Abstract
Hypoxia is a key limiting factor for seedling emergence and establishment of direct-seeded rice (DSR) under submergence. The anaerobic germination (AG) tolerance, therefore, is one of the most critical traits in DSR breeding programs. However, the genetic basis of AG is still unclear. In this study, 229 single segment substitution lines (SSSLs) were arranged in an augmented design in field planting. We evaluated the anaerobic germinability by measuring the SSSLs’ coleoptile length (CL) under submergence, and then identified AG QTLs. AG was a typical quantitative trait based on the results of five cropping seasons. We repeatedly detected 19 SSSLs with putative AG QTLs in at least two cropping seasons. Moreover, substitution mapping identified 11 integrated AG QTLs on nine chromosomes, including six positive and five negative QTLs with additive effect contributions ranging from 6.8 to 18.5%. Among the 11 QTLs, qAG4 and qAG11 were novel ones, qAG9 was cloned in a previous study, and the remaining eight QTLs overlapped with reported AG QTLs. Overall, we identified some genetic loci controlling AG in multiple environments, which may be useful for developing varieties suitable for DSR.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Rice (Oryza sativa L.) is an essential food crop for billions of people and is usually grown by transplanting in Asian countries. The shortages of fresh water and labor are compelling farmers to shift from transplanting to direct seeding, which is a more efficient and economical method (Angaji 2008; Sun et al. 2022). For example, the area of DSR has increased from 2% in 2007 to more than 44.6% in 2016 in China (Luo et al. 2019). However, DSR has a substantially higher risk of encountering unfavorable climates in the field than transplanted rice (Cao et al. 2018). Low temperature in early spring and hypoxia caused by flooding are the two main adverse environmental factors affecting the growth of DSR. Hypoxia during germination will lead to low emergence rate, weak seedlings, and lack of uniformity, and eventually decreased rice quality and yield.
It is proved that identifying AG QTLs and breeding new DSR varieties is an effective way to cope with early flooding stress in rice (Mondal et al. 2020). In previous studies, CL, as well as survival rate (SR), seedling height, and anoxic response index were used as phenotypes of flooding tolerance to explore the genetic basis of AG (Ma et al. 2020; Pucciariello 2020). Under anaerobic conditions caused by flooding, rice coleoptiles will elongate to the water surface and then transport oxygen to hypoxic tissues for starch mobilization (Magneschi and Perata 2009; Yu et al. 2021). The anaerobic adaptive capacity of rice during the germination stage depends on the growth rate and length of coleoptiles (Pucciariello 2020). Therefore, the morphological and physiological variation of coleoptile is an important indicator of rice AG tolerance trait in anaerobic environments.
In the past few years, CL has been widely used as a phenotypic trait to evaluate the flooding tolerance of rice seed. Several studies have been conducted and AG QTLs controlling CL have been mapped through approaches like genome-wide association study (GWAS), biparental mapping etc. (Hus et al. 2015; Nghi et al. 2019; Su et al. 2021; Yang et al. 2019; Zhang et al. 2017). In addition to CL, SR is also a phenotypic indicator broadly used to identify AG QTLs, especially in numerous studies conducted at International Rice Research Institute (Angaji 2008; Angaji et al. 2010; Ghosal et al. 2019, 2020; Kim and Reinke 2018; Septiningsih et al. 2013). qAG-9-2, a major AG QTL, explained 33.49% of the phenotypic variation. Its target gene was cloned as OsTPP7 (LOC_Os09g20390), a trehalose-6-phosphate phosphatase gene (Kretzschmar et al. 2015). OsTPP7 can promote starch mobilization to drive growth kinetics of the germinating embryo and elongating coleoptile to enhance AG tolerance. Recently, two GWAS studies cloned two key genes controlling AG in rice, OsGF14h (LOC_Os11g39540) and OsUGT75A (LOC_Os11g25990). OsGF14h encodes a 14-3-3 protein, and OsUGT75A encodes a UDP-glucosyltransferase, both of which mediate coleoptile elongation under flooding conditions through phytohormone pathways (ABA, GA, JA) (He et al. 2023; Sun et al. 2022).
SSSL contains a single homozygous marker-defined chromosome segment from a donor parent in a recurrent parent background (Zhang 2021). It is a permanent advanced mapping population due to its advantages of simple genetic background and repeatable cultivation compared to the traditional biparental mapping populations. Thus, SSSL shows higher efficiency and better accuracy in the genetic dissection of complex agronomic traits in crops. Recent studies have demonstrated that SSSL plays a crucial role in QTL mapping and gene cloning of seed-related traits, such as grain shape (Wang et al. 2012), grain chalkiness (Yang et al. 2021), seed dormancy (Zhou et al. 2017), and low-temperature germinability (Pan et al. 2021).
Although there are increasing reports about AG, its genetic mechanism remains unclear. Further exploration of new AG QTLs is an effective strategy to develop and improve DSR. In this research, the genetic dissection of the CL under the anaerobic condition was carried out with 229 SSSLs in the genetic background of Huajingxian 74 (HJX74) (Li et al. 2021) across five cropping seasons. The objects of this research were (1) to identify AG QTLs that control CL under submergence in a HJX74-SSSL population; (2) to integrate and narrow down QTLs by substitution mapping using SSSLs with overlapping segments corresponding to a putative QTL; (3) to compare and colocalize with previously reported AG QTLs to characterize the novel or stable QTLs. The AG QTLs mapped in the present study lay a foundation for cloning new hypoxic germination genes and provide a reference for the breeding of elite DSR cultivars.
Materials and methods
Plant materials
An HJX74-SSSL library containing 2360 SSSLs was constructed by crossing between the receptor parent of HJX74, an elite India rice variety, and 43 donor parents from seven AA-genome species (Zhang 2021). In this study, we selected 229 SSSLs, most of which were described in a previous study (Pan et al. 2021), from the HJX74-SSSL library to conduct a genetic analysis of AG. The substitution segments of the 229 SSSLs are distributed across 12 chromosomes of rice and originate from 24 cultivated rice donors, including 14 indica and 10 japonica accessions. The length of substitution segments was 0.1–69.9 cM, giving a genome coverage of 70.5%.
Field planting
All rice materials were grown in a paddy field located at South China Agricultural University, Guangzhou, China (23° N, 113° E, approximately). Each SSSL line was grown in a single plot, while the control line HJX74 was grown in 9–12 plots. Each plot consists of two rows with ten plants per row and all plots were arranged in an augmented design. The rice plants were grown using a wide-narrow row method, 33.33 cm for the wide row and 16.67 cm for the narrow row, with regular management. Each SSSL line was planted in one to five cropping seasons in 2015E, 2015L, 2016E, 2016L, or 2017E. “E” represents the early cropping season from March to July, and “L” represents the late cropping season from July to November. Three to ten individual plants per SSSL line were harvested from the middle of each plot based on the heading date, about 32–35 days after the first panicle appeared. The seeds were threshed, cleaned, and oven-dried at 50 ℃ for seven days to break seed dormancy, and then stored at − 20 ℃ until phenotypic assays.
Genotypic verification
Fresh leaves from rice seedlings were collected between one and two weeks after transplantation to paddy fields. Genomic DNA was then extracted from the leaf tissues of 229 SSSLs and HJX74 by the TPS method (Mu et al. 2010). Genotyping of each SSSL was performed by PCR using SSR markers to detect the substitution segment in each growing season. A total of 326 SSR markers were used in this study. The markers labeled “RM” were selected from the online resources and those labeled “PSM” were developed in our laboratory based on published sequences (Table S1).
Phenotypic evaluation of AG
AG was evaluated using CL as the phenotypic trait according to the previously described procedure by Cao et al. (2018) with some changes. Approximately 30–100 well-developed seeds from three to ten individual plants of each SSSL were used for phenotypic evaluation. The seeds were randomly distributed into some plastic boxes (14.2 × 14.2 × 9.5 cm) which have 100 holes (10 × 10) with five seeds per hole. Meanwhile, 8 ml distilled water was added into each hole to make a hypoxic condition with about 5 cm water above the seeds, while 0.5 ml water was added for the control normoxic condition (Fig. 1a, b). Subsequently, these boxes were covered and transferred to some large airtight boxes (34.7 × 24.5 × 12.5 cm) with two layers of wet filter papers on the bottom, and finally placed in a 30 ℃ incubator in 16 h light/8 h dark. After seven days of culture, we photographed the seedlings with a scanner and measured the CL under hypoxia and control conditions by ImageJ software.
QTL analysis
We conducted a QTL analysis according to Zhou et al. (2017) with slight modifications. First, the variation among different HJX74 plots was calculated by one-way ANOVA in Microsoft Excel 2019 to assess the effect of field environment. Second, the CL data were used in the student’s t-test to calculate the significant difference between SSSL and HJX74. Based on the ANOVA and t-test results, if the P value of CL is less than 0.001 (2015E, 2016L, and 2017E) or 0.01 (2015L and 2016E) in at least two cropping seasons, we consider that a putative AG QTL exists on the substitution segment of the corresponding SSSL. The nomenclature of identified QTLs follows the general rules in rice. The additive effect (a) and additive effect contribution (R2) of a putative QTL were calculated as described by Pan et al. (2021).
Substitution mapping of QTLs
Substitution mapping was conducted based on previous reports (Zhou et al. 2017) to reduce the QTL interval. In brief, if a QTL is detected simultaneously in two SSSLs with overlapping substitution segments, it is considered to lie on the overlapping interval of the two substitution segments. In contrast, a QTL is regarded as lying on a non-overlapping interval of two substitution segments if it is detected only in one of the two SSSLs derived from the same donor parent.
Results
Variation in CL of the SSSLs
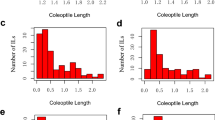
To explore the phenotypic variation of AG, we investigated the CL upon submerged germination of HJX74 and 229 SSSLs in five cropping seasons from 2015 to 2017 (Fig. 1c). The control parent HJX74 was planted in 10, 12, 12, 12, and 9 plots, and the average CL was 2.58, 2.20, 2.28, 2.38, and 2.28 cm in 2015E, 2015L, 2016E, 2016L, and 2017E, respectively (Fig. 2). One-way ANOVA analysis of CL in different HJX74 plots showed no significant difference in 2015L and 2016E, but slight variations of field environment in 2015E, 2016L, and 2017E (0.01 ≤ P ≤ 0.05) (Table S2). Therefore, when conducting the QTL analysis, the P value of Student’s t-test for 2015E, 2016L, and 2017E was increased by an order of magnitude to 0.001 to remove the field environmental differences. On the other hand, the CL in SSSLs displayed wide and continuous variation under submergence, ranging from 1.59–3.24, 1.49–2.87, 1.06–2.87, 1.61–3.06, and 1.05–2.65 cm in 2015E, 2015L, 2016E, 2016L, and 2017E, respectively (Figs. 1c and 2). Thus, our data from multiple environments demonstrate that AG is a typical quantitative trait. In addition, our results revealed that most of the SSSLs headed within five days and exhibited a similar heading date compared to HJX74 (Fig. S1), indicating that the seeds for the AG test were developed in a consistent field environment.
Phenotypic distribution of CL for 229 rice SSSLs in five cropping seasons. 2015E (a), 2015L (b), 2016E (c), 2016L (d), and 2017E (e) represent the early (E) and late (L) cropping seasons in 2015, 2016, and 2017, respectively. Parameters are the number of SSSLs (N) and mean (M) and standard deviation (S) for CL under submergence. The arrow indicates the CL of the recipient parent HJX74
Identification of SSSLs with putative AG QTLs
For QTL analysis, we employed Student’s t-test to analyze the CL difference between SSSLs and the recipient parent HJX74. Our results demonstrated that, compared with HJX74, 19 SSSLs exhibited significantly different CL (Tables 1 and S3), implying that putative AG QTLs were harbored in the substitution segments of these 19 SSSLs.
As shown in Table 1, the 19 SSSLs originated from eight donor parents, including Zhong4188 (W03), Zihui 100 (W05), Katy (W06), Suyunuo (W07), IR64 (W08), Basmati 370 (W11), IRAT261 (W18) and Chenglongshuijingmi (W20), most of which are indica accessions. Two indica donors, Zihui 100 and IR64, contributed six and four SSSLs, respectively, accounting for more than half of the 19 SSSLs. The 19 substitution segments varied in length from 5.3 to 60.1 cM, with an average length of 23.5 cM, and were distributed across nine chromosomes, except for 3, 10, and 12. Notably, some SSSLs shared overlapping segments and therefore they embraced the same QTL with the same or different alleles depending on the donor parent. Accordingly, six QTLs corresponding to nine SSSLs, qAG1, qAG2-2, qAG4, qAG5, qAG7-1, and qAG8, showed positive effects with additive effects ranging from 0.10 to 0.25 and additive effect contributions ranging from 4.3 to 11.2%. Meanwhile, five QTLs corresponding to ten SSSLs, qAG2-1, qAG6, qAG7-2, qAG9, and qAG11, showed negative effects. The additive effects were between − 0.17 and − 0.42, and additive effect contributions were between 7.2 and 18.5% (Table 1).
Five QTLs, qAG2-1 (S2, S4), qAG5 (S8), qAG7-1 (S15), qAG8 (S17), and qAG11 (S19), were stably detected in both the early and late cropping seasons. It should be noted, however, that other QTLs exhibited season-specific patterns. For instance, qAG2-1 (S3), qAG2-2 (S5), qAG4 (S7), qAG5 (S9), qAG6 (S10–S13), qAG7-1 (S14), qAG7-2 (S16), and qAG9 (S18) were only identified in the early cropping season. By contrast, qAG1 (S1) and qAG2-2 (S6) specifically existed in the environment during late cropping season (Table 1).
Substitution mapping of AG QTLs
Table 1 shows that six QTLs, qAG1, qAG4, qAG7-2, qAG8, qAG9, and qAG11, were detected by a single SSSL and thus located on their corresponding substitution segments. However, the remaining five QTLs were detected simultaneously by two to four SSSLs sharing overlapping chromosome fragments. Therefore, we employed substitution mapping to merge these putative QTLs into integrated QTLs and thereby locate them to smaller genomic intervals (Fig. 3; Table 2).
Substitution mapping of qAG2-1 (a), qAG2-2 (b), qAG5 (c), qAG6 (d), and qAG7-1 (e). The long horizontal dark bars represent the chromosomes with the molecular markers above them. The white bars are the background of HJX74, while the black bars represent the substitution segments of the SSSLs detected with AG QTLs. The blue bar of S20 means that no QTL was detected. The intervals located AG QTLs are shown between two vertical dotted lines with the names of QTLs under the substitution segments. Red names are for positive-effect QTLs, and black names are for negative-effective QTLs
qAG2-1 was detected simultaneously on S2, S3, and S4; thus, it was mapped in the overlapping region in the vicinity of PSM124-RM525 with an interval length of 25.3 cM (Fig. 3a). The integrated qAG2-1 had an additive effect of − 0.29 and an additive effect contribution of 12.8%. S5 and S6 carried qAG2-2, which was hence delimited to a 14.5-cM overlapping region flanked by RM525 and RM530 (Fig. 3b). The integrated qAG2-2 had an additive effect of 0.17, giving an additive effect contribution of 7.6%. It should be noted that there is a small overlapping region between qAG2-1 and qAG2-2, however, considering that these two QTLs have opposite additive effects, we therefore identified them as two independent QTLs. The simultaneous detection of qAG5 on S8 and S9, but not on S20, which shares an overlapping region with the former two SSSLs, suggests that qAG5 was located near RM430 with a narrow interval length of 3.1 cM (Fig. 3c). The additive effect of the integrated qAG5 was 0.16 and the additive effect contribution was 7.0%. qAG6 was detected simultaneously on four SSSLs from S10 to S13, and it was hence located in an overlapping region close to RM589 and RM190 with a short interval length of 5.8 cM (Fig. 3d). The integrated qAG6 had an additive effect of − 0.34 and a 14.8% additive effect contribution. S14 and S15 were demonstrated to carry qAG7-1 in their overlapping segments close to RM11 with a short interval length of 4.1 cM (Fig. 3e). The integrated qAG7-1 had an additive effect of 0.21 and the additive effect contribution was 9.1%.
After substitution mapping, 11 QTLs controlling AG were finally mapped on nine chromosomes except on chromosomes 3, 10, and 12 (Fig. 4). Six positive QTLs were located on 1, 2, 4, 5, 7, and 8 chromosomes, while five negative QTLs were located on 2, 6, 7, 9, and 11 chromosomes, respectively. Their chromosomal intervals were reduced to smaller genetic regions ranging from 3.1 to 25.3 cM, including eight QTLs with interval lengths less than 10 cM. The additive effects changed from − 0.42 to 0.25, giving the additive effect contributions ranging from 6.8 to 18.5% (Table 2). Co-localization analysis showed that nine QTLs in our study were also identified in previous association or linkage mapping analysis, while qAG4 and qAG11 were novel AG genetic loci identified for the first time in the present study (Table 3).
Chromosome location of 11 QTLs for AG in rice. The linkage map of QTLs was constructed by using MapChart 2.2 (Voorrips 2002). SSR markers are indicated on the right of the chromosomes. Genetic distance (cM) is shown as rulers on the left margin. Red and black bars on each chromosome’s right are the location intervals of positive-effect and negative-effect QTLs, respectively, with their names on the right. Chr chromosome
Phenotypic confirmation of SSSLs with AG QTLs
To verify the effect of the mapped AG QTLs, we additionally tested the dynamic growth phenotype of the coleoptiles of two positive and two negative QTLs in a novel hypoxic environment (10-ml centrifuge tubes) (Fig. 5). In general, SSSLs carrying positive QTLs, S1-qAG1 and S7-qAG4 exhibited rapid germination and faster coleoptile growth compared to HJX74, indicating strong tolerance to flooding. On the contrary, SSSLs carrying negative QTLs, S12-qAG6 and S19-qAG11, showed slower germination and coleoptile growth rates compared to HJX74. For example, the CL of S1-qAG1 was 0.45, 1.73, and 2.09 cm on days 3, 5, and 7 after submergence, respectively, while that of HJX74 was 0.21, 1.33, and 1.69 cm. Significant differences were observed at all three time points (Fig. 5a). However, the CL of S7-qAG4 was significantly different from that of HJX74 only on day 7 after submergence (Fig. 5b). The CL of S12-qAG6 on days 3, 5, and 7 after submergence was 0.09, 0.44, and 0.62 cm, respectively, significantly shorter than 0.31, 1.69, and 1.75 cm of HJX74 at each time point (Fig. 5c). Similar results to S12-qAG6 were also observed for S19-qAG11 (Fig. 5d). In terms of the temporal dimension, the coleoptile growth showed a slow-fast-slow growth pattern. The coleoptile grew slowly in the stage of 0–3 days, rapidly in the stage of 3–5 days, and slowly again in the stage of 5–7 days.
Effects of AG QTLs on coleoptile elongation. a, b, c, and d show CL comparison between the SSSLs carrying AG QTL and HJX74 under hypoxia stress. The seeds were flooded in 10-ml centrifuge tubes with 8 cm water above them and then incubated in a growth chamber under photoperiodic cycles of 16 h light and 8 h dark at 30 ℃. Pictures were taken on the 3rd, 5th, and 7th days after submergence germination. Scale bars, 1 cm. At each time point, columns and bars represent the means and standard deviations of CL calculated based on ten seedlings. Significance is determined using t-test (**P < 0.01)
Discussion
Global flood events have occurred mainly in Asian countries such as China and India, major rice-growing regions, and the impact of floods is predicted to increase further by 2030 (Tellman et al. 2021). Identifying the natural variation of AG is the key to enhancing flood germination and seedling development of modern cultivated rice, especially DSR. Although many studies have been carried out on AG, current knowledge of its genetic basis is still not well understood. To explore additional loci controlling AG, we examined the CL of 229 SSSLs under submergence in five different growing seasons and performed a comprehensive QTL analysis and identified 11 AG QTLs (Table 2).
Rice is the only crop that can germinate in a flooded environment. Its strategy in response to flooding is to rapidly elongate the coleoptile to reach the water surface for oxygen, making seedlings adapt to anaerobic stress. Because of this, coleoptile-related traits are frequently used as phenotypes for genetic studies of AG in rice (Hsu and Tung 2015; Jiang et al. 2004; Nghi et al. 2019; Nishimura et al. 2020; Rauf et al. 2019; Su et al. 2021; Yang et al. 2019; Zhang et al. 2017). This method is simple and suitable for large-scale phenotypic screening in the laboratory. Our data demonstrate that flooding CL is normally distributed, consistent with the characteristics of a quantitative trait (Fig. 2). However, most AG QTLs we identified were only detected in two to three environments across five growing seasons. The stability of CL in different environments is not as good as seed dormancy and low-temperature germinability that we have studied before (Pan et al. 2021; Zhou et al. 2017). It suggests that AG is a complex trait that is easily affected by the external environment, leading to our limited understanding. Interestingly, Zhang et al. (2017) found that the coleoptiles of some indica cultivars did not elongate but were inhibited after submergence, similar to the quiescence strategy conferred by the Sub1 gene (Xu et al. 2006). In natural conditions, these quiescent seedlings may resume growth when the flood recedes. In addition, the SR of seedlings under flooded conditions is also commonly used in AG study (Angaji et al. 2008, 2010; Baltazar et al. al. 2019; Ghosal et al. 2019, 2020; Kim and Reinke 2018; Septiningsih et al. 2012, 2013). Compared with CL, the SR of seedlings is closer to natural field conditions, which is beneficial for applications in real agricultural production. However, this method requires more labor and material resources, and phenotype identification also requires up to 21 days (Ghosal et al. 2020). Notably, despite differences in phenotypic indicators and mapping populations, nine of our 11 AG QTLs colocalized with previously reported ones, including some QTLs identified by SR (Table 3). This not only suggests that our study is reliable, but also indicates that there is a certain correlation between the AG QTLs identified by CL in the laboratory and those identified by SR in the field. Therefore, we should comprehensively apply different phenotypic indicators to complement and validate each other to improve research accuracy.
OsTPP7, the target gene underlying a major QTL, qAG-9-2, is the first map-based cloned locus controlling AG in rice (Angaji et al. 2010; Kretzschmar et al. 2015). Genome sequencing revealed that a 20.9-kb chromosomal segment encompassing OsTPP7 was deleted in the AG-sensitive indica rice IR64 and functional allele was found in most japonica varieties. The interval of qAG9 in this study covered OsTPP7 (Table 3), but qAG9 was a negative-effect QTL, probably because it is derived from the indica donor Zihui 100 (W05) (Table 2). qAG1 was the QTL with the largest positive effect in this study, with an additive effect of 0.25 and an additive effect contribution of 11.2%, which showed co-localization with four reported AG loci (Tables 2 and 3). Previous studies showed that the alleles with increasing effects of this locus were derived from japonica rice Khao Hlan On and Nipponbare (Angaji et al. 2010; Hsu and Tung 2015). In agreement with this, we also found that the donor parent of qAG1 is Katy (W06), a japonica cultivar (Table 2). Additionally, the interval of qAG1 contains OsCBL10 (LOC_01g51420), a possible AG gene encoding rice calcineurin B-like protein10 (Ye et al. 2018). qAG6 represented the most stable QTL in present study, as it was detected simultaneously in four SSSLs (S10-S13) derived from three different donors (Table 1). We, therefore, narrowed it down to a smaller genetic interval of 5.8 cM by substitution mapping (Fig. 3d). Moreover, qAG6 showed large additive effect contributions, ranging from 9.4 to 18.5%. The interval of qAG6 contained a gene (LOC_Os06g03520) encoding a DUF domain-containing protein which was previously considered as the candidate gene of two AG loci (Zhang et al. 2017). Surprisingly, among the 11 haplotypes of LOC_Os06g03520, only Hap.2 exhibited flood tolerance, while the remaining ten haplotypes were all flood intolerant. This may explain why our identified qAG6 was a negative-effect QTL, as it is a high-probability event. Another interesting observation is that the four SSSLs corresponding to qAG6 can only be detected in the early season, indicating its environment-specific regulation. Similarly, we noticed that many other QTLs in current study also showed seasonal specificities, such as qAG1, qAG2-2, qAG4, qAG7-2, and qAG9 (Table 1). Together these results remind us to pay attention to the stability of these QTLs when we applied them in different environments.
More recently, researchers identified a 14-3-3 protein-encoding gene OsGF14h in weedy rice, which confers tolerance to AG and anaerobic seedling development (Sun et al. 2022). By comparison, we found that the novel locus identified in our study, qAG11, was located on the short arm of chromosome 11 (Fig. 4), while OsGF14h was located on the long arm of chromosome 11, suggesting they are different loci. OsGF14h interacts with transcription factors OsHOX3 and OsVP1 to balance ABA signaling and GA biosynthesis and thereby confers AG tolerance in weedy rice. Cloning of the OsGF14h gene once again demonstrates that GA and ABA are the core regulators of seed germination, even under flooded conditions. Su et al. (2021) also identified four possible candidate genes related to AG by integrating GWAS, transcriptome, and metabolome, among which Os05g0560900 and Os01g0911700 were involved in GA metabolism and ABA signaling, respectively. However, the mechanism by which OsTPP7 regulates AG appears to be independent of these two kinds of phytohormones.
Obviously, there is plenty of evidence supporting that japonica rice has a stronger AG tolerance than indica rice. For example, several studies have demonstrated that japonica rice has a greater ability than indica rice to elongate coleoptile upon flooding. Under dark submergence, the average CL of 432 indica cultivars was 1.27 cm (Zhang et al. 2017), while that of 273 japonica accessions was 5.15 cm (Nghi et al. 2019), showing a huge difference. In this investigation, SSSLs derived from both indica and japonica donors showed an intermediate average CL ranging from 2.05 to 2.56 cm (Fig. 2). Similarly, Hsu and Tung (2015) found that the average anaerobic response index of the japonica subgroup was 2.18 cm, which was significantly higher than that of the indica subgroup of 1.66 cm. They also noted that some tropical japonica varieties exhibited particularly strong AG. Consistently, evolutionary analysis uncovered that functional OsGF14hWR04-6 primarily existed in tropical japonica, as well as wild ancestors and temperate japonica weedy rice (Sun et al. 2022). Tropical regions have abundant rainfall and are prone to floods. Consequently, strong natural selection pressures may force tropical japonica to retain the AG capabilities of its ancestors. A new study adds more evidence to this idea. He et al. (2023) found that the CL of japonica rice was longer than that of indica rice when submerged, and the elite haplotype of OsUGT75A was mostly found in tropical japonica rice.
Although numerous studies have demonstrated the strong AG of japonica rice, other studies have shown different experimental evidence. An assessment of 256 rice core accessions found no significant difference in AG tolerance between indica and japonica (Chen et al. 2012). Quite surprisingly, our investigation clearly showed that an indica cultivar from China, Zihui 100 (W05), performed well in our AG test. Among all 19 SSSLs with putative QTLs, six SSSLs were derived from Zihui 100, and four positive-effect QTLs, qAG2-2, qAG4, qAG5, and qAG7-1, were identified, contributing most of the positive-effect QTLs (Tables 1 and 2). Therefore, some indica varieties may also have good AG tolerance, at least Zihui 100 is an excellent germplasm. In agreement with this point, the favorable alleles of the reported AG QTLs were not only from japonica accessions, but also from indica accessions (Angaji et al. 2008; Jiang et al. 2004; Septiningsih et al. 2013; Yang et al. 2019). However, another indica variety, IR64 (W08), was further demonstrated to be an AG-sensitive accession in this study. Only negative-effect QTLs (qAG2-1 and qAG6) were detected in S2, S3, S12, and S13 derived from IR64 (Table 1). For those negative QTLs, they can be utilized through marker-assisted selection breeding after exploring their excellent haplotypes, or they can be edited for utilization in DSR breeding.
Conclusion
In conclusion, the 11 AG QTLs identified in this study enrich the genetic basis of AG in rice and also provide new options for DSR breeding practices. Additional studies are required to clone the target genes of these QTLs and understand the underlying mechanisms of AG tolerance for more efficient DSR breeding strategies and sustainable agriculture.
Data availability
All data generated or analyzed during this study are included in this published article.
References
Angaji SA (2008) Mapping QTLs for submergence tolerance during germination in rice. Afr J Biotechnol 7(15):2551–2558
Angaji SA, Septiningsih EM, Mackill DJ, Ismail AM (2010) QTLs associated with tolerance of flooding during germination in rice (Oryza sativa L.). Euphytica 172:159–168. https://doi.org/10.1007/s10681-009-0014-5
Baltazar MD, Ignacio JCI, Thomson MJ, Ismail AM, Mendioro MS, Septiningsih EM (2019) QTL mapping for tolerance to anaerobic germination in rice from IR64 and the aus landrace Kharsu 80A. Breed Sci 69(2):227–233. https://doi.org/10.1270/jsbbs.18159
Cao W, Wang Y, Tan B, Liu W, Chu L, Pan Z, Zhao G, Cao G, Zhou Y (2018) Evaluation of seed germination vigor of rice cultivars under low temperature or hypoxic condition related with direct seeding rice. Mol Plant Breed 16(10):3259–3268 ((in Chinese with English abstract))
Chen S, Wang J, Pan Y, Ma J, Zhang J, Zhang H, Teng S (2012) Genetic analysis of seed germinability under submergence in rice. Chin Bull Bot 47(1):28–35 ((in Chinese with English abstract))
Ghosal S, Casal C, Quilloy FA, Septiningsih EM, Mendioro MS, Dixit S (2019) Deciphering genetics underlying stable anaerobic germination in rice: phenotyping, QTL identification, and interaction analysis. Rice 12:50. https://doi.org/10.1186/s12284-019-0305-y
Ghosal S, Quilloy FA, Casal C, Septiningsih EM, Mendioro MS, Dixit S (2020) Trait-based mapping to identify the genetic factors underlying anaerobic germination of rice: phenotyping, GXE, and QTL mapping. BMC Genet 21:6. https://doi.org/10.1186/s12863-020-0808-y
He Y, Sun S, Zhao J, Huang Z, Peng L, Huang C, Tang Z, Huang Q, Wang Z (2023) UDP-glucosyltransferase OsUGT75A promotes submergence tolerance during rice seed germination. Nat Commun 14:2296. https://doi.org/10.1038/s41467-023-38085-5
Hsu SK, Tung CW (2015) Genetic mapping of anaerobic germination-associated QTLs controlling coleoptile elongation in rice. Rice 8:38. https://doi.org/10.1186/s12284-015-0072-3
Jiang L, Hou M, Wang C, Wan J (2004) Quantitative trait loci and epistatic analysis of seed anoxia germinability in rice (Oryza sativa). Rice Sci 11(5–6):238–244
Kim SM, Reinke RF (2018) Identification of QTLs for tolerance to hypoxia during germination in rice. Euphytica 214:160. https://doi.org/10.1007/s10681-018-2238-8
Kretzschmar T, Pelayo MAF, Trijatmiko KR et al (2015) A trehalose-6-phosphate phosphatase enhances anaerobic germination tolerance in rice. Nat Plants 1:15124. https://doi.org/10.1038/nplants.2015.124
Li F, Gao Y, Wu B, Cai Q, Zhan P, Yang W, Shi W, Li X, Yang Z, Tan Q, Luan X, Zhang G, Wang S (2021) High-quality de novo genome assembly of Huajingxian 74, a receptor parent of single segment substitution lines. Rice Sci 28(2):109–113
Luo X, Wang Z, Zeng S, Zang Y, Yang W, Zhang M (2019) Recent advances in mechanized direct seeding technology for rice. J South China Agric Univ 40(5):1–13 ((in Chinese with English abstract))
Ma M, Cen W, Li R, Wang S, Luo J (2020) The molecular regulatory pathways and metabolic adaptation in the seed germination and early seedling growth of rice in response to low O2 stress. Plants 9(10):1363. https://doi.org/10.3390/plants9101363
Magneschi L, Perata P (2009) Rice germination and seedling growth in the absence of oxygen. Ann Bot 103(2):181–196. https://doi.org/10.1093/aob/mcn121
Mondal S, Khan MIR, Dixit S, Cruzb PCS, Septiningsihd EM, Ismail AM (2020) Growth, productivity and grain quality of AG1 and AG2 QTLs introgression lines under flooding in direct-seeded rice system. Field Crop Res 248:107713. https://doi.org/10.1016/j.fcr.2019.107713
Mu C, Zhang F, Li W, Sun Q, Ding Z, Wang L, Meng Z (2010) A method of genomic DNA extraction of maize. J Maize Sci 18(03):170–172 ((in Chinese with English abstract))
Nghi KN, Tondelli A, Vale G, Tagliani A, Mare C, Perata P, Pucciariello C (2019) Dissection of coleoptile elongation in japonica rice under submergence through integrated genome-wide association mapping and transcriptional analyses. Plant Cell Environ 42(6):1832–1846. https://doi.org/10.1111/pce.13540
Nishimura T, Sasaki K, Yamaguchi T, Takahashi H, Yamagishi J, Kato Y (2020) Detection and characterization of quantitative trait loci for coleoptile elongation under anaerobic conditions in rice. Plant Prod Sci 23(3):374–383. https://doi.org/10.1080/1343943X.2020.1740600
Pan Z, Tan B, Cao G, Zheng R, Liu M, Zeng R, Wang S, Zhu H, Ye H, Zhao G, Cao W, Liu G, Zhang G, Zhou Y (2021) Integrative QTL identification, fine mapping and candidate gene analysis of a major locus qLTG3a for seed low-temperature germinability in rice. Rice 14:103. https://doi.org/10.1186/s12284-021-00544-2
Pucciariello C (2020) Molecular mechanisms supporting rice germination and coleoptile elongation under low oxygen. Plants 9(8):1037. https://doi.org/10.3390/plants9081037
Rauf M, Choi YM, Lee S, Lee MC, Oh S, Hyun DY (2019) Evaluation of anaerobic germinability in various rice subpopulations: identifying genotypes suitable for direct-seeded rice cultivation. Euphytica 215:19. https://doi.org/10.1007/s10681-019-2341-5
Septiningsih EM, Sanchez DL, Singh N, Sendon PMD, Pamplona AM, Heuer S, Mackill DJ (2012) Identifying novel QTLs for submergence tolerance in rice cultivars IR72 and Madabaru. Theor Appl Genet 124:867–874. https://doi.org/10.1007/s00122-011-1751-0
Septiningsih EM, Ignacio JCI, Sendon PMD, Sanchez DL, Ismail AM, Mackill DJ (2013) QTL mapping and confirmation for tolerance of anaerobic conditions during germination derived from the rice landrace Ma-Zhan Red. Theor Appl Genet 126:1357–1366. https://doi.org/10.1007/s00122-013-2057-1
Su L, Yang J, Li D et al (2021) Dynamic genome-wide association analysis and identification of candidate genes involved in anaerobic germination tolerance in rice. Rice 14:1. https://doi.org/10.1186/s12284-020-00444-x
Sun J, Zhang G, Cui Z et al (2022) Regain flood adaptation in rice through a 14–3-3 protein OsGF14h. Nat Commun 13:5664. https://doi.org/10.1038/s41467-022-33320-x
Tellman B, Sullivan JA, Kuhn C, Kettner AJ, Doyle CS, Brakenridge GR, Erickson TA, Slayback DA (2021) Satellite imaging reveals increased proportion of population exposed to floods. Nature 596:80–86. https://doi.org/10.1038/s41586-021-03695-w
Voorrips RE (2002) MapChart: software for the graphical presentation of linkage maps and QTLs. J Hered 93:77–78. https://doi.org/10.1093/jhered/93.1.77
Wang Y, Guo Y, Hong D (2010) QTL analysis of the anoxic tolerance at the seedling stage in rice. Chin J Rice Sci 24(1):18–24 ((in Chinese with English abstract))
Wang S, Wu K, Yuan Q et al (2012) Control of grain size, shape and quality by OsSPL16 in rice. Nat Genet 44:950–954. https://doi.org/10.1038/ng.2327
Xu K, Xu X, Fukao T, Canlas P, Maghirang-Rodriguez R, Heuer S, Ismail AM, Bailey-Serres J, Ronald PC, Mackill DJ (2006) Sub1A is an ethylene-response-factor-like gene that confers submergence tolerance to rice. Nature 442:705–708. https://doi.org/10.1038/nature04920
Yang J, Sun K, Li D et al (2019) Identification of stable QTLs and candidate genes involved in anaerobic germination tolerance in rice via high-density genetic mapping and RNA-Seq. BMC Genom 20:355. https://doi.org/10.1186/s12864-019-5741-y
Yang W, Liang J, Hao Q et al (2021) Fine mapping of two grain chalkiness QTLs sensitive to high temperature in rice. Rice 14:33. https://doi.org/10.1186/s12284-021-00476-x
Ye N, Wang F, Shi L et al (2018) Natural variation in the promoter of rice calcineurin B-like protein10 (OsCBL10) affects flooding tolerance during seed germination among rice subspecies. Plant J 94(4):612–625. https://doi.org/10.1111/tpj.13881
Yu SM, Lee HT, Lo SF, Ho TD (2021) How does rice cope with too little oxygen during its early life? New Phytol 229(1):36–41. https://doi.org/10.1111/nph.16395
Zhang G (2021) Target chromosome-segment substitution: a way to breeding by design in rice. Crop J 9(3):658–668. https://doi.org/10.1016/j.cj.2021.03.001
Zhang M, Lu Q, Wu W et al (2017) Association mapping reveals novel genetic loci contributing to flooding tolerance during germination in indica rice. Front Plant Sci 8:678. https://doi.org/10.3389/fpls.2017.00678
Zhou Y, Xie Y, Cai J et al (2017) Substitution mapping of QTLs controlling seed dormancy using single segment substitution lines derived from multiple cultivated rice donors in seven cropping seasons. Theor Appl Genet 130:1191–1205. https://doi.org/10.1007/s00122-017-2881-9
Acknowledgements
We thank Guiquan Zhang for providing SSSLs.
Funding
This work was supported by the National Natural Science Foundation of China (31771888), the double first-class discipline promotion project (2021B10564001), and the Natural Science Foundation of Guangdong Province (2022A1515010843, 2019A1515010237).
Author information
Authors and Affiliations
Contributions
YZ designed the research. WC carried out the experiments with the help of BT, GZ, HK, YW, and WL. HL assisted in the field experiments. BT and YZ analyzed the data and wrote the manuscript. YZ revised the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no relevant financial or non-financial interests to disclose.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Tan, B., Cao, W., Zhao, G. et al. Identification and substitution mapping of QTLs for anaerobic germination in rice. Euphytica 219, 124 (2023). https://doi.org/10.1007/s10681-023-03252-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10681-023-03252-8