancer 18, 343–348 (2019). https://doi.org/10.1007/s10689-019-00130-y
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10689-019-00130-y