Abstract
The cultivation of Theobroma grandiflorum in the Brazilian Amazon is mainly conducted by family farmers who use a range of different management strategies. Thus, breeding programs of the species must address the challenge of developing cultivars that are adapted and stable in a variety of cultivation environments. In this context, this study aimed to estimate the optimum number of harvests for genetic selection of T. grandiflorum progenies and identify the most promising ones in terms of productivity, stability, and adaptability. The trials were implemented in three environments, using a randomized complete block design, with 25 full-sib progenies, five replications, and three plants per plot. The traits mean number of fruits/plant, mean fruit production/plant, and rate of infection with witches’ broom (Moniliophthora perniciosa) were evaluated over 11 harvests. The Restricted Maximum Likelihood/Best Linear Unbiased Prediction (REML/BLUP) procedure was used to estimate genetic parameters and predict genetic values, which were then applied to assess stability and adaptability. The results show that there is genetic variability among the studied T. grandiflorum progenies and that accurate genetic selection aiming at recombination and recommendation is effective after three and eleven harvests, respectively. Six progenies were selected that met the requirements for productivity, stability, and adaptability to different cultivation environments. These results can be used to optimize and advance T. grandiflorum breeding programs.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The allogamous tree Theobroma grandiflorum (Willd. Ex Spreng.) Schum. (Malvaceae family), commonly known as cupuassu tree, is native to Southeast Pará and Northwest Maranhão States in the Brazilian Amazon (Alves et al. 2003). Due to the movement of indigenous peoples throughout the interior of the Amazon region, the species is now dispersed across all Amazonian states (Alves et al. 2007), and plantations of T. grandiflorum have been established in 97 (67 %) of the 144 municipalities in Pará State (Pará 2020). These plantations are generally small-scale seed orchards of less than one hectare, planted by family farmers (Homma 2014). The expansion of the crop and its adaptation to different environments in Pará is an indicator of the genetic plasticity of the species (Alves et al. 2007).
The economic importance of T. grandiflorum has grown in recent years as the main products derived from the tree, including its seeds and the pulp covering them, have attracted increased attention in national and international markets (Silva et al. 2020). The pulp, with high acidity and strong aroma, is used to produce juices, sweets, and jellies, among other food products (Pugliese et al. 2013). The almonds, which have antioxidant properties, are used in the pharmaceutical and cosmetic industries (Oliveira and Genovese 2013), as well as to produce cupuassu chocolate, a product known as “cupulate” (Genovese and Lannes 2009). The municipality of Tomé Açu, Northeast Pará, was a pioneer in the cultivation of this fruit tree. The region has become a model for production as farmers have organized an agricultural cooperative that processes all cupuassu products, which is essential for expanding its production and use in the region (Alves et al. 2014). To ensure the development and sustainability of the crop, communities have continuously sought research support, particularly in terms of developing varieties that are well adapted to local conditions.
At the end of the 1980 s, Embrapa Amazonia Oriental initiated a T. grandiflorum breeding program and developed genetic resources to produce genotypes with high levels of fruit production and tolerance to the fungus Moniliophthora perniciosa (Stahel) Aime & Phillips-Mora, etiological agent of the witches’ broom disease, a pathogen that can affect the cultivation of all species of the Theobroma genus, including T. grandiflorum and T. cacao (Gramacho et al. 2016; Patrocínio et al. 2017). However, the previously developed genotypes have inconsistent fruit production when subjected to different environments.
Currently, the Restricted Maximum Likelihood/Best Linear Unbiased Prediction (REML/BLUP) mixed model method is the standard for analyses of genotype x environment (GE) interaction (Coelho et al. 2020; Evangelista et al. 2021) and repeated measures (Resende et al. 2014; Ferreira et al. 2020). There are numerous reasons for its use, including the fact that it enables the simultaneous estimates of variance components and prediction of genetic values. The method also deals well with unbalanced data, describes the heterogeneity of genetic covariances and residual variances across environments, and models spatial trends (Resende 2016).
The evaluation of different genotypes in a variety of environments enables the quantification of the GE interaction effects (Resende 2015) and the analysis of genotypic stability and adaptability (Chipeta et al. 2017). Understanding stability and adaptability enables the identification of productive, stable, and adaptable genotypes (Silva et al. 2019). However, evaluating GE interaction is one of the most costly aspects of a breeding program (Dias et al. 2018), especially for perennial fruit trees such as T. grandiflorum, where the breeding cycle can last up to 15 years (Alves et al. 2021). This may explain why studies on GE interaction in T. grandiflorum are extremely rare.
Variation throughout years can create different environments, which, in turn, will influence genotypes differently (Jaimez et al. 2020). The evaluation of genotypes across several harvests is crucial in perennial fruit trees as it enables the quantification of the genotype x measurement (GM) interaction effects and estimates of the repeatability coefficient to determine the optimal number of harvests necessary to conduct effective genetic selection (Resende 2015; Ferreira et al. 2020).
In this context, this study aimed to estimate the optimum number of harvests for genetic selection of T. grandiflorum progenies and identify the most promising progenies in terms of productivity, stability, and adaptability.
Materials and methods
Experimental data
Full-sib T. grandiflorum progeny tests were established in three farms in Northeastern Pará State, Brazil; two located in the municipality of Tomé Açu and one in the municipality of São Francisco do Pará, approximately 210 km apart. The three environments represent a sample of the different cultivation systems used to produce T. grandiflorum in Pará. This experimental system enables the evaluation and selection of genotypes for conditions similar to those in which they are often cultivated. The differences between the three environments are mainly the different cropping systems used for each trial, in terms of temporary and definitive shading or full sun, and spacing.
Each T. grandiflorum progeny test was installed in consortium with other tree species, all of which were planted in February 2005. The field arrangement affected conditions of luminosity and competition over and under the soil. In trial 1, T. grandiflorum was maintained in shade during the productive phase, while in the other two trials (trials 2 and 3) the trees were kept in full sun (Table 1). In trial 1, T. grandiflorum progenies were part of an agroforestry system (AFS), together with Passiflora edulis Sims. (passion fruit) and Swietenia macrophylla King. (Brazilian mahogany), at initial densities of 400, 800, and 100 plants/ha, respectively. After the third year, the passion fruit was removed from the AFS as it had completed its cycle. Therefore, through all production stages, T. grandiflorum was shaded with S. macrophylla. Trial 2 was also installed as an AFS and consisted of T. grandiflorum progenies, Piper nigrum L. (Black pepper), and Bertholletia excelsa Bonpl. (Brazil nut), at densities of 303, 1800, and 75 plants/ha, respectively. As B. excelsa developed a very prolific crown, producing too much shade for T. grandiflorum in the first years, it was removed from the AFS in the fifth year. Piper nigrum cultivation occurred over the first seven years of the trial. Thus, after P. nigrum tree mortality in the seventh year, T. grandiflorum was left in full sun. In trial 3, T. grandiflorum progenies were intercropped with Musa spp. (banana tree), both with a density of 400 plants/ha. As in trial 2, after the fifth year, the Musa spp. was removed from the AFS, with T. grandiflorum progenies remaining in full sun until the end of the study (Table 1). It is important to highlight that these different field arrangements, involving full sun and temporary and definitive shading, were designed to reproduce a similar environment to what farmers cultivate cupuassu tree in the state of Pará.
The progeny test, carried out in three trials, was established using a randomized complete block design, with 25 T. grandiflorum full-sib progenies, five replications, and three plants per plot. The 25 full-sib progenies were obtained through controlled pollination. Phenotypic data were measured over 11 consecutive annual harvests, based on a total plot. The harvest opening coincides with the beginning of the rainy season and extends over the entire period of about six months. Therefore, each harvest was divided into four evaluations with 45-day intervals between them. Response variables included the mean number of fruits/plant (NF) and mean fruit production (kg/plant), obtained by multiplying the NF by the average weight of the fruit of each genotype. We also assessed tolerance to witches’ broom disease (M. perniciosa) based on the rate of symptomatic plants per progeny (WB, %). Plants were deemed symptomatic when at least one branch presented misshapen phyllotaxis, compared to a normal branch, and after one month appeared desiccated (Alves et al. 2009). For selection, a tolerance threshold of 30 % was adopted as the maximum rate of symptomatic plants per progeny.
Statistical analyses
Due to the unbalance caused by the mortality of some trees, which is common in long-term trials involving perennial crops, the mixed model methodology was adopted for statistical analysis. With such an approach, the variance components and genetic parameters are estimated by the restricted maximum likelihood (REML) (Patterson and Thompson 1971), and genetic values are predicted by the best linear unbiased prediction (BLUP) (Henderson 1975). The mixed linear model associated with the analysis of progeny, with a complete randomized block design, three locations, at the plot level, and with repeated measures, was defined as:
where y is the vector of phenotypic data; f is the vector of the effects of the combination repetition-environment-measurement (assumed to be fixed), added to the general mean; g is the vector of genotypic effects (assumed to be random), g ~ NID (0,\({{\upsigma }}_{\text{g}}^{2}\)), where \({{\upsigma }}_{\text{g}}^{2}\) is the genotypic variance; ge is the vector of genotypes × environments (GE) interaction effects (assumed to be random), ge ~ NID (0,\({{\upsigma }}_{\text{ge}}^{2}\)), where \({{\upsigma }}_{\text{ge}}^{2}\) is the GE interaction variance; gm is the vector of genotypes × measurements (GM) interaction effects (random), gm ~ NID (0,\({{\upsigma }}_{\text{gm}}^{2}\)), where \({{\upsigma }}_{\text{gm}}^{2}\) is the GM interaction variance; gem is the vector of the genotypes x environments x measurements (GEM) interaction effects (random), gem ~ NID (0,\({{\upsigma }}_{\text{gem}}^{2}\)), where \({{\upsigma }}_{\text{gem}}^{2}\) is the GEM interaction variance; p is the vector of the permanent plot effects within locations (assumed to be random), p ~ NID (0, \({{\upsigma }}_{\text{perm}}^{2}\)), where \({{\upsigma }}_{\text{perm}}^{2}\) is the permanent plot effect variance; and e is the vector of residuals (random), e ~ NID (0,\({{\upsigma }}_{\text{e}}^{2}\)), where \({{\upsigma }}_{\text{e}}^{2}\) is the residual variance. The capital letters (X, Z, Q, T, W and S) represent the incidence matrices for the correspondents’ effects.
The significance of the random effects of the statistical model was tested by the likelihood ratio test (LRT), given by the following equation (Wilks 1938):
where \(LogL\) is the logarithm of the maximum point of the residual likelihood function (L) of the complete model; and \({LogL}_{R}\) is the logarithm of the maximum point of the residual likelihood function (\({L}_{R}\)) of the reduced model (without the effect under test). The chi-square statistic with one degree of freedom and a probability level equal to 1 % was used to test the random effects significance.
From the variance components (\({{\upsigma }}_{\text{g}}^{2}\), \({{\upsigma }}_{\text{ge}}^{2}\), \({{\upsigma }}_{\text{gm}}^{2}\), \({{\upsigma }}_{\text{gem}}^{2}\), \({{\upsigma }}_{\text{perm}}^{2}\), and \({{\upsigma }}_{\text{e}}^{2}\)), we estimated (Resende et al. 2014):
Phenotypic variance:\({{{\upsigma }}}_{\text{p}}^{2}={{\upsigma }}_{\text{g}}^{2}+{{\upsigma }}_{\text{ge}}^{2}+{{\upsigma }}_{\text{gm}}^{2}+{{\upsigma }}_{\text{gem}}^{2}+{{\upsigma }}_{\text{perm}}^{2}+{{\upsigma }}_{\text{e}}^{2};\)
Mean phenotypic variance:\({{{\upsigma }}}_{\text{mp}}^{2}={{\upsigma }}_{\text{g}}^{2}+\frac{{{\upsigma }}_{\text{ge}}^{2}}{\text{e}}+\frac{{{\upsigma }}_{\text{gm}}^{2}}{\text{m}}+\frac{{{\upsigma }}_{\text{perm}}^{2}}{\text{b}}+\frac{{{\upsigma }}_{\text{e}}^{2}}{\text{emb}}\)where e, m, and b are the number of environments, measurements, and blocks, respectively;
Individual broad-sense heritability:\({{\text{h}}}_{\text{g}}^{2}=\frac{{{\upsigma }}_{\text{g}}^{2}}{{\widehat{{\upsigma }}}_{\text{p}}^{2}};\)
Mean broad-sense heritability:\({{\text{h}}}_{\text{mg}}^{2}=\frac{{{\upsigma }}_{\text{g}}^{2}}{{\widehat{{\upsigma }}}_{\text{mp}}^{2}};\)
Accuracy of genotype selection:\({\text{r}}_{{\widehat{\text{g}}}{\rm g}}=\sqrt{{{\text{h}}}_{\text{mg}}^{2}};\)
Coefficient of determination of GE interaction effects:\({\text{c}}_{\text{ge}}^{2}=\frac{{{\upsigma }}_{\text{ge}}^{2}}{{\widehat{{\upsigma }}}_{\text{p}}^{2}};\)
Coefficient of determination of GM interaction effects:\({\text{c}}_{\text{gm}}^{2}=\frac{{{\upsigma }}_{\text{gm}}^{2}}{{\widehat{{\upsigma }}}_{\text{p}}^{2}};\)
Coefficient of determination of GEM interaction effects:\({\text{c}}_{\text{gem}}^{2}=\frac{{{\upsigma }}_{\text{gem}}^{2}}{{\widehat{{\upsigma }}}_{\text{p}}^{2}};\)
Coefficient of determination of plot effects:\({\text{c}}_{\text{perm}}^{2}=\frac{{{\upsigma }}_{\text{perm}}^{2}}{{{{\upsigma }}}_{\text{p}}^{2}};\)
Coefficient of individual repeatability:\({\uprho }=\frac{{{\upsigma }}_{\text{g}}^{2}+{{\upsigma }}_{\text{gm}}^{2}+{{\upsigma }}_{\text{perm}}^{2}}{{{{\upsigma }}}_{\text{p}}^{2}};\)
Genotypic correlation among environments:\({\text{r}}_{\text{ge}}=\frac{{{\upsigma }}_{\text{g}}^{2}}{{{\upsigma }}_{\text{g}}^{2}+{{\upsigma }}_{\text{ge}}^{2}};\)
Genotypic correlation among measurements:\({\text{r}}_{\text{gm}}=\frac{{{\upsigma }}_{\text{g}}^{2}}{{{\upsigma }}_{\text{g}}^{2}+{{\upsigma }}_{\text{gm}}^{2}};\) and
Genotypic correlation among environments and measurements:\({\text{r}}_{\text{gem}}=\frac{{{\upsigma }}_{\text{g}}^{2}}{{{\upsigma }}_{\text{g}}^{2}+{{\upsigma }}_{\text{gr}}^{2}+{{\upsigma }}_{\text{gm}}^{2}+{{\upsigma }}_{\text{gem}}^{2}}\).
To determine the optimal number of harvests for genetic selection, accuracy was calculated considering the use of m harvests (\({\text{r}}_{\text{m}}\)) (Resende et al. 2014):
We also considered the efficiency (E) of the use of m harvests in relation to the use of only one harvest (Resende et al. 2014):
To select progenies with greater adaptability, stability, and productivity, the Harmonic Mean of Relative Performance of Genotypic Values (HMRPGV) method was used. This method provides a genotypic value that is affected negatively by instability and positively by adaptability (Resende 2004):
where, n is the number of environments; \({RPGV}_{ij}\) is the Relative Performance of Genotypic Values, estimated as
where \({GV}_{ij}\) is the genotypic value of the ith genotype in the jth environment, and µ is the phenotypic mean of the jth environment. All statistical analyses were performed using the SELEGEN REML/BLUP software (Resende 2016).
Results
The genotypic effects were significant for both the mean number of fruits/plant and mean fruit production/plant, indicating genetic variability among progenies. Furthermore, the presence of GE and GEM interaction was verified for these traits. As expected for polygenic traits, there was a high level of influence of uncontrolled environmental factors, which is reflected in the residual variance that corresponds to the largest proportion of phenotypic variance (Table 2).
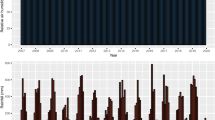
Only mean broad-sense heritability of genotypes (\({\text{h}}_{\text{mg}}^{2}\)) can be considered high. Both mean and individual broad-sense heritability (\({\text{h}}_{\text{g}}^{2}\)) were higher for mean number of fruits/plant than for mean fruit production/plant. Selective accuracy (\({\text{r}}_{\widehat{\text{g}}{\rm g}}\)) followed the same pattern. The coefficient of repeatability (ρ) showed similar magnitudes for mean number of fruits/plant and mean fruit production/plant, a positive aspect in selection when considering both traits simultaneously since the optimal number of measurements will coincide. With the use of a greater number of measurements, compared to only one measurement, the selective accuracy increases (Fig. 1). With the use of data from three harvests, the selective accuracy exceeds 0.70; with the use of 11 harvests, the selective accuracy exceeds 0.90 (Fig. 1 A). The efficiency associated with the use of m measures indicates smaller increments as the number of harvests increases (Fig. 1B). These increments become almost constant as of the ninth harvest, with only 2 % increase compared to the previous harvest.
The coefficient of determination of GE interaction effects (\({\text{c}}_{\text{ge}}^{2}\)), GM interaction effects (\({\text{c}}_{\text{gm}}^{2}\)), and plot effects (\({\text{c}}_{\text{perm}}^{2}\)) were all low for both traits (ranging from 0.004 to 0.15). The genotypic correlations across environments (rge) and environments and measurements (rgm) were higher for mean number of fruits/plant, while the genotypic correlation across measurements (rgm) was slightly higher for mean fruit production/plant.
For mean number of fruits/plant, the coincidence was high (90 %) between HMRPGV, genotypic values (µ + g), and genotypic values plus the mean effect of the GE interaction (µ + g + gem), considering the selection of the 10 best progenies. This demonstrates that the most productive progenies are also the most adapted and stable. The selection of the ten best progenies (36, 37, 11, 5, 49, 23, 6, 43, 19, and 25) provided a predicted selection gain of 2.61 fruits/plant (~ 23 %) (Table 3).
For mean fruit production/plant (kg), which is the trait of greatest economic importance, there was an 80 % coincidence between µ + g, µ + g + gem, and HMRPGV. Progenies 56 and 43, despite having higher values for µ + g and µ + g + gem compared to genotypes 25 and 38 (ranked 9th and 10th, respectively), did not show satisfactory stability. The selection of the ten best progenies (36, 5, 37, 11, 23, 49, 21, 6, 25, and 38) led to a predicted selection gain of 2.03 kg per plant (~ 11.3 %; Table 3). However, for genetic selection, the incidence rate of witches’ broom must be taken into account. Considering a rate of 30 % symptomatic plants per progeny as a tolerance threshold, progenies 36, 6, and 38 should not be selected. Thus, the choice of the seven remaining progenies (5, 37, 11, 23, 49, 21, and 25, in bold) provided a predicted selection gain of 2.05 kg (~ 11.4 %).
Discussion
LRT shows genetic variability among progenies for both the traits mean number of fruits/plant and mean fruit production/plant. Although the GE and GEM interactions were significant, the genotypic correlation across environments (\({\text{r}}_{\text{ge}}\)) was high (0.77) only for mean number of fruits/plant, according to the classification proposed by Resende and Alves (2020). These results indicate that for this trait the performance of progenies is moderately similar among the trials and some of the same progenies can be selected for them all.
The significance of the GE interaction associated with moderate genotypic correlation across environments (rge= 0.356) for mean fruit production/plant indicate that the cultivation system of T. grandiflorum can have a significant influence on the productive performance of different genotypes in different environments. Given that the varied needs of the stakeholders and intended end users must be considered when developing cultivars, it is important to highlight that the vast majority of producers in Northeast Pará use cultivation systems similar to those studied herein. Therefore, it is essential to select genotypes that have satisfactory adaptability, stability, and productivity in a range of management scenarios (Diouf et al. 2020). In studying T. cacao, Salazar et al. (2018) found that the level of shade, one of the distinguishing characteristics of each environment studied herein, can have an effect on photosynthesis and, thus, productive capacity.
Based on the coefficient of determination of the GE interaction (\({\text{c}}_{\text{ge}}^{2}\)) and genotypic correlation across environments (rge), mean fruit production/plant was more heavily influenced by the environment than mean number of fruits/plant. Resende and Alves (2020) highlighted that a useful indicator is the ratio between the variances attributed to the GE interaction and the genotype (\(\text{P}={{\upsigma }}_{\text{ge}}^{2}/{{\upsigma }}_{\text{g}}^{2}\)). Variables with P < 0.5, as is the case with mean number of fruits/plant (0.33), will not be problematic for selection; while traits with P > 0.5, such as mean fruit production/plant (1.80), tend to generate problems due to the complex GE interaction, especially when the intention is to carry out indirect selection.
Such pattern is expected since mean fruit production/plant is a quantitative trait composed of two other traits that are also polygenic: the mean number of fruits/plant and the mean fruit weight. Traits of this nature are influenced by the activity of numerous genes, combined with significant environmental effects (Kumar et al. 2017). This highlights the impact the type of management can have on the phenotypic manifestation of the evaluated traits, especially mean fruit production/plant. This fact, combined with the variability resulting from the species’ self-incompatibility (Venturieri 2011), is reflected in uncertainties about the cultivation of genetic materials that have not been evaluated in a range of environments and emphasizes the importance of studies of this nature.
On the other hand, the GM interaction was not significant. According to Ferreira et al. (2020), this result is an indication that there is consistency in the performance of genotypes across the years of evaluation. This was confirmed by the high values of the genotypic correlation across measurements for both mean number of fruits/plant and mean fruit production/plant (> 0.90). The GM interaction is mainly related to the reaction of progenies to climate change and its consequences. The development of improved genetic materials must take into account the variation of the climate between and within years, aiming to increase the resilience of the cultivars offered to producers (Atlin et al. 2017). The genotypes evaluated in this study fulfil this requirement. When evaluating both components mentioned above jointly through the triple interaction (GEM), differential behavior was observed across trials and years. However, given the non-significance of GM, it appears that most of the GEM interaction is due to the GE interaction.
According to the classification presented by Resende (2015), individual broad-sense heritability showed a low magnitude (\({\text{h}}_{\text{g}}^{2}\) < 0.15) for both traits. In contrast, estimates of mean heritability showed a high magnitude (\({\text{h}}_{\text{mg}}^{2}\)> 0.50). The low individual broad-sense heritability (\({\text{h}}_{\text{g}}^{2}\)) refers to the quantitative nature of both traits, as discussed above, making the process of selection more complex (Sousa et al. 2019). Through the interpretation of these heritability values, along with the high mean heritability (\({\text{h}}_{\text{mg}}^{2}\)) for both traits, we can infer that priority should be given to the selection of progenies, rather than the selection of ortets. This is due to the relationship between heritability and genetic gains with selection, in which the latter is a direct function of the former. Thus, heritability estimates can reveal the selection efficacy, before it is done (Schmidt et al., 2019). Therefore, smaller-scale heritabilities, as observed at the individual level, can jeopardize the genetic gains. A previous study (Bezerra et al. 2020) considered this fact to recommend the selection of full-sib families in guava (Psidium guajava L.) to achieve greater genetic gains. Another attribute of heritability is related to selection accuracy. According to Resende and Duarte (2007), values above 0.7 are considered high, as is the case for mean fruit production/plant, and above 0.9 they are considered very high, as is the case with mean number of fruits/plant. The selective accuracy, or the correlation between the true and predicted genotypic value, enables us to infer the selection precision (Resende 2002; Cavalcante et al. 2017).
Heritability is also related to the repeatability coefficient, which is the maximum value that individual broad-sense heritability can achieve (Resende 2002). Estimating the repeatability coefficient using the mixed model method has greater flexibility when compared to analysis of variance (ANOVA), as it can be used even when the assumptions required for the ANOVA are not met (Ferreira et al. 2020). Previous research has highlighted the importance of this parameter for perennial species, which have a long breeding cycle (Sanchéz et al. 2017). In that study, the authors obtained a repeatability coefficient of 0.35 for fruit production in Annona muricata L., classifying it as moderate, based on Resende (2002). Taking into account this same classification, the estimates of repeatability coefficients for both traits studied herein were of low magnitude (< 0.3), which indicates that the evaluation of several harvests is necessary for genetic selection. To achieve an accuracy of 0.70, the minimum value for selection aiming at recombination (Resende and Duarte, 2007), data from three consecutive initial harvests are enough, which is consistent with what was found for yield components in T. cacao (Carvalho et al. 2002; Mustiga et al. 2018). If the objective is only to recombine and advance the cycle, subsequent measurements are unnecessary as they would offer limited increases in efficiency (Fig. 1B) but require more time and incur higher costs related to the measurement of each harvest. If the intention is to identify genetic materials for cultivation, the evaluation of 11 harvests is recommended, since conducting such measurements can offer a selective accuracy of 0.90, a value suggested by Resende and Duarte (2007). Within the Theobroma genus, analyses similar to the present study have only been conducted for T. cacao (Carvalho et al. 2002; Dias and Kageyama 1998) and have offered substantial and fundamental results for breeding programs. For T. grandiflorum, the results presented herein will enable optimization of the breeding program, increasing gains by decreasing the time of field assessment from 15 to six years, considering the three years needed for initial establishment (juvenility period). Studies of this nature are rare for T. grandiflorum, thus demonstrating the pioneering nature of this work.
The use of HMRPGV proved to be a viable alternative for T. grandiflorum, as it allows us to infer the adaptability and stability of genotypic values. We found high levels of coincidence between the best genotypes in HMRPGV and \({\upmu }+\text{g}\), what indicates the efficiency of the method (Evangelista et al. 2021). Comparing both studied traits, it is clear that there is no perfect match as to the best genotypes. This is due to the low levels of correlation between the number of fruits and the average weight of fruits, that is, plants that produce heavy fruits and in large quantities are exceptions. The identification of these genotypes is essential for advancing the improvement of the species (Alves et al. 2021). In this context, progenies 36, 37, 11, 5, 49, 23, 6, and 25 are the most suitable, as they stand out for both traits simultaneously.
Combining the analysis of productive traits with the resistance to witches’ broom (M. perniciosa) disease, progenies 6, 36, and 38 were excluded, as they did not present satisfactory tolerance, and thus, can increase the pathogen pressure on resistant individuals. Thus, of the 25 studied progenies, 5, 37, 11, 23, 49, 21, and 25 were selected. Breeding programs for T. grandiflorum must always take into account the plants’ reaction to the fungus since it is the main pathogen that affects the cultivation of both T. grandiflorum and T. cacao (Mournet et al. 2020). Therefore, through breeding of the species, genetic materials can be developed that combine high levels of productivity and resistance to M. perniciosa, thus reducing production costs related to phytosanitary pruning and the application of fungicide. This, in turn, can mitigate the risks and effects of chemical contamination for humans, animals, and the environment (Willocquet et al. 2017).
Conclusions
Data from three and 11 harvests should be used in selection aiming at recombination and identification of genotypes for recommendation, respectively.
Seven progenies (5, 37, 11, 23, 49, 21 and 25) were identified and selected with high adaptability, stability, productivity, and resistance to witches’ broom disease.
Data availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
References
Alves RM, Sebbenn AM, Artero AS, Clement C, Figueira A (2003) Mating system in a natural population of Theobroma grandiflorum (Willd. ex Spreng.) Schum., by microsatellite markers. Genet Mol Biol 26(3):373–379. doi: https://doi.org/10.1590/S0100-29452013000300019
Alves RM, Sebbenn AM, Artero AS, Clement C, Figueira A (2007) High levels of genetic divergence and inbreeding in populations of cupuassu (Theobroma grandiflorum). Tree Genetics Genomes 3(4):289–298. doi: https://doi.org/10.1007/s11295-006-0066-9
Alves RM, Resende MDV, Bandeira BDS, Pinheiro TM, Farias DCR (2009) Evolução da vassoura-de-bruxa e avaliação da resistência em progênies de cupuaçuzeiro. Rev Bras Frutic 31(4):1022–1032. doi: https://doi.org/10.1590/S0100-29452009000400015
Alves RM, Filgueiras GC, Homma AKO (2014) Aspectos socioeconômicos do cupuaçuzeiro na Amazônia: do extrativismo a domesticação. In: Santana AC (ed) Mercado, cadeias produtivas e desenvolvimento rural na Amazônia, 1st edn. UFRA, Belém, pp 197–223
Alves RM, Chaves SFS, Alves RS, Santos TG, Araújo DG, Resende MDV (2021) Cupuaçu tree genotype selection for an agroforestry system environment in the Amazon. Pesq Agropec Bras 56:e02139. doi: https://doi.org/10.1590/S1678-3921
Atlin GN, Cairns JE, Das B (2017) Rapid breeding and varietal replacement are critical to adaptation of cropping systems in the developing world to climate change. Glob. Food Sect. 12:31–37. doi: https://doi.org/10.1016/j.gfs.2017.01.008
Bezerra CM, Ribeiro RM, Viana AP, Cavalcante NR, Silva FAD, Ambrósio M, Amaral Junior ATD (2020) Guava breeding via full-sib family selection: conducting selection cycle and divergence between parents and families. Crop Breed Appl Biotechnol 20(1):e256520112. doi: https://doi.org/10.1590/1984-70332020v20n1a12
Carvalho CGP, Cruz CD, Almeida CMVC, Machado PFR (2002) Yield repeatability and evaluation period in hybrid cocoa assessment. Crop Breed Appl Biotechnol 2(1):149–156. doi: https://doi.org/10.12702/1984-7033.v02n01a19
Cavalcante NR, Krause W, Viana AP, Silva CA, Porto KKX, Martinez RAS (2017) Anticipated selection for intrapopulation breeding of passion fruit. Acta Sci Agron 39(2):143–148. doi: https://doi.org/10.4025/actasciagron.v39i2.31022
Chipeta MM, Melis R, Shanahan P, Sibiya J, Benesi RMI (2017) Genotype x environment interaction and stability analysis of cassava genotypes at different harvest times. J Anim Plant Sci 27:901–919
Coelho IF, Peixoto MA, Evangelista JSPC, Alves RS, Sales S, Resende MDV et al (2020) Multiple-trait, random regression, and compound symmetry models for analyzing multi-environment trials in maize breeding. PLoS ONE 15(11):e0242705. doi: https://doi.org/10.1371/journal.pone.0242705
Dias LAS, Kageyama PY (1998) Repeatability and minimum harvest period of cacao (Theobroma cacao L.) in Southern Bahia. Euphytica 102(1):29–35. doi: https://doi.org/10.1023/A:1018373211196
Dias PC, Xavier A, Resende MDV, Barbosa MHP, Bierkaski FA, Estopa RA (2018) Genetic evaluation of Pinus taeda clones from somatic embryogenesis and their genotype x environment interaction. Crop Breed Appl Biotechnol 18:55–64. doi: https://doi.org/10.1590/1984-70332018v18n1a8
Diouf I, Derivot L, Koussevitzky S, Carretero Y, Bitton F, Moreau L, Causse M (2020) Genetic basis of phenotypic plasticity and genotype × environment interactions in a multi-parental tomato population. J Exp Bot 71(18):5365–5376. doi: https://doi.org/10.1093/jxb/eraa265
Evangelista JSPC, Alves RS, Peixoto MA, Resende MDV, Teodoro PE, Silva FL, Bhering LL (2021) Soybean productivity, stability, and adaptability through mixed model methodology. Cienc Rural 51(2):e20200406. doi: https://doi.org/10.1590/0103-8478cr20200406
Ferreira FM, Alves RS, Elizeu AM, Benites FRG, Resende MDV, Souza Sobrinho F, Bhering LL (2020) Estimates of repeatability coefficients and optimum number of measures for genetic selection of Cynodon spp. Euphytica 216(5):1–11. doi: https://doi.org/10.1007/s10681-020-02605-x
Genovese MI, Lannes SCS (2009) Comparison of total phenolic content and antiradical capacity of powders and “chocolates” from cocoa and cupuassu. Food Sci Technol 29:810–814. doi: https://doi.org/10.1590/S0101-20612009000400017
Gramacho KP, Luz EDMN, Silva FS, Lopes UV, Pires JL, Pereira L (2016) Pathogenic variability of Moniliophthora perniciosa in three agroecological zones of the cacao region of Bahia, Brazil. Crop Breed Appl Biotechnol 16:7–13. doi: https://doi.org/10.1590/1984-70332016v16n1a2
Henderson CR (1975) Best linear unbiased estimation and prediction under a selection model. Biometrics 31:423–447. doi: https://doi.org/10.2307/2529430
Homma AKO (2014) Extrativismo vegetal na Amazônia: história, ecologia, economia e domesticação, 2nd edn. Brasília, Embrapa
Jaimez RE, Vera DI, Mora A, Loor RG, Bailey BA (2020) A disease and production index (DPI) for selection of cacao (Theobroma cacao) clones highly productive and tolerant to pod rot diseases. Plant Pathol 69:698–712. doi: https://doi.org/10.1111/ppa.13156
Kumar J, Gupta DS, Gupta S, Dubey S, Gupta P, Kumar S (2017) Quantitative trait loci from identification to exploitation for crop improvement. Plant Cell Rep 36(8):1187–1213. doi: https://doi.org/10.1007/s00299-017-2127-y
Mournet P, Albuquerque PSB, Alves RM, Silva-Werneck JO, Rivallan R, Marcellino LH, Clément D (2020) A reference high-density genetic map of Theobroma grandiflorum (Willd. ex Spreng) and QTL detection for resistance to witches’ broom disease (Moniliophthora perniciosa). Tree Geneti Genomes 16(6):1–13. doi: https://doi.org/10.1007/s11295-020-01479-3
Mustiga GM, Gezan SA, Phillips-Mora W, Arciniegas-Leal A, Mata-Quirós A, Motamayor JC (2018) Phenotypic description of Theobroma cacao L. for yield and vigor traits from 34 hybrid families in Costa Rica based on the genetic basis of the parental population. Front Plant Sci 9:808. doi: https://doi.org/10.3389/fpls.2018.00808
Oliveira TB, Genovese MI (2013) Chemical composition of cupuassu (Theobroma grandiflorum) and cocoa (Theobroma cacao) liquors and their effects on streptozotocin-induced diabetic rats. Food Res Int 51:929–935. doi: https://doi.org/10.1016/j.foodres.2013.02.019
Para [Internet] Secretaria de Estado de Desenvolvimento Agropecuário e da Pesca: Indicadores Agropecuários 2020 [cited 2021 Apr 21]. Available from: http://www.sedap.pa.gov.br/content/cupua%C3 %A7u
Patrocinio NGRB, Ceresini PC, Gomes LZ, Resende ML, Mizubbuti ESG, Gramacho KP (2017) Population structure and migration of the witches’ broom pathogen Moniliophthora perniciosa from cacao and cultivated and wild solanaceous hosts in southeastern Brazil. Plant Pathol 66:900–911. doi: https://doi.org/10.1111/ppa.12636
Patterson HD, Thompson R (1971) Recovery of inter-block information when block sizes are unequal. Biometrika 58:545–554. doi: https://doi.org/10.1093/biomet/58.3.545
Pugliese AG, Tomas-Barberan FA, Truchado P, Genovese MI (2013) Flavonoids, proanthocyanidins, vitamin C, and antioxidant activity of Theobroma grandiflorum (Cupuassu) pulp and seeds. J Agric Food Chem 61:2720–2728. doi: https://doi.org/10.1021/jf304349u
Resende MDV (2002) Genética biométrica e estatística no melhoramento de plantas perenes, 1st edn. Embrapa Informação Tecnológica: Brasília
Resende MDV (2004) Métodos estatísticos ótimos na análise de experimentos de campo. Embrapa Florestas:Colombo
Resende MDV, Duarte JB (2007) Precisão e controle de qualidade em experimentos de avaliação de cultivares. Pesqui Agropecu Trop 37(3):182–194
Resende MDV, Silva FF, Azevedo CF (2014) Estatística Matemática, Biométrica e Computacional, 1st edn. Visconde do Rio Branco, Suprema
Resende MDV (2015) Genética quantitativa e de populações, 1st edn. Visconde do Rio Branco, Suprema
Resende MDV (2016) Software Selegen-REML/BLUP: a useful tool for plant breeding. Crop Breed Appl Biotechnol 16:330–339. doi: https://doi.org/10.1590/1984-70332016v16n4a49
Resende MDV, Alves RS (2020) Linear, Generalized, Hierarchical, Bayesian and Random Regression mixed models in genetics/genomics in plant breeding. Funct Plant Breed J 2:1–31. doi: https://doi.org/10.35418/2526-4117/v2n2a1
Salazar JCS, Melgarejo LM, Casanoves F, Rienzo JA, DaMatta FM, Armas C (2018) Photosynthesis limitations in cacao leaves under different agroforestry systems in the Colombian Amazon. PLoS ONE 13(11):e0206149. doi: https://doi.org/10.1371/journal.pone.0206149
Sanchéz CFB, Alves RS, Garcia A, Teodoro PE, Peixoto LA, Silva LA, Bhering LL, Resende MDV (2017) Estimates of repeatability coefficients and the number of the optimum measure to select superior genotypes in Annona muricata L. Genet. Mol Res 16(3):gmr16039753. doi: https://doi.org/10.4238/gmr16039753
Schmidt P, Hartung J, Bennewitz J, Piepho H (2019) Heritability in plant breeding on a genotype-difference basis. Genetics 212:991–1008. doi: https://doi.org/10.1534/genetics.119.302134
Silva EM, Nunes EWLP, Costa JM, Ricarte AO, Nunes GHS, Aragão FAZ (2019) Genotype x environment interaction, adaptability and stability of ‘Piel de Sapo’ melon hybrids through mixed models. Crop Breed Appl Biotechnol 19:402–411. doi: https://doi.org/10.1590/1984-70332019v19n4a57
Silva LMR, Sousa PHM, Sabino LBS, Prado GM, Torres LBV, Maia GA, Figueiredo RW, Ricardo NMPS (2020) Brazilian (North and Northeast) fruit by products. In: Campos-Vega R, Oomah BD, Vergara-Castañeda HA (eds) Food Wastes and By-products: Nutraceutical and Health Potential. John Wiley and Sons, New Jersey, pp 127–158
Sousa TV, Caixeta ET, Alkimim ER, Oliveira ACB, Pereira AA, Sakiyama NS, Zambolim L, Resende MDV (2019) Early selection enabled by the implementation of genomic selection in Coffea arabica breeding. Front. Plant Sci 9:1934
Venturieri GA (2011) Flowering levels, harvest season and yields of cupuassu (Theobroma grandiflorum). Acta Amaz 41:143–152. doi: https://doi.org/10.1590/S0044-59672011000100017
Wilks SS (1938) The Large-Sample Distribution of the Likelihood Ratio for Testing Composite Hypotheses. Ann Math Stat 9:60–62. doi: https://doi.org/10.1214/aoms/1177732360
Willocquet L, Savary S, Yuen J (2017) Multiscale phenotyping and decision strategies in breeding for resistance. Trends Plant Sci 22(5):420–432. doi: https://doi.org/10.1016/j.tplants.2017.01.009
Acknowledgements
The authors would like to thank the rural producers Emerson Tokumaru, Paulino Taguchi, Michinori Konagano, Seia Takaki, Elton Takaki, and Maria do Socorro Lima for generously providing their properties to install and conduct the trials. We also thank the agricultural technician José Raimundo Quadros Fernandes for assistance with fieldwork. Saulo F. S. Chaves was supported by the master’s degree fellowship granted by CAPES, Rodrigo S. Alves was supported by post-doctoral fellowship granted by FAPEMIG, and Alexandre M. Sebbenn and Luiz A. S. Dias were supported by research productivity fellowships granted by CNPq.
Funding
This research was supported by CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico), CAPES (Coordenação de Aperfeiçoamento de Pessoal de Nível Superior), FAPEMIG (Fundação de Amparo à Pesquisa do Estado de Minas Gerais) and Embrapa Amazônia Oriental.
Conflict of interestThe authors declare that they have no conflict of interests.
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Saulo F. S. Chaves, Rafael M. Alves, Rodrigo S. Alves, Alexandre M. Sebbenn, Marcos D. V. Resende and Luiz A. S. Dias performed material preparation, data collection and analysis. Saulo F. S. Chaves wrote the first draft of the manuscript and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Chaves, S.F.d., Alves, R.M., Alves, R.S. et al. Theobroma grandiflorum breeding optimization based on repeatability, stability and adaptability information. Euphytica 217, 211 (2021). https://doi.org/10.1007/s10681-021-02944-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10681-021-02944-3