Abstract
Research to control yield losses from Sclerotinia (Sclerotinia sclerotiorum) in oilseed rape (Brassica napus) has focused on stem resistance. However, resistance to leaf infection against this pathogen would also be beneficial, both in limiting additional plant leaf damage and in reducing inoculum build up within-crop and resultant spread onto stems. Three B. napus breeding populations, C2 (NC-8 × RQ-001-NCA-8 NC2-7), C5 (cv. Charlton × RQ-001-NCA-8 NC2-7) and C6 (cv. Charlton × NC4-5), were screened for leaf resistance (based on mean lesion diameter) under controlled environment conditions. Each population consisted of parents (P1 and P2), F1, F2, BC1P1 and BC2P2, except for C5 that lacked BC1P1. Moderate broad sense heritability for leaf resistance (0.45) to S. sclerotiorum was only found in population C6, where genetic variance was mostly non-additive. Analyses of generation means and variances indicated that both dominance and complex epistatic interactions were present in C6. Bivariate analysis revealed a positive genetic covariance between the non-additive effects for mean leaf lesion and cotyledon lesion diameters, and significant negative covariance of residuals, which supports a common genetic control of cotyledon and leaf resistance to S. sclerotiorum. These results will guide breeders in selection and development of genotypes with both cotyledon and leaf resistance against this important pathogen worldwide.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Sclerotinia stem rot (Sclerotinia sclerotiorum) is a devastating disease affecting host oilseed rape (Brassica napus L.) and mustard (B. juncea L.) worldwide, including in Australia, North America, China and Europe. In Australia, it causes up to 24% yield loss (Garg et al. 2008), translating into an estimated AU$10 million yield loss per annum (Murray and Brennan 2012). The mode of germination for this pathogen is usually myceliogenic or carpogenic; the former of these causes direct stem infection, generally from soil, while the latter infects through airborne ascospores (Singh et al. 2008a, b, 2010). In Western Australia, up to 29% of oilseed rape crops are infected by this pathogen in any one season (Khangura et al. 2014). While most losses come directly from S. sclerotiorum attack on adult plant stems, infection of leaves of seedlings and older Brassicaceae plants also occurs and is also important (Garg et al. 2008, 2010b; Hims 1979; Laemmlen 2001; Delourme et al. 2012; Uloth et al. 2013, 2014; Barbetti et al. 2014; You et al. 2016). The expression of disease severity differs across different isolates/pathotypes (Ge et al. 2012, 2015), different plant age/growth stages (e.g., Li et al. 2006b; Uloth et al. 2014), and different plant components e.g., cotyledon, leaf and stem (e.g., Garg et al. 2008, 2010b; You et al. 2016; Uloth et al. 2013; Ge et al. 2015).
Mitigation strategies include cultural practices, fungicide application and host resistance. Cultural control is less-than-ideal for avoiding infested stubbles and carry-over inoculum from previous crops (Garg et al. 2008, 2010d; Barbetti et al. 2015b; Uloth et al. 2015a; Barbetti 2019). Fungicides (e.g., prothioconazole + tebuconazole) are widely used for control of Sclerotinia, but they are relatively expensive (Barbetti et al. 2015b) and unreliable. It is difficult to synchronise application timing with dispersal of ascospores (Bolton et al. 2006). The most common recommendation is that fungicides should be applied during flowering when infected petals drop (Lee 2014), but petal infestation is often unrelated to stem infection, particularly in Australia (Uloth et al. 2013; You et al. 2016). These limitations of cultural and chemical practices emphasise the need for effective host resistance. However, even then, the use of Sclerotinia-resistant varieties should be integrated with other management options (Barbetti et al. 2014, 2015b; Barbetti 2019).
There are significant challenges in developing resistant varieties. These include differences in expression of resistance as associated with plant age or growth stage (Singh et al. 2008a; Uloth et al. 2013), with different isolates (Ge et al. 2012, 2015), with temperature differences (Uloth et al. 2015b), and, most importantly, with pathotype of S. sclerotiorum. Defined pathotypes of S. sclerotiorum are recommended for controlled screening for resistance, and pathogen infection is very sensitive to humidity and mycelia convert quickly towards resting stage once growth conditions (e.g. nutrition and appropriate environmental conditions) are not appropriate, even for a short time (Khan et al. 2020, Ge et al. 2012, 2015; Ge and Barbetti 2019; Uloth et al. 2013, 2014, 2015a,b; Barbetti et al. 2014, 2015a; Purnamasari et al., 2015; You et al. 2016).
Many host genotypes with stem resistance to S. sclerotiorum have been identified. Examples have been reported in India (Rana et al. 2017, 2019; Atri et al. 2019), China (Zhao and Meng 2003; Zhao et al. 2004, 2006; Zhang et al. 2011; Wu et al. 2013), and in Western Australia (Li et al. 2006a, 2009; Uloth et al. 2013; Barbetti et al. 2014, 2015a). Wild and weedy species (Uloth et al. 2013) and/or their introgressions into crop Brassica species have also been targeted. Examples include introgressions into B. juncea (Garg et al. 2010a; Barbetti et al. 2014; You et al. 2016; Rana et al. 2017, 2019; Atri et al. 2019) as well as resistances introgressed from the B-genome of B. carinata into B. napus (Barbetti et al. 2014; You et al. 2016). In contrast to stem resistance, there has been little attention on leaf resistance in leafy horticultural or in broadacre oilseed Brassica crops. Exceptions include seedling resistance to S. sclerotiorum in Brassica seedling mixtures grown as salad vegetable mixes (Hims 1979), and in seedling-transplant production systems (Laemmlen 2001).
More recently, there has been interest in locating leaf resistance in oilseed rape to reduce within-crop inoculum that readily transfers into stem infections (You et al. 2016). Uloth et al. (2013) demonstrated high-level leaf resistances in the field from within a range of diverse forage and/or vegetable crucifers, particularly B. rapa var. chinensis ‘Ivory’ and R. sativus ‘Oriental radish’. In that study, leaf and stem resistances were not correlated and appeared to be under separate genetic control. Similarly, You et al. (2016) showed no correlation between expressions of stem versus leaf resistances in a field study of 52 Chinese genotypes of B. oleracea var. capitata, 14 Indian B. juncea genotypes carrying wild weedy Brassicaceae introgression(s) and four carrying B-genome introgressions, 22 Australian commercial B. napus varieties, and another 12 B. napus and B. juncea genotypes, which suggests independent inheritance of stem and leaf resistance. Disi et al. (2014) confirmed the independent inheritance of leaf and stem resistance in a population derived from B. oleracea × B. incana. Similarly, Uloth et al. (2014) showed a lack of correlation between lesion size from S. sclerotiorum on the cotyledon with the severity of disease initiated by stem inoculation or natural infections in a previous field test. While cotyledon resistance often does not correlate with stem resistance in the field (Uloth et al. 2014), there are exceptions in controlled environmental conditions (Garg et al. 2008). You et al. (2016) found one Indian B. juncea line with introgressions from weedy Brassica species that displayed a significant level of both stem and leaf resistance.
Various quantitative trait loci (QTL) have been reported for leaf and/or stem resistance to S. sclerotiorum (Zhao and Meng 2003; Mei et al. 2013; Wei et al. 2014; Wei et al. 2016; Wu et al. 2013, 2016; Rana et al. 2017, 2019; Atri et al. 2019; Qasim et al. 2020), across wild Brassica types (B. oleracea, B.juncea etc.) and B. napus genotypes, suggesting a key role of quantitative (additive) effects in inheritance of the disease. However, genotypes of B. napus have shown a dramatically variable performance (Wu et al. 2016) and this needs a lot more attention. The quantitative nature of this trait has been demonstrated by a moderate to high (57–84%) broad sense heritability of stem resistance in B. napus (Wu et al. 2013; Wei et al. 2014; Wei et al. 2016; Qasim et al. 2020). Epistatic genetic control of resistance is also common across different host species or diseases i.e. non-additive genetics was observed for downy mildew (Pseudoperonospora cubensis) in muskmelon (Cucumis melo) (Shashikumar et al. 2010). Similarly, multiple epistatic interactions (Disi et al. 2014) for resistance to S. sclerotiorum were found at one or more different plant growth stages for Brassica incana and Brassica oleracea. More recently, a study that aimed at understanding the complex nature of resistance in B. napus against S. sclerotiorum at the cotyledon stage in three genetic populations was shown to be under non-additive genetic control (Khan et al. 2020). The same three populations were investigated here for leaf resistance and potential genetic correlations of leaf and cotyledon resistance against S. sclerotiorum in B. napus. Thus, the potential existence of correlated genetic factors involved in leaf and cotyledon resistance, will guide breeders in selection and development of genotypes with resistance that is effective across both tissues.
Materials and methods
Crossing populations
The background of four inbred parent lines of B. napus used in this genetic study (Charlton, NC8, NC4-5, and RQ-001-NCA-8 NC2-7), and three cross populations C2, C5, and C6 derived from these parents, were described previously (Khan et al. 2020). Among the parents, NC-8 is known to have stem resistance to S. sclerotiorum (Uloth et al. 2013), and Charlton has both cotyledon (Garg et al. 2008; 2010b, c; 2013) and stem (Uloth et al. 2013) resistance. RQ-001-NCA-8 NC2-7 is known to have cotyledon-susceptibility (Khan et al. 2020), while NC4-5 is highly susceptible to stem rot (Uloth et al. 2013). Control check varieties were B. napus cv. Mystic and B. napus cv. Rainbow, as used previously (Khan et al. 2020). B. napus cv. Mystic was reported to have both cotyledon (Garg et al. 2008; 2010b) and stem resistance (Uloth et al. 2013; 2015a) to S. sclerotiorum, and B. napus cv. Rainbow was cotyledon-susceptible (Garg et al. 2008) and stem-susceptible (Uloth et al. 2015a) to S. sclerotiorum. Seeds from the three cross populations, including the inbred parents (P1 and P2), F1, F2, and first backcross (BC1) of the F1 to each parent, were available (Khan et al. 2020). Plants previously infected at the cotyledon stage by S. sclerotiorum (Khan et al. 2020) were inoculated 3 weeks later on leaves 1, 2, 3, 4 and 5 in this experiment.
Plants were regularly fertilised and watered as previously described (Khan et al. 2020). Seeds of parents, F1, F2, BC1P1, BC1P2 and check varieties were sown in small pots, measuring 6.6 cm length, 6.6 cm width and 9.8 cm depth, and thinned to a single plant in each pot. Plants were grown in a controlled environment room with a temperature maintained at 18 ± 1 °C during day (12 h) and 13 ± 1 °C during night (12 h), and with light intensity of 450 µE/m2 s− 1, following the protocol of Garg et al. (2008).
Experimental design
Each of the populations C2, C5 and C6 were maintained on separate benches. In each population, there were 13–15 plants (replicates) of each parent, 12–15 plants (replicates) of the F1, 80–95 F2 individuals, and 11–15 individuals of each BC1 for C2 and C6. There were 20 plants for BC1P2 of C5 and 5 plants (replicates) each of the check varieties Mystic and Rainbow. Thus, in total, there were 167, 156 and 146 plants for C2, C5 and C6, respectively. Pots within each population were randomised in an experiment 12 columns deep by 20 rows wide, in an arrangement based on a p-rep design generated by DiGGer (Coombes 2009).
S. sclerotiorum isolate and culturing
A single isolate of S. sclerotiorum (isolate MBRS-1; available at The University of Western Australia) was used for inoculation as previously described (Khan et al. 2020). This isolate belongs to the highly virulent pathotype 76, the prevailing pathotype in Western Australia comprising approximately 74% of the S. sclerotiorum population (Ge et al. 2012). This same isolate has been used in other studies to identify cotyledon (Garg et al. 2008; Garg et al. 2010b; Uloth et al. 2014), leaf (Uloth et al. 2013) and stem (Li et al. 2006a, 2007, 2009; Ge et al. 2012; Uloth et al. 2013, 2015a, b, 2016) resistances in B. napus and/or B. juncea genotypes. The isolate was cultured according to the methods used by Garg et al. (2008), but with slight modification. Briefly, a single sclerotium of MBRS-1 was surface sterilized in 1% (v/v) sodium hypochlorite and 70% ethanol for 4 min followed by double washing in sterile distilled water for 1 min as described by Clarkson et al. (2003). The sclerotium was cut in half and placed cut side down onto potato dextrose agar (PDA; Merck). S. sclerotiorum was then sub-cultured and maintained at room temperature (23 ± 2 °C) on PDA.
Inoculum production
The method used for inoculum production was as previously described by Garg et al. (2008). Seven colonised agar plug disks (5 mm2) were cut from growing edges of three-day-old S. sclerotiorum colonies and added to 250 mL flasks each having 75 mL of sterilized potato dextrose liquid broth medium (PDB: 24 g potato dextrose, 10 g peptone and water to make 1 L). Flasks were shaken on an Innova 2300 platform shaker (New Brunswick Scientific, Edison, NJ) at 120 rpm/min. Three days later, colonies of S. sclerotiorum were harvested and thoroughly washed twice using sterilised deionised water. The fungal mats obtained were transferred to 125 mL of the same liquid medium and mycelia macerated in a Breville® food grinder for 3 min. Macerated mycelial suspension was then filtered through four layers of muslin cloth and concentration adjusted with the same liquid medium to 105 hyphal fragments mL− 1 using a haemocytometer (SUPERIOR, Berlin, Germany) counting chamber.
Leaf inoculation and disease assessment
Inoculations were made 5 weeks after sowing on the same plants as used by Khan et al. (2020) for cotyledon resistance assessment. Leaf inoculations followed the technique described by Garg et al. (2008). A single droplet (10 µL) of mycelial suspension was deposited by micropipette onto the left and right lobes of fully expanded leaves of plants. Up to five fully expanded leaves were inoculated (maximum ten inoculation sites). The mycelial suspension was regularly mixed by shaking to prevent clumping of hyphae. The temperature remained constant (13 ± 1 °C) and high humidity was maintained by hand misting inoculated plants with deionized water and covering trays of 20 plants with a polythene cover to maintain high humidity for 4 days post-inoculation. After inoculation, to ensure reliable and consistent infection, low light intensity of 65 µE/m2 s− 1 was maintained for 2 days, then plants were returned to the original light intensity on the third day after inoculation. Those conditions were then maintained until the end of the experiment. Four days after inoculation, lesion diameter was recorded to the nearest millimetre on each leaf lobe. The mean leaf lesion diameter (mm) at all inoculation sites was used as the leaf resistance score for each seedling.
Data analysis
A linear mixed model for mean leaf lesion diameter was fitted using ASReml-R (v. 3.0) software (Butler et al. 2009), which produced residual maximum likelihood (REML) estimates of the variance parameters and best linear unbiased prediction (BLUP) of the random effects. In each population, a pedigree file was used for generating genetic relationships. Both additive (predicted breeding values) and non-additive genetic effects were included in the mixed model analysis of mean leaf lesion diameter.
Analysis of generation means and variance for mean leaf lesion diameter followed the method of Mather and Jinks (1982), and was applied to populations C2 and C6 where all generations (P1, P2, F1, F2, and BC1P1 and BC1P2) were available. This analysis was conducted in Indo-stat, version 7.5 software (http://www.indostat.org, Hyderabad, India) and SAS (SAS Institute 1999).
Scaling tests (Mather and Jinks 1982) were undertaken for populations C2 and C6 (in which all generations were present), to assess the validity of the additive-dominance model for leaf lesion diameter. The values of A, B, C and D were used to determine if a simple additive-dominance model was adequate to explain the observations, or if non-allelic interactions were present.
The parameters m (mid-point), [d] (additive), and [h] (dominance) and terms for additive × additive [i], additive × dominance [j] and dominance × dominance [l] non-allelic interactions were fitted following Mather and Jinks (1982). The significance of each parameter was assessed using a t-test.
We utilised bivariate analysis in ASREML-R, as described in Ganesalingam et al. (2013), to test for genetic covariance between leaf resistance scores (mean leaf lesion diameter) and cotyledon resistance scores (mean cotyledon lesion diameter) which were previously reported in Khan et al. (2020) in population C6. Firstly, a univariate model was conducted with a diagonal variance structure for the effects of genotype on each trait. This was followed by an unstructured covariance bivariate model that allowed for covariance of non-additive genetic effects and residuals.
Results
Analysis of variance
A wide range of mean leaf lesion diameter was observed across the parents, F1, F2, and available backcrosses to each parent. Susceptible genotypes showed necrosis and water soaked lesions while more resistant genotypes confined the lesion to the inoculation droplet location. Moderate broad-sense heritability for mean leaf lesion diameter was evident in population C6, and the genetic variance was mostly non-additive (Table 1).
Scaling tests (test of the additive-dominance model)
Populations C2 and C5 did not show any significant genetic effects for mean leaf lesion diameter, so we report results for C6 only (Table 2). The significant value of parameter C for population C6 shows the inadequacy of the additive-dominance model, and indicates the presence of epistasis.
Effects and magnitude of epistatic interactions
The additive, dominance and interaction parameters for mean leaf lesion diameter were further explored for population C6 (Table 3). Highly significant dominance [h] effects were observed and additive × additive interaction effects [i] were also significant for this population. The type of dominance for mean leaf lesion diameter expressed in the F1 was above the mid-point in the direction of susceptibility (Table 1; Fig. 1).
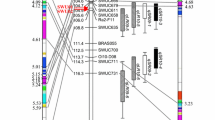
Predicted values for mean leaf lesion diameter and mean cotyledon lesion diameter (mm) in population C6 from bivariate analysis, allowing for covariance of genetic effects and residuals. Parent Charlton (closed diamond), parent NC4-5 (open diamond), F1 (cross), control variety Mystic (open triangle), control variety Rainbow (closed triangle), F2 progeny (open circles), and BC1 progeny (closed circles). Average standard error of mean cotyledon and leaf lesion diameter in F2 and BC1 single plants was 1.65 mm and 1.64 mm, respectively
Bivariate analysis comparing cotyledon with leaf disease
The loglikelihood for the diagonal variance model (−675.2676) significantly increased in the unstructured covariance bivariate model (−673.3301) (P < 0.05) which included covariance of non-additive genetic effects and residuals for mean leaf lesion and cotyledon lesion diameter in population C6. As expected (following Khan et al. 2020) for C6, there was a significant non-additive genetic variance for cotyledon lesion diameter (5.1 ± 1.7). The non-additive genetic variance for leaf lesion diameter (4.5 ± 2.3) was also significant. The benefit of the bivariate analysis arose from a positive genetic covariance between the non-additive effects for mean leaf lesion and cotyledon lesion diameter (1.9 ± 1.5, correlation r = 0.40), and significant negative covariance of residuals (−2.4 ± 1.2, r = −0.30). The predicted total genetic values for mean leaf lesion diameter and mean cotyledon lesion diameter for population C6 in the unstructured covariance bivariate model are shown in Fig. 1.
Discussion
This study has provided critical new understanding of patterns of inheritance for leaf resistance against S. sclerotiorum in B. napus. For the first time, we show genetic correlation of resistance in cotyledon and leaf tissue, which for resistance in leaf and cotyledon tissue. There have been previous studies comparing expression of leaf and stem resistances (e.g., Bradley et al. 2006). However, the current study is the first to include comparisons between expression of leaf and cotyledon resistances, and, importantly, these were based on the same plants in the same genetic populations. Previous studies were either based on detached and/or excised leaves (Mei et al. 2011; Wu et al. 2013; Disi et al. 2014) or natural infections as part of a stem rot experiment (Uloth et al. 2013; You et al. 2016). Both leaf resistance (this study) and cotyledon resistance (Khan et al. 2020) in population C6 were non-additive; leaf resistance was mostly due to dominance and additive × additive epistasis (Table 3), whereas cotyledon resistance was due to dominance × dominance epistasis (Khan et al. 2020).
Population C6 showed moderate heritability (broad sense) for leaf resistance to S. sclerotiorum, supporting the notion that oilseed rape has significant potential for selection, based on leaf resistance, to this devastating pathogen (Uloth et al. 2013, 2014; You et al. 2016). Population C2, however, had a low broad sense heritability (Table 1) and since, neither one of the parents (NC-8 or RQ-001-NCA-8 NC2-7) had leaf resistance reported before, a further testing for this population is recommended. One of the major reasons for higher variance in F1 might be, the fact that both the parents in C2 cross were introgression lines which can bring in degree of variation in F1 due to structural heterozygosity inherent in the parents. Further, it would be prudent to research their behaviour for stem resistance also, as NC-8 (P1) is reported to have stem resistance (Uloth et al. 2013). Since, no significant genetic effects observed for C2 or C5, we did not discuss these further. Importantly, the resistances identified in this study and that of Khan et al. (2020) are highly relevant, as pathotype 76 that we used in both studies makes up to three quarters of the total S. sclerotiorum isolates in Western Australia (Ge et al. 2012). Critically, Barbetti et al. (2014) showed that the stem resistance could be either isolate-dependent or isolate-independent, the latter being ideal for commercial exploitation of host resistance to multiple prevailing pathotypes.
This study supports previous work on cotyledons (Khan et al. 2020) that host resistance is non-additive and a simple additive-dominance model does not adequately explain leaf and cotyledon resistance to S. sclerotiorum. For population C6 (leaf resistance), the same sign (±) for dominance × dominance [l] and dominance effects [h] indicates a complementary type of epistatic genetic control. However, a significant component of genetic variance was due to additive × additive [i] interactions which are the result of interactions between homozygous loci and can be fixed as selfing progresses beyond F2. Non-additive genetic control for inheritance of resistance has been reported for other diseases, for example, against downy mildew (Pseudoperonospora cubensis) in muskmelon (Cucumis melo) (Shashikumar et al. 2010). Hence, while our findings are not unique across all diseases, they are unique for explaining leaf resistance to S. sclerotiorum in oilseed rape (B. napus) and corroborate our earlier reports for inheritance patterns of cotyledon resistance (Khan et al. 2020).
The current study showed covariance of non-additive genetic effects and residuals for mean lesion diameter on cotyledons and leaves in population C6. Based on the same pathotype 76, Uloth et al. (2014) reported a correlation (r = 0.22) between cotyledon and leaf resistances across diverse crucifer species (Uloth et al. 2014). Other studies for leaf and stem resistances show either no correlation (e.g., Uloth et al. 2013; You et al. 2016) or low to moderate correlation e.g., r = 0.15 (Zhao and Meng 2003), r = 0.18–0.46 (Wu et al. 2013) and r = 0.31 (Disi et al. 2014). The practical significance of testing different plant growth stages in controlled environments for resistance to this pathogen in oilseed Brassicas, and its relevance to field application, was also demonstrated by Singh et al. (2008a).
Our results concur with those for partial resistance identified by Zhao and Meng (2003) in an F2 population of B. napus at the seedling stage against S. sclerotiorum. Additive control of resistance against S. sclerotiorum was reported by Mei et al. (2013) in a B. oleracea F2 population and by Wu et al. (2016) in B. napus accessions. Further, multiple epistatic control (additive × additive, additive × dominance, and dominance × dominance) for leaf or stem resistance to this pathogen in B. napus has also been reported (Disi et al. 2014). Partial resistance to this pathogen is found in other crops, such as in soybean (Glycine max) (e.g., Kim & Diers 2000) and in sunflower (Helianthus annuus) (e.g., Amoozadeh et al. 2013, 2015). Additive genes for resistance were also found in dry bean (Phaseolus vulgaris) (Fuller et al. 1984), cauliflower (B. oleracea var. botrytis) (Baswana et al. 1991) and more recently (for leaves/stem) in B. napus (Wu et al. 2013). This evolving nature of resistance to S. sclerotiorum demands continued efforts, such as challenging new host genotypes with prevailing pathotypes, if we are to have more practical and effective solutions to include in existing plant breeding pipelines. Moreover, dominance and additive × additive epistatic interactions for leaf and stem resistance to this pathogen were reported by Disi et al. (2014) in Brassica interspecific cross progeny. Importantly, we used the same isolate (pathotype 76) as previously used for cotyledon (Garg et al. 2008, 2010b; Uloth et al. 2014; Khan et al. 2020), leaf (Uloth et al. 2013; You et al. 2016) and stem (Li et al. 2006a, 2007, 2009; Ge et al. 2012; Uloth et al. 2013, 2015a, b, 2016) resistances in B. napus and/or B. juncea genotypes. This allows a comparison of resistance across these studies, and therefore potential common genetic and/or phenotypic expression in host genotypes across growth stages (e.g., cotyledon, leaves and stem). Different phenotypic expression at different growth stages is common for other diseases, for example such as blackleg (Leptosphaeria maculans) in B. napus (Li et al. 2006b), downy mildew (Peronospora parasitica) in broccoli (Brassica oleracea L. italica group) (Dickson and Petzoldt 1993), and Pseudomonas syringae (pv. tomato or maculicola) (Kus et al. 2002) and Cauliflower mosaic virus (Leisner et al. 1993) in Arabidopsis.
The positive covariance of resistance across cotyledons and leaves in population C6 is in contrast with previous observations of the independent inheritance of seedling vs. mature plant resistance either through intensive phenotypic evaluations (Uloth et al. 2013; Disi et al. 2014; You et al. 2016) or through molecular studies (Zhao and Meng 2003; Zhang et al. 2011). Molecular evidence for unique resistance QTLs at the seedling stage relative to the adult plant stages (Zhao and Meng 2003; Zhang et al. 2011) was in agreement with phenotypic expressions observed by Uloth et al. (2013), You et al. (2016) and Taylor et al. (2018), all of whom failed to find any significant correlation in resistance between these two plant stages. Furthermore, You et al. (2016), who used a diverse set of 52 Chinese genotypes of B. oleracea var. capitata, 14 Indian B. juncea genotypes carrying wild weedy Brassicaceae introgression(s) and four carrying B-genome introgression, 22 Australian commercial B. napus varieties, and another 12 B. napus and B. juncea genotypes, found independent inheritance of cotyledon vs. adult plant resistances. Also, Disi et al. (2014) confirmed the generally independent nature of inheritance for both leaf and stem resistances in a study of a population derived from B. oleracea × B. incana. Similarly, Uloth et al. (2014) reported a lack of correlation between lesion size from S. sclerotiorum on the cotyledon with the severity of stem disease initiated either by stem inoculation or by ‘natural’ ascospores in the field. However, in contrast, there are examples that complement our study, showing low (e.g., Uloth et al. 2014) to significant moderate (e.g., Garg et al. 2008; Wu et al. 2013) correlation between cotyledon and adult disease ratings in independent trials. Importantly, You et al. (2016), found an Indian B. juncea line carrying weedy introgression that displayed a significant level of both stem and leaf resistance. More recent evidence demonstrates Sclerotinia resistance is under the influence of uncharacterized multiple (quantitative) genes (Rana et al. 2017). Rana et al. (2017, 2019) and Atri et al. (2019) focused on introgression of such resistances, first into B. juncea using either B. fruticulosa or E. cardaminoides (Erucastrum cardaminoides) and, later, proposed introduction of such resistances into B. napus. Furthermore, the integration of resistance QTL identified by Li et al. (2015) could also help to locate genes for early/and or adult plant stage resistances.
Similar as with Zhao et al. (2006) and Khan et al. (2020), we found some transgressive segregants in population C6 that showed particular potential as breeding material. Such resistance could be fixed by selfing in later generations, as done for Chinese cabbage (B. rapa ssp. pekinensis) in the F5 for resistance against club root (Plasmodiophora brassicae) (Niemann et al. 2017). One advantage of screening for resistance to S. sclerotiorum at the cotyledon or leaf stages is that such screening is more rapid and efficient than field screening on adult plants (Taylor et al. 2015). However, such an approach is dependent upon there being significant correlation between the relevant growth stages.
In conclusion, the positive correlation we found between resistances expressed at the leaf stage in the current study to that of cotyledons in the same genotypes in the Khan et al. (2020) study could be utilised for further genetic studies. Leaf resistance against S. sclerotiorum, along with cotyledon resistance, would be beneficial not only in limiting plant leaf damage and in protecting young plants, but also would reduce inoculum build up within-crop and reduce spread onto stems. We believe this study, for the first time, will guide breeders in selection and development of genotypes with combined cotyledon and leaf resistance against prevailing/characterised pathotypes of S. sclerotiorum. Overall, the use of characterised pathogens, selectable genetic variation, and combined cotyledon/leaf resistances makes the outcome of this study readily available and of practical relevance for B. napus breeders in developing effective host resistances.
Data availability
All critical data supporting the conclusions of this manuscript will be made available by the authors, without undue reservation, to any qualified researcher.
References
Amoozadeh A, Darvishzadeh R, Haddadi P, Abdollahi MB, Rezaee DY (2013) Genetic analysis of partial resistance to basal stem rot (Sclerotinia sclerotiorum) in Sunflower. Genetika 45:737–748
Amoozadeh M, Darvishzadeh R, Davar R, Abdollahi MB, Haddadi P, Basirnia A (2015) Quantitative trait loci associated with isolate specific and isolate non-specific partial resistance to Sclerotinia sclerotiorum in sunflower. J Agric Sci Technol 17:213–226
Atri C, Akhtar J, Gupta M, Gupta N, Goyal A, Rana K, Kaur R, Mittal M, Sharma A, Singh MP, Sandhu PS (2019) Molecular-genetic analysis of defensive responses of Brassica juncea – B. fruticulosa introgression lines to Sclerotinia infestation. Sci Rep 9:17089 |. https://doi.org/10.1038/s41598-019-53444-3
Barbetti MJ, Banga SK, Fu TD, Li YC, Singh D, Liu SY, Ge XT, Banga SS (2014) Comparative genotype reactions to Sclerotinia sclerotiorum within breeding populations of Brassica napus and B. juncea from India and China. Euphytica 197:47–59
Barbetti MJ, Li CX, Banga SS, Banga SK, Singh D, Sandhu PS, Singh R, Liu SY, You MP (2015a) New host resistances in Brassica napus and Brassica juncea from Australia, China and India: Key to managing Sclerotinia stem rot (Sclerotinia sclerotiorum) without fungicides. Crop Prot 78:127–130
Barbetti MJ, Uloth M, Banga SS, Liu SY, Salisbury PA, You MP (2015b) Managing Sclerotinia stem rot in oilseed Brassicas using pathotype-independent and pathotype-dependent host resistances. In: Proceedings of 13th international rapeseed congress, Saskatoon, July 2015
Barbetti MJ (2019) Breakthrough in Brassica disease (Sclerotinia). Austgrain, Moree, New South Wales, p 29
Baswana KS, Rastogi KB, Sharma PP (1991) Inheritance of stalk rot resistance in cauliflower (Brassica oleracea var. Botrytis L.). Euphytica 57:93–96
Bolton MD, Thomma BP, Nelson BD (2006) Sclerotinia sclerotiorum (Lib.) de Bary: biology and molecular traits of a cosmopolitan pathogen. Mol Plant Pathol 7:1–16
Bradley CA, Henson RA, Porter PM, LeGare DG, Khot SD (2006) Response of canola cultivars to Sclerotinia sclerotiorum in controlled and field environments. Plant Dis 90:215–219
Butler DG, Cullis BR, Gilmour AR, Gogel BJ (2009) ASReml-R Reference Manual.V.3.0. Department of Primary Industries and Fisheries, Brisbane
Clarkson JP, Staveley J, Phelps K, Young CS, Whipps JM (2003) Ascospore release and survival in Sclerotinia sclerotiorum. Mycol Res 107:213–222
Coombes NE (2009) DiGGer design search tool in R. http://nswdpibiom.org/austatgen/software/. Accessed April 2017
Delourme R, Barbetti MJ, Snowdon R, Zhao J, Manzanares-Dauleux M (2012) Genetics and Genomics of Resistance. In: Edwards D, Batley J, Parkin IAP, Kole C (eds) Genetics, genomics and breeding of oilseed Brassicas. Science Publishers, CRC Press, Boca Raton, pp 276–318
Dickson MH, Petzoldt R (1993) Plant age and isolate source affect expression of downy mildew resistance in broccoli. Hortscience 28:730–731
Disi JO, Mei J, Wei D, Ding Y, Qian W (2014) Inheritance of leaf and stem resistance to Sclerotinia sclerotiorum in a cross between Brassica incana and Brassica oleracea var. alboglabra. J Agric Sci 152:146–152
Fuller PA, Coyne DP, Steadman JR (1984) Inheritance of resistance to white mold disease in a diallel cross of dry beans. Crop Sci 24:929–933
Ganesalingam A, Smith AB, Beeck CP, Cowling WA, Cullis BR (2013) A bivariate mixed model approach for the analysis of plant survival data. Euphytica 190:371–383
Garg H, Sivasithamparam K, Banga SS, Barbetti MJ (2008) Cotyledon assay as a rapid and reliable method of screening for resistance against Sclerotinia sclerotiorum in Brassica napus genotypes. Australas Plant Pathol 37:106–111
Garg H, Atri C, Sandhu PS, Kaur B, Renton M, Banga SK, Singh H, Singh C, Barbetti MJ, Banga SS (2010a) High level of resistance to Sclerotinia sclerotiorum in introgression lines derived from hybridization between wild crucifers and the crop Brassica species B. napus and B. juncea. Field Crop Res 117:51–58
Garg H, Kohn LM, Andrew M, Li H, Sivasithamparam K, Barbetti MJ (2010b) Pathogenicity of morphologically different isolates of Sclerotinia sclerotiorum with Brassica napus and B. juncea genotypes. Eur J Plant Pathol 126:305–315
Garg H, Li H, Sivasithamparam K, Kuo J, Barbetti MJ (2010c) The infection processes of Sclerotinia sclerotiorum in cotyledon tissue of a resistant and a susceptible genotype of Brassica napus. Ann Bot 106:897–908
Garg H, Sivasithamparam K, Barbetti MJ (2010d) Scarification and environmental factors that enhance carpogenic germination of sclerotia of Sclerotinia sclerotiorum. Plant Dis 94:1041–1047
Garg H, Li H, Sivasithamparam K, Barbetti MJ (2013) Differentially expressed proteins and associated histological and disease progression changes in cotyledon tissue of a resistant and susceptible genotype of Brassica napus infected with Sclerotinia sclerotiorum. PLoS ONE 8:e65205
Ge XT, Li YP, Wan ZJ, You MP, Finnegan PM, Banga SS, Sandhu PS, Garg H, Salisbury PA, Barbetti MJ (2012) Delineation of Sclerotinia sclerotiorum pathotypes using differential resistance responses on Brassica napus and B. juncea genotypes enables identification of resistance to prevailing pathotypes. Field Crops Res 127:248–258
Ge XT, You MP, Barbetti MJ (2015) Virulence differences among Sclerotinia sclerotiorum isolates determines host cotyledon resistance responses in Brassicaceae genotypes. Eur J Plant Pathol 143:527–541
Ge XT, Barbetti MJ (2019) Host response of Arabidopsis thaliana ecotypes is determined by Sclerotinia sclerotiorum isolate type. Eur J Plant Pathol 153:51–65
Hims MJ (1979) Damping-off of Brassica napus (‘mustard and cress’) by Sclerotinia sclerotiorum. Plant Pathol 28:201–202
Khan MA, Cowling W, Banga SS, You MP, Tyagi V, Bharti B, Barbetti MJ (2020) Patterns of inheritance for cotyledon resistance against Sclerotinia sclerotiorum in Brassica napus. Euphytica 216:79–89. https://doi.org/10.1007/s10681-020-02612-y
Khangura R, Burgel AV, Salam M, Aberra M, MacLeod WJ (2014) Why Sclerotinia was so bad in 2013? Understanding the disease and management options. Available from URL: http://www.giwa.org.au/pdfs/2014/Presented_Papers/Khangura%20et%20al%20presentation%20paper%20CU2014%20-DR.pdf (accessed June 2017)
Kim HS, Diers BW (2000) Inheritance of partial resistance to Sclerotinia stem rot in soybean. Crop Sci 40:55–61
Kus JV, Zaton K, Sarkar R, Cameron RK (2002) Age-related resistance in Arabidopsis is a developmentally regulated defence response to Pseudomonas syringae. Plant Cell 14:479–490
Laemmlen F (2001) Damping-off diseases. Publ No 8041. University of California, Davis
Lee N (2014) Prompt action on sclerotinia will reduce yield losses. https://grdc.com.au/news-and-media/news-and-media-releases/west/2014/07/prompt-action-on-sclerotinia-will-reduce-yield-losses. Accessed 10 July 2020
Leisner SM, Turgeon R, Howell SH (1993) The effects of host plant development and genetic determinants on the long-distance movement of cauliflower mosaic virus in infected turnip plants. Plant Cell 5:191–202
Li CX, Li H, Sivasithamparam K, Fu TD, Li YC, Liu SY, Barbetti MJ (2006a) Expression of field resistance under Western Australian conditions to Sclerotinia sclerotiorum in Chinese and Australian Brassica napus and Brassica juncea germplasm and its relation with stem diameter. Aust J Agric Res 57:1131–1135
Li H, Smyth F, Barbetti MJ, Sivasithamparam K (2006b) Relationship between Brassica napus seedling and adult plant responses to Leptosphaeria maculans is determined by plant growth stage at inoculation and temperature regime. Field Crops Res 96:428–437
Li CX, Li H, Siddique AB, Sivasithamparam K, Salisbury P, Banga SS, Banga S, Chattopadhyay C, Kumar A, Singh R, Singh D, Agnihotri A, Liu SY, Li YC, Tu J, Fu TD, Wang YF, Barbetti MJ (2007) The importance of the type and time of inoculation and assessment in the determination of resistance in Brassia napus and B. juncea to Sclerotinia sclerotiorum. Aust J Agric Res 58:1198–1203
Li CX, Liu SY, Sivasithamparam K, Barbetti MJ (2009) New sources of resistance to Sclerotinia stem rot caused by Sclerotinia sclerotiorum in Chinese and Australian Brassica napus and B. juncea germplasm screened under Western Australian conditions. Australas Plant Pathol 38:149–152
Li J, Zhao Z, Hayward A, Cheng H, Fu D (2015) Integration analysis of quantitative trait loci for resistance to Sclerotinia sclerotiorum in Brassica napus. Euphytica 205:483–489
Mather K, Jinks JL (1982) Biometrical Genetics, 3rd edn. Chapman and Hall, London, New York, ISBN-10: 0412228904
Mei J, Qian L, Disi JO, Yang X, Li Q, Li J (2011) Identification of resistant sources against Sclerotinia sclerotiorum in Brassica crops with emphasis on B. oleracea. Euphytica 177:393–399
Mei J, Ding Y, Lu K, Wei D, Liu Y, Disi JO, Li J, Liu L, Liu S, McKay J, Qian W (2013) Identification of genomic regions involved in resistance against Sclerotinia sclerotiorum from wild Brassica oleracea. Theor Appl Genet 126:549–556
Murray GM, Brennan JP (2012) The current and potential costs from diseases of oilseed crops in Australia. (Grains Research and Development Corporation No. CER00002.)
Niemann J, Kaczmarek J, Ksiazczyk T, Wojciechowski A, Jedryczka M (2017) Chinese cabbage (Brassica rapa ssp pekinensis) - a valuable source of resistance to clubroot (Plasmodiophora brassicae). Eur J Plant Pathol 147:181–198
Purnamasari MI, Cawthray GR, Barbetti MJ, Erskine W, Croser JS (2015) Camalexin production in Camelina sativa is independent of cotyledon resistance to Sclerotinia sclerotiorum. Plant Dis 99(11):1544–1549
Qasim MU, Zhao Q, Shahid M, Samad RA, Ahmar S, Wu J, Fan C, Zhou Y (2020) Identification of QTLs containing resistance genes for Sclerotinia stem rot in Brassica napus using comparative transcriptomic studies. Front Plant Sci 11:776. https://doi.org/10.3389/fpls.2020.00776
Rana K, Atri C, Gupta M, Akhatar J, Sandhu PS, Kumar N, Jaswal R, Barbetti MJ, Banga SS (2017) Mapping resistance responses to Sclerotinia infestation in introgression lines of Brassica juncea carrying genomic segments from wild Brassicaceae B. fruticulosa. Sci Rep 7:5904
Rana K, Atri C, Akhatar J, Nagra R, Goyal A, Singh MP, Kumar N, Sharma A, Sandhu P, Kaur G, Barbetti MJ, Banga SS (2019) Detection of first marker trait associations for resistance against Sclerotinia sclerotiorum in Brassica juncea–Erucastrum cardaminoides introgression lines. Front Plant Sci 10:1015
SAS Institute Inc (1999) SAS Online Doc®, Version 8, Cary, NC
Shashikumar KT, Pitchaimuthu M, Rawal RD (2010) Generation mean analysis of resistance to downy mildew in adult muskmelon plants. Euphytica 173:121–127
Singh R, Singh D, Barbetti MJ, Wade SMS, Singh H, Banga SS, Kumar A, Caixia L, Sivasithamparam K, Salisbury P, Buraton W, FU T (2008a) Sclerotinia rot tolerance in oilseed Brassica. In: Proceedings of the 12th international rapeseed congress, March 2007, Wuhan, China, pp 94–97
Singh R, Singh D, Li H, Sivasithamparam K, Yadav NR, Salisbury, Barbetti MJ (2008b) Management of Sclerotinia rot of oilseed Brassicas- a focus on India. Brassica 10:1–27
Singh R, Singh D, Salisbury P, Barbetti MJ (2010) Field evaluation of Indian and exotic oilseed Brassica napus and B. juncea germplasm against Sclerotinia stem rot. Indian J Agr Sci 80:1067–1071
Taylor A, Coventry E, Jones JE, Clarkson JP (2015) Resistance to a highly aggressive isolate of Sclerotinia sclerotiorum in a Brassica napus diversity set. Plant Pathol 64:932–940. https://doi.org/10.1111/ppa.12327
Taylor A, Rana K, Handy C, Clarkson JP (2018) Resistance to Sclerotinia sclerotiorum in wild Brassica species and the importance of Sclerotinia subarctica as a Brassica pathogen. Plant Pathol 67:433–444. https://doi.org/10.1111/ppa.12745
Uloth MB, You MP, Finnegan PM, Banga SS, Banga SK, Sandhu PS, Yi H, Salisbury PA, Barbetti MJ (2013) New sources of resistance to Sclerotinia sclerotiorum for crucifer crops. Field Crop Res 154:40–52
Uloth M, You MP, Finnegan PM, Banga SS, Yi H, Barbetti MJ (2014) Seedling resistance to Sclerotinia sclerotiorum as expressed across diverse cruciferous species. Plant Dis 98:184–190
Uloth MB, You MP, Barbetti MJ (2015a) Host resistance to Sclerotinia stem rot in historic and current Brassica napus and B. juncea varieties: critical management implications. Crop Pasture Sci 66:841–848
Uloth MB, You MP, Cawthray G, Barbetti MJ (2015b) Temperature adaptation in isolates of Sclerotinia sclerotiorum affects their ability to infect Brassica carinata. Plant Pathol 64:1140–1148
Uloth MB, Clode PL, You MP, Barbetti MJ (2016) Attack modes and defence reactions in pathosystems involving Sclerotinia sclerotiorum, Brassica carinata, B. juncea and B. napus. Ann Bot 117:79–95
Wei D, Mei J, Fu Y, Disi JO, Li J, Qian W (2014) Quantitative trait loci analyses for resistance to Sclerotinia sclerotiorum and flowering time in Brassica napus. Mol Breed 34:1797–1804
Wei L, Jian H, Lu K, Filardo F, Yin N, Liu L, Qu C, Li W, Du H, Li J (2016) Genome-wide association analysis and differential expression analysis of resistance to Sclerotinia stem rot in Brassica napus. Plant Biotechnol J 14:1368–1380
Wu J, Cai G, Tu J, Li L, Liu S, Luo X, Zhou L, Fan C, Zhou Y (2013) Identification of QTLs for resistance to sclerotinia stem rot and BnaC.IGMT5.a as a candidate gene of the major resistant QTL SRC6 in Brassica napus. PLoS One 8(7):e67740
Wu J, Zhao Q, Liu S, Shahid M, Lan L, Cai G (2016) Genome-wide association study identifies new loci for resistance to Sclerotinia stem rot in Brassica napus. Front Plant Sci 7:1418. doi:https://doi.org/10.3389/fpls.2016.01418
You MP, Uloth MB, Li XX, Banga SS, Banga SK, Barbetti MJ (2016) Valuable new resistances ensure improved management of sclerotinia stem rot (Sclerotinia sclerotiorum) in horticultural and oilseed Brassica species. J Phytopath 164:291–299
Zhang J, Qi C, Pu H, Chen S, Chen F (2011) Genetic map construction and Sclerotinia resistance QTLs identification in rapeseed (Brassica napus L.). In: Proceedings of the 13th international rapeseed congress, June 2011, Prague, Czech Republic, GCIRC, Paris, France, pp 713–715
Zhao JW, Meng JL (2003) Genetic analysis of loci associated with partial resistance to Sclerotinia sclerotiorum in rapeseed (Brassica napus L.). Theor Appl Genet 106:759–764
Zhao J, Peltier AJ, Meng J, Osborn TC, Grau CR (2004) Evaluation of Sclerotinia stem rot resistance in oilseed Brassica napus using a petiole inoculation technique under greenhouse conditions. Plant Dis 88:1033–1039
Zhao J, Udall JA, Quijada PA, Grau CR, Meng J, Osborn TC (2006) Quantitative trait loci for resistance to Sclerotinia sclerotiorum and its association with a homeologous non-reciprocal transposition in Brassica napus L. Theoret Appl Genet 112:509–516
Acknowledgements
The first author gratefully acknowledges a UWA-UAF Scholarship jointly from the University of Agriculture, Faisalabad (38000) in Pakistan and the University of Western Australia. The authors gratefully acknowledge the exceptional technical support from Robert Creasy and Bill Piasini in the UWA Plant Growth Facilities.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Khan, M.A., Cowling, W., Banga, S.S. et al. Inheritance of leaf resistance to Sclerotinia sclerotiorum in Brassica napus and its genetic correlation with cotyledon resistance. Euphytica 216, 188 (2020). https://doi.org/10.1007/s10681-020-02717-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10681-020-02717-4