Abstract
The nucleotide-binding site (NBS)-leucine-rich repeat (LRR) gene family comprises the largest number of known disease resistance (R) genes and is one of the largest gene families in plants. In the present study, the full-length cDNA of ZmNL (GenBank Accession Number KF765443) was isolated using Rapid Amplification of cDNA Ends. The nucleotide sequence of ZmNL contains an open reading frame of 3156 bp that encodes the ZmNL protein, which is comprised of 1051 amino acid residues. This putative protein has high homology to other known resistance proteins (84% to Triticum aestivum LR10) and belongs to the CC–NBS–LRR type R gene family. The ZmNL gene was introduced into the maize inbred line of Huangzao4 which was highly susceptible to head smut under the control of the maize ubiquitin promoter by Agrobacterium-mediated transformation. The head smut disease incidence of 3 T2 transgenic lines was significantly reduced (by 18.38–29.40%) compared with the wild type, which indicated that the overexpression of ZmNL gene in maize enhanced the resistance to the fungus Sporisorium reilianum (Kühn) Clint of these plants.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Fewer disease resistance (R) genes have been reported in maize (Zea mays) than in other grains, such as rice (Oryza sativa) and common wheat (Triticum aestivum). Maize R genes that have been studied include Hm1, which confers Cochliobolus carbonum race 1 resistance; Rp1-D, which confers common rust (Puccinia sorghi) resistance; and Rxo1, which controls resistance to Burkholderia andropogonis (Johal and Briggs 1992; Collins et al. 1999; Zhao et al. 2005). Therefore, the isolation and study of R genes from maize is important in terms of understanding the resistance mechanisms against these diseases and facilitate methods for development of disease-resistant maize varieties.
R genes in plant encode receptors that specifically recognize pathogen avirulence (avr) gene products (Lamb et al. 1989), in a manner consistent with Flor’s gene-for-gene hypothesis (Flor 1942). The interaction of proteins encoded by R and avr genes has been suggested to trigger a series of signaling cascades including influxes of ions, increases in oxidative activity that generate reactive oxygen intermediates, production of nitric oxide, activation of the hypersensitive response (HR; programmed cell death at infection sites that inhibits growth of pathogens), and production of toxic antimicrobial substances, all of which result in disease resistance (Dangl and Jones 2001). Most R genes encode proteins containing a nucleotide-binding site (NBS) domain and C-terminal leucine-rich repeats (LRR) (Dangl and Jones 2001) that mediate plant defenses against fungal, bacterial, viral, and nematode pathogens (Hammond-Kosack and Jones 1997; Ellis and Jones 1998; Meyers et al. 1999; Williamson 1999). Binding of ATP and GTP by the NBS domain helps to transduce signals during disease resistance responses (Walker et al. 1982; Saraste et al. 1990; Tameling et al. 2006). The C-terminal LRR domains, which are often present in proteins that perceive signals, mediate interactions between proteins or between peptides and their ligands (Dangl and Jones 2001; Holt et al. 2002). The LRR domain specifically recognizes pathogens that express a corresponding avr gene, binds the NBS domain, and keeps the R protein in an inactive state (Ellis et al. 2000; Tameling et al. 2002).
Since the R gene Hm1 was cloned (Johal and Briggs 1992), more than 70 other R genes have been cloned from numerous monocot and dicot crops and model plant species such as Arabidopsis, Zea mays, Sorghum bicolor, Solanum tuberosum, Oryza sativa, etc. (Wei et al. 2013). Most of plant R genes have been cloned using positional information from linkage studies or by tagging with transposons (Collins et al. 1999; Zhou et al. 2006; Lee et al. 2009). In the first approach, a high-resolution linkage map of the area around the gene is used to isolate the R genes for which there is only phenotypic and map location information. In the second method, the target of R gene is inactivated by insertion of a transposon into its coding or regulatory sequences. However, few transposons are suitable for use in tagging, and the likelihood that they will insert within a target R gene is very low (Brutnell 2002). Thus, for the above reasons and due to the complexity of the maize genome, progress in map- and transposon-based cloning of R genes has been relatively difficult and slow. One solution is to use degenerate primers to target and amplify the sequences encoding conserved NBS domains of NBS-containing maize R genes. This method has been used to clone the NBS-containing R genes from soybean (Glycine max) (Kanazin et al. 1996; Yu et al. 1996; Penuela et al. 2002), potato (Solanum tuberosum) (Leister et al. 1996), lettuce (Lactuca sp.) (Shen et al. 1998), bean (Vicia sp.) (Creusot et al. 1999; Rivkin et al. 1999), the Arabidopsis (Aarts et al. 1998), maize (Collins et al. 1998), rice (Oryza sativa) (Yuan et al. 2011), barley (Hordeum vulgare) (Leister et al. 1998), and other angiosperms. The present study builds on earlier work in which we identified a putative NBS–LRR gene that was expressed differentially in different maize lines under artificial inoculation of the fungus Sporisorium reilianum (Kühn) Clint.
In the present study, we cloned the full-length cDNA of this NBS–LRR gene, the ZmNL, from maize using 5′-RACE and 3′-RACE, analyzed its expression profile by qRT-PCR in maize after infection with the Sphacelotheca reiliana. We used established the Agrobacterium-mediated methods to introduce the ZmNL gene into the maize inbred line and subsequently evaluated the Sphacelotheca reiliana incidence of the transgenic plants. Here, we aimed to understand the function of the ZmNL gene in the resistance to the head smut.
Materials and methods
Plant materials
The elite Chinese inbred maize (Zea mays) line (Huangzao4) is highly susceptible to head smut with about 75% of plants showing susceptibility in the field (Chen et al. 2008; Weng et al. 2012). In addition to this, the line of L282 was developed from a cross between the (resistant) line Qi319 and the Huangzao4 and subsequent backcrosses with the Huangzao4. Evaluations of resistance for the head smut disease under artificial inoculation in the field were revealed that the L282 was highly resistant to head smut with disease incidence of 0% (Di et al. 2015a).
Extraction of genomic DNA and amplification of genomic NBS fragments
Genomic DNA was extracted from leaves of the L282 using the DNeasy Kit (Qiagen, CA, USA) according to the manufacturer’s instructions. DNA quality was assessed by electrophoresis in 1.0% agarose gel (Biowest, Madrid, Spain) in 1 × TAE buffer at 120 V for 30 min, and concentrations were determined by measuring absorbance at 260 nm using an ultraviolet (UV) spectrophotometer.
A pair of degenerate primers, mZmNL-F and mZmNL-RP1, targeting the conserved P-loop and GLPL domains, respectively, was designed to amplify an internal conserved fragment of the NBS domain from the line L282 (See Table 1). PCR reactions were performed in 25 μL containing 2.5 μL of 10 × PCR Buffer (Mg2+ Plus), 2 μL of dNTPs (2.5 mM each), 1 μL of each primer (10 μM), 17.5 μL of sterile ddH2O, 0.5 μL (5 U/μL) of LA Taq DNA polymerase (TaKaRa, Dalian, China), and 0.2 μg of DNA template. Temperature cycling conditions were: 94 °C for 5 min; 35 cycles of 94 °C for 45 s, 58 °C for 45 s, and 72 °C for 60 s; followed by 72 °C for 10 min. PCR products were then separated by size on a 1.0% agarose gel in 1 × TAE buffer and the appropriately sized band was recovered using a Quick Gel Extraction Kit (TransGen, Beijing, China) and cloned into the pEasy-T1Simple vector (TransGen). Recombinant clones were isolated, and then sequenced at Shenggong Biotechnology Company (Shanghai, China).
Cloning of the full-length of ZmNL cDNA
Total RNA was isolated from leaves of the L282 with a Plant RNA Kit (TransGen) following the manufacturer’s procedure and treated with DNase I (Omega, GA, USA) to remove residual DNA. The first-strand of cDNA was synthesized from 1 μg of total RNA using 1 μL (200 U) PrimeScript™ RT Enzyme Mix I (TaKaRa) according to the manufacturer’s protocol. To clone the full-length of cDNA of ZmNL from maize, 5′ RACE and 3′ RACE were carried out using the SMART™ RACE Amplification Kit (Clontech, Palo Alto, USA) with 5′ RACE and 3′ RACE gene-specific primers (GSP) (See Table 1). RACE reactions were set up in volumes of 50 μL, as recommended by the manufacturer. The temperature cycling conditions were: 5 cycles of 94 °C for 30 s and 72 °C for 3 min; 5 cycles of 94 °C for 30 s, 70 °C for 30 s, and 72 °C for 3 min; followed by 25 cycles of 94 °C for 30 s, 68 °C for 30 s, and 72 °C for 3 min. All PCR products were cloned into the pEasy-T1Simple vector (TransGen) and sequenced by Shenggong Biotechnology Company (Shanghai, China). The sequence of this ZmNL cDNA clone has been submitted to GenBank (GenBank Accession No. KF765443).
Sequence analysis
The sequence analysis as below was conducted in 2012. Analysis of the ZmNL cDNA ORF was performed using ORFfinder (http://www.ncbi.nlm.nih.gov/gorf/gorf.html). The amino acid sequence of the putative ZmNL protein was deduced using the Expert Protein Analysis System (http://www.expasy.org/). Determinations of the amino acid composition and the secondary and tertiary structure of the ZmNL protein were carried out using DNA tool 6.0, SOPMA (https://npsa-prabi.ibcp.fr/cgi-bin/npsa_automat.pl?page=/NPSA/npsa_sopma.html), and Phyre2 (http://www.sbg.bio.ic.ac.uk/phyre/). Conserved motifs and signal sequences of the ZmNL protein were predicted at the Conserved Domain Search Service (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi/), and SignalP 4.1 (http://www.cbs.dtu.dk/services/SignalP/). A phylogenetic tree of the aligned protein sequences was constructed using the neighbor-joining (NJ) method in MEGA 5.0 (http://www.megasoftware.net/) with the bootstrap value set to 1000.
Artificial inoculation of plants with the S. reilianum
The compatible of S. reilianum mating-type strains SRZ1 and SRZ2 (Schirawski et al. 2005) were inoculated into 2 mL potato dextrose broth (2.4%) (PDB, Difco, Detroit, USA) and incubated with shaking at 200 rpm at 28 °C for 8–12 h. Each culture was then used to inoculate 50 mL of PDB (Difco, Detroit, USA) and allowed to grow overnight to an optical density (OD) of 0.5–1.0 at 600 nm. The cultures were then centrifuged at 3500 rpm for 5 min. Cell pellets from each culture were then re-suspended in water to an OD of 2.0 at 600 nm. The SRZ1 and SRZ2 suspensions were then mixed and used as inoculum for 7-days-old maize seedlings (Gillissen et al. 1992). Similar inoculation was performed with deionized water as a control. At 7 days after inoculation (DAI), infection of maize leaves by the S. reilianum was observed by light microscopy, scanning electron microscopy, and transmission electron microscopy, as described by Martinez et al. (1999).
Inducible expression analysis by real-time quantitative PCR
The expression of the ZmNL gene was assayed by quantitative real-time PCR (qRT-PCR) on a Chromo4™ Real-Time Detector (Bio-Rad, CA, USA) using the TransStart Top Green qPCR SuperMix (TransGen). Three biological replicate leaf samples were collected for qPCR. Total RNA was extracted from infected leaves collected from the L282 and Huangzao4 at 0, 1, 2 3, 4, 5, or 6 DAI using the Plant RNA Kit (TransGen) according to the manufacturer’s instructions. RNA quality was assessed by electrophoresis on 1.0% agarose (Biowest, Madrid, Spain) gels in 1 × TAE buffer at 120 V for 15 min, and concentrations were determined by measuring absorbance at 260 nm in a UV spectrophotometer. First-strand cDNA was synthesized using the PrimeScript™ RT Enzyme Mix I (TaKaRa) for RT-PCR. The primers used in these PCR reactions are shown in Table 1. The qRT-PCR template consisted of 1 μL (100 ng) of cDNA in reactions containing 10 μL 2 × SYBR Green PCR Master Mix, 0.5 μL (10 μM) each of forward and reverse primer, and 8 μL water. The thermal cycling conditions were 95 °C for 1 min, followed by 40 cycles of 95 °C for 10 s, 60 °C for 12 s, and 72 °C for 18 s. The maize Actin1 gene (Gene ID: 100282267) was amplified with two primers (See Table 1) as an internal control.
Binary plant expression vector construction and maize transformation
The whole length cDNA of ZmNL was subcloned into the plasmid pTF101-Ubi-Nos which contained a maize constitutive ubiquitin promoter, the Bar gene driven by the CaMV 35S promoter and a Nos terminator according to the standard protocol using the In-Fusion Cloning Kit [Takara Biomedical Technology (Beijing) Co., Ltd]. The freeze–thaw technique method was used to introduce the recombined plasmid pTU-ZmNL into the Agrobacterium tumefaciens strain EHA101 (Höfgen and Willmitzer 1988).
Seeds of maize inbred line Huangzao4, which was used as the genetic materials in this study, were surface sterilized and were incubated on autoclaved solid MS medium at 28 °C in darkness until the etiolated seedlings had grown to 3.0–4.0 cm in length. After immersed in the suspension of transformed Agrobacterium tumefaciens (OD600 = 0.8) for 10 min under 0.05 MPa pressure, maize shoot tips with exposed meristems were co-cultivated on solid MS medium in the dark at 24 °C for 3 days. Then the plantlets were transplanted into pots and cultivated in the greenhouse. The herbicide Basta with concentration of 0.1% was sprayed on the seedlings (Li et al. 2008). Then the seeds of survived seedlings and PCR-positive plants, which were assayed for the presence of the transgene by PCR, were harvested. The T2 generation seedlings were confirmed the integration and stability of the ZmNL gene with RT-PCR, Southern blotting, and RT-PCR (Di et al. 2015b). These experiments were conducted in the Molecular Breeding Experimental Base of Northeast Agricultural University located in Harbin City, Heilongjiang Province of China. A special room was used to store the plants. The related law of China were abided to burn the dedicated spot when the experiments were over.
PCR, RT-PCR and southern blotting analysis
The CTAB procedure was used to extract Genomic DNA of maize plants (Murray and Thompson 1980). PCR assays were performed with specific primers NF2 (5′- CGCGAATTTGGGAATGAC -3′) and NR2 (5′- CGACGGAAGAGTTTGTAGGAG -3′). Due to the ZmNL gene was an endogenous gene of maize, the primer designed by the sequences of ZmNL gene and Nos terminator. The Bar gene was assayed by the specific primers BF1 (5′-CCATCGTCAACCACTACATCG -3′) and BR1 (5′- AGCTGCCAGAAACCCACGT -3′). Total RNAs were extracted by the Trizol reagent (Tiangen, Beijing, China), and the RNA were reverse transcribed using the RT Reagent Kit (Transgene, Beijing, China). The gene-specific primers same as PCR assay from sequences of the conserved region of the Bar gene were used to amplify a 439 bp transcript. The adigoxigenin (DIG)-labelled probe consisting of the Bar gene fragment was used as template of Southern blotting, as described in the DIG System Manual (Roche, Inc., Basel, Switzerland). The EcoR V and Hind III were taken to digest the Genomic DNA from each of the PCR-positive maize plants, the restriction enzymes that cut a single site in the T-DNA and cut no sites within the Bar sequence. Digested genomic DNA was electrophoresis on 0.8% agarose gels and transfer onto a nylon membrane with positively charged (Roche, USA). After drying at 80 °C for 2 h, as described in the DIG System Manual, the membrane was hybridized to digoxigenin (DIG)-labelled probe.
Evaluation of resistance against head smut disease in the field
The randomized complete blocks with three replicates was used to evaluate the resistance to the S. reiliana of both plant materials in disease ‘hot spot’, Harbin, Heilongjiang Province, China (45.8°N, 126.5°E) during April to September in 2016. Each plot consisted of two 5 m rows with 0.67 m between rows, which were over-planted and later thinned to 25 plants per row. The inoculum was collected from the previous crop season and stored in cloth bags in a dry and well-ventilated environment. Artificial inoculation was applied by sowing seed together with the 0.1% S. reiliana inoculum and soil, with normal agronomic practices in field management. When disease symptoms were fully expressed, individual plants were scored at crop maturity, and the mean of three replicates per plot was determined.
Results
Isolation and characterization of the ZmNL cDNA
In our preliminary study, the ZmNL DNA sequences of B73 RefGen_v2 in bin 2.09 were confirmed by the presences of conserved nucleotide binding site (NBS) and leucine rich repeat (LRR) R gene domains using Fgenesh software (www.softberry.com/berry.phtml), ORF Finder (http://www.ncbi.nlm.nih.gov/projects/gorf/) and InterProScan search (https://www.ebi.ac.uk/Tools/pfa/iprscan/). The specific primer mZmNLF and mZmNLR was designed according to the coding sequence of ZmNL. The RT-PCR reaction was carried out using RNA isolated from leaf of the line L282 as template for the synthesis of cDNA. A cDNA fragment showed a 678-bp band in a 1% agarose gel (Fig. 1a) was cloned into pEasy-T1Simple vector. Sequencing results of various clones showed the same nucleic acid sequence which was fully consistent with the candidate sequence in MaizeGDB (http://www.maizegdb.org/). Based on the obtained fragment, RACE strategies were conducted at cloning the full-length cDNA sequence of the ZmNL. The 5′-end (Fig. 1b) and the 3′-end (Fig. 1c) of cDNA with the specific primers and universal primers were amplified using first strand cDNA derived from total RNA as template. Then the ZmNL gene was assembled according to overlapping sequences from the three fragments above (Fig. 1e). The reconstructed full length of the cDNA sequence, 3572-bp long (Supplementary Fig. 1), was identified from the sequence of the ZmNL clone, including 3156 bp CDS (Fig. 1d), 163 bp of 5′-UTR and 253 bp of 3′-UTR. The ZmNL sequence was deposited in GenBank with Accession Number of KF765443.
Cloning of the full length cDNA of ZmNL gene from maize line L282. M, DL2000 DNA marker; Lane 1, ZmNL cDNA fragment; a The ZmNL cDNA was amplified from maize line L282 by a pair of primers mZmNL-F and mZmNL-R designed according to coding sequence of ZmNL. b 5′-end of ZmNL cDNA obtained by 5′RACE GSP-1 and 5′ RACE GSP-2. c 3′-end of ZmNL cDNA obtained by 3′RACE GSP-1 and 3′ RACE GSP-2. d The ZmNL clone obtained by RT–PCR with a pair of gene specific primers pZmNL-F and pZmNL-R designed according to ORF sequence. e Diagram of overlapping relationship of subclone fragments and positions of corresponding primers
The ZmNL gene encodes a putative protein of 1051 amino acids with a predicted molecular mass of 116.6 kD and a predicted PI of 8.47. Table 2 showed the comparison of the amino acid sequence of ZmNL protein via ExPASy Protparam. Its putative secondary structure includes 587 α-helices (55.85%), 341 random coils (32.35%), 100 extended strands (9.90%), and 23 β-helices (1.90%). CD-Search results indicated that the ZmNL protein contains conserved NBS and LRR domains (Fig. 2).
Conserved domains of the ZmNL protein predicted by CD-Search. The light and dark gray rectangles indicate the major domains predicted by CD-Search, including an RX-CC-like domain (from Ala11 to Glu134), a P-loop_NTPase super family (Asp207-305 Ile), an NB-ARC domain (from Ile183 to Gly531), and an LRR domain (from Leu647 to Gly755)
Figure 3 indicated the comparison of the amino acid sequence of NBS-containing R protein in some higher plant. The typical conserved motif were presented in ZmNL protein and other plant R proteins, including P-loop(GMGGVGKTT), Kinase-2(LIVLDDVW), Kinase-3a(GSR/KILVTTR), and hydrophobic domain HD(GLPLAL) (Fig. 3a). Based on nucleotide and amino acid primary structure homology, the ZmNL showed higher homology to the sequence of the plant NBS-containing R gene (nucleotide comparison not shown). Evolutionary relationships between the ZmNL and R proteins from other plant species demonstrated that ZmNL is 84% similar to the Triticum aestivum resistance protein LR10 and showed extensive amino acid sequence similarity to other coiled-coil-NBS–LRR (CC–NBS–LRR) proteins (Fig. 3b). The protein sequences used for comparison and phylogenic trees were from Linum usitatissimum (AAA91021.1, AAB47618.1, AAK28806.1, AAC83165.1), Arabidopsis thaliana (AAC72977.1, AAB58295.1, Q42484.1, NP_172686.1, NP_187360.1, NP_196820.1, NP_001077962.1), Triticum aestivum (EMS68464.1, AAQ01784.1), Nicotiana glutinosa (Q40392.1), Oryza sativa (BAA93618.1, O48647), Solanum lycopersicum (AAB63274.1, AAC49408.1), Solanum tuberosum (CAB50786.1), and Lactuca sativa (AAD03156.1).
Comparison of encoding amino acid sequence of ZmNL protein with other plants. a ClustalW was cuducted at Comparison of encoding amino acid sequence of ZmNL protein with other plants. b Phylogenetic tree based on alignment of the deduced amino-acid sequences of ZmNL and other known R proteins from different plant species. The scale bar represents conversion of branch length to genetic distance between clades (0.1 = 10% genetic distance). The protein sequences used for phylogenetic analysis include: LuL6 (AAA91021.1) from Linum usitatissimum, LuM (AAB47618.1) from Linum usitatissimum, AtRPP1 (AAC72977.1) from Arabidopsis thaliana, AtRPP5 (AAB58295.1) from Arabidopsis thaliana, TuRPP13 (EMS68464.1) from Triticum urartu, AtRPS2 (Q42484.1) from Arabidopsis thaliana, AtRPS5 (NP_172686.1) from Arabidopsis thaliana, NgN (Q40392.1) from Nicotiana glutinosa, TaLR10 (AAQ01784.1) from Triticum aestivum, OsPib (BAA93618.1) from Oryza sativa, LuP2 (AAK28806.1) from Linum usitatissimum, AtRPM1 (NP_187360.1) from Arabidopsis thaliana, AtRPP8 (AAC83165.1) from Arabidopsis thaliana, SlI2C-1 (AAB63274.1) from Solanum lycopersicum, OsXA1 (O48647) from Oryza sativa, SuRx (CAB50786.1) from Solanum tuberosum, SlPRF (AAC49408.1) from Solanum lycopersicum, LsRGC2B (AAD03156.1) from Lactuca sativa, AtPBS1 (NP_196820.1) from Arabidopsis thaliana, and AtRCY1 (NP_001077962.1) from Arabidopsis thaliana
Inducible expression of ZmNL in response to the S. reilianum
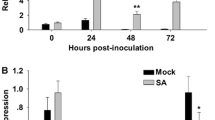
To confirm that artificial inoculation was successful, hyphae of the S. reilianum were observed in the leaves at 7 DAI. Comparing the mock- and the S. reilianum-infected L282 leaves, chlorotic flecks were only observed on the newly emerged leaves of infected Huangzao4 (Fig. 4a). Hyphae in the infected leaves were stained with cotton blue and observed under light microscopy (Fig. 4b). Scanning electron micrographs showed numerous hyphae growing around the stomata (Fig. 4c). Cross-sections of the leaves showed infection hyphae growing in the intercellular spaces inside the leaves (Fig. 4d, e). Thus, the hyphae of S. reilianum could penetrate into the host tissues and cause disease symptoms. To determine its contribution to resistance to S. reilianum infection, the expression of ZmNL was analyzed using qRT-PCR. As shown in Fig. 5, ZmNL expression was up-regulated in the L282 and Huangzao4 after inoculation with the S. reilianum and reached a peak on the second DAI, but peak expression was significantly higher in the L282 (4.29) than in the Huangzao4 (2.99). The expression of ZmNL in the both L282 and Huangzao4 then decreased continually from 3 to 6 DAI. These results indicate that the expression of ZmNL is likely induced by the S. reilianum infection.
Symptoms of infection with S. reilianum and hyphae of S. reilianum-infected Huangzao4 leaves observed under light microscopy (LM), scanning electron microscopy (SEM), and transmission electron microscopy (TEM). a S. reilianum-infected plants (right) show chlorotic flecks (arrow) on leaves; no symptoms were observed on leaves of mock-infected plants (left). b Hyphae (h) on the leaf surfaces (ls) of the S. reilianum-infected plants. Magnification × 400. c Stomata surrounded by numerous long hyphae, bar = 2 μm. d Hyphae (h) in the intercellular spaces within the leaves (lc), bar = 10 μm. e Hyphae (h) in the intercellular space between two leaf cells, bar = 10 μm
qRT-PCR to monitor ZmNL transcript expression in L282 and Huangzao4 leaves inoculated with S. reilianum at 0, 1, 2, 3, 4, 5 and 6 days. The relative expression of ZmNL transcripts was normalized to the expression of maize Actin1 mRNA. Leaves were injected with water as the mock-inoculation control. The error bars denote standard deviations of the qRT-PCR signals (n = 3). The expression levels of ZmNL transcript in L282 and Huangzao4 were significantly different (**p < 0.01, Student’s t test)
Molecular characterization of transgenic maize plants
Transgenic maize plants overexpressing the ZmNL gene were successfully generated by the Agrobacterium-mediated transformation of binary vector pTF101-ZmNL into the inbred line Huangzao4. More than 20 T0 independent transgenic plant lines were established and propagated in the greenhouse. A 398-bp PCR product amplified with ZmNL gene-specific primers is shown in Fig. 6. Three transgenic maize lines (the ZmNL-1, ZmNL-4 and ZmNL-6) with single or double transgenes integration events were verified via Southern blotting analysis, and no integration of Bar was detected in the WT plants (Fig. 7).
PCR analysis of transformed samples using primers specific for the ZmNL gene and Nos terminator. A 398-bp PCR product was amplified from three transgenic maize lines (ZmNL-1, ZmNL-4, and ZmNL-6) with primers specific for the ZmNL gene and Nos terminator. M, DNA Marker DL2000 (TaKaRa); P, pTU-ZmNL plasmid; N, wild-type plants of Huangzao4; W, water (no template DNA) control; ZmNL-1, ZmNL-4, and ZmNL-6, transgenic maize lines
Southern blot analysis of transgenic plants and the wild type. Total genomic DNA of three transgenic maize lines (ZmNL-1, ZmNL-4, and ZmNL-6) and WT was digested with EcoR V and HindIII. A single-copy insertion of the Bar gene was detected in transgenic maize lines, and no integration of Bar was detected in the corresponding WT plants. M, DNA Marker DL15000 (TaKaRa); WT, wild-type plants of KF513; AKF58, AKF61, and AKF65, transgenic lines
The expression of the ZmNL gene in T2 plants leaves was monitored by reverse transcriptase-polymerase chain reaction (RT-PCR), and the results are shown in Fig. 8. The RT-PCR did not produce a product in wild type plant samples.
RT-PCR analysis of transformed samples using Bar gene-specific primers. A 439-bp PCR product was amplified with Bar gene-specific primers from three transgenic maize lines (ZmNL-1, ZmNL-4, and ZmNL-6) by reverse transcriptase-polymerase chain reaction (RT-PCR). In WT plants, no expression of the Bar gene was detected. WT, wild-type plants of Huangzao4; ZmNL-1, ZmNL-4, and ZmNL-6, transgenic lines
Over-expressing the ZmNL gene increased the resistance of maize to head smut
The mean disease incidence of 3 transgenic maize lines and acceptor line Huangzao4 was observed (Table 3). The highest level was 81.52% in Huangzao4 line, and the lowest level was 0 in control line L282. The disease incidence of transgenic lines ZmNL-1, ZmNL-4, and ZmNL-6 decreased from wild line by 29.40, 29.51 and 18.38% respectively. The introduction of ZmNL gene declined the head smut disease incidence by an average of 25.76 percent.
Discussion
The ORF of the 3156-bp ZmNL cDNA that we isolated from the S. reilianum-resistant maize line L282 encodes a deduced protein of 1051 amino acids with high sequence similarity to other CC–NBS–LRR R proteins. The NBS–LRR proteins can be further distinguished by the particular motif located at the N-terminus into two subclasses: Toll and human interleukin-1 receptor (TIR)- NBS–LRR proteins and CC–NBS–LRR proteins. It is noteworthy that the CC domains of the NRG1 from Nicotiana benthamiana, and ADR1 (Collier et al. 2011), RPS2, RPS5, and RPM1 from the Arabidopsis (Tao et al. 2000; Ade et al. 2007; Gao et al. 2011) can trigger the PCD. Their NBS domains help transduce signals for plant defense responses. The LRR domain can determine the specificity of pathogen recognition via protein–protein and protein–ligand interactions with effector molecules derived from pathogens (Jia et al. 2013). The protein encoded by the ZmNL gene might recognize the corresponding avirulence protein expressed the head smut pathogen, bind with ATP or GTP, and activate a kinase or G protein involved in protein phosphorylation to amplify the disease resistance signal. Its LRR domain might then interact with intracellular disease-related proteins to transmit the disease resistance signal and trigger the expression of downstream defense genes.
The 2000 bp upstream sequence of ZmNL of B73 RefGen_v2 was analyzed via P1antCARE to explore the cis-acting elements in the promoter region (Supplementary Fig. 2). Except TATA-box and CAAT-box, the multiple cis-acting elements related to pathogens responses, such as MeJA response elements TGACG-motif (− 882 to − 886 bp) and CGTCA-motif (− 1292 to − 1296 bp). Defense response element TC-rich repeat (− 1025 to − 1033) was also found in the promoter region of ZmNL. In our study, the ZmNL gene was introduced into maize line of Huangzao4, which was highly susceptible to head smut. The transgenic lines showed the increased disease resistance under artificial inoculation in the field. These results proved further the strong correlation between the expression of ZmNL gene and disease responses of maize.
As for most plant R genes, the ZmNL gene was expressed at a low constitutive level in uninfected plants. When plants were treated with the S. reilianum, the expression of ZmNL increased in the L282 more than the Huangzao4. During earlier stages of the infection, ZmNL expression increased. At later stages of infection with the S. reilianum, the expression ZmNL decreased. The higher expression of ZmNL in the smut-resistant line L282 than in the Huangzao4 suggests a positive role for the expression of ZmNL in response to the S. reilianum infection in the L282 and Huangzao4. Further, the delayed response and lower level of ZmNL expression in the Huangzao4 upon S. reilianum infection correlates with the weaker resistance of the Huangzao4 to the S. reilianum infection.
A wall-associated kinase gene ZmWAK was reported in the same region of ZmNL (Zuo et al. 2015). The expression of the two genes was analyzed after inoculation with S. reilianum using qRT-PCR (Fig. 5, Supplementary Fig. 3). The similar up-regulated changed trend was observed in the L282 and the Huangzao4 and the expression of two genes both reached a peak on the second DAI. These results indicate that the ZmNL protein might cooperate with other proteins associated with resistance to the S. reilianum, such as the ZmWAK, to stimulate HR around attempted pathogen infection sites in maize and limit the further expansion of the pathogen via PCD. Future studies will be more precisely address the ZmNL function.
References
Aarts MG, Hekkert BT, Holub EB, Beynon JL, Stiekema WJ, Pereira A (1998) Identification of R-gene homologous DNA fragments genetically linked to disease resistance loci in Arabidopsis thaliana. Mol Plant Microbe Interact 11:251–258. https://doi.org/10.1094/MPMI.1998.11.4.251
Ade J, DeYoung BJ, Golstein C, Innes RW (2007) Indirect activation of a plant nucleotide binding site-leucine-rich repeat protein by a bacterial protease. Proc Natl Acad Sci USA 104:2531–2536. https://doi.org/10.1073/pnas.0608779104
Brutnell TP (2002) Transposon tagging in maize. Funct Integr Genom 2(1):4–12. https://doi.org/10.1007/s10142-001-0044-0
Chen YS, Chao Q, Tan GQ, Zhao J, Zhang MJ, Ji Q, Xu ML (2008) Identification and fine-mapping of a major QTL conferring resistance against head smut in maize. Theor Appl Genet 117:1241–1252. https://doi.org/10.1007/s00122-008-0858-4
Collier SM, Hamel LP, Moffett P (2011) Cell death mediated by the N-terminal domains of a unique and highly conserved class of NB-LRR protein. Mol Plant Microbe Interact 24:918–931. https://doi.org/10.1094/MPMI-03-11-0050
Collins NC, Webb CA, Seah S, Ellis JG, Hulbert SH, Pryor A (1998) The isolation and mapping of disease resistance gene analogs in maize. Mol Plant Microbe Interact 11:966–978
Collins N, Drake J, Ayliffe M, Sun Q, Ellis J, Hulbert S, Pryor T (1999) Molecular characterization of the maize Rpl-D rust resistance haplotype and its mutants. Plant Cell 11:1365–1376
Creusot F, Macadre C, Ferrier-Cana E, Riou C, Geffroy V, Sevignac M, Dzron M, Langin T (1999) Cloning and molecular characterization of three members of the NBS–LRR subfamily located in the vicinity of the Co-2 locus for anthracnose resistance in Phaseolus vulgaris. Genome 42:254–264. https://doi.org/10.1139/g98-134
Dangl JL, Jones JDG (2001) Plant pathogens and integrated defence responses to infection. Nature 411(6839):826–833. https://doi.org/10.1038/35081161
Di H, Liu XJ, Wang QK, Weng JF, Zhang L, Li XH, Wang ZH (2015a) Development of SNP-based dCAPS markers linked to major head smut resistance quantitative trait locus qHS2.09 in maize. Euphytica 202:69–79. https://doi.org/10.1007/s10681-014-1219-9
Di H, Tian ZuHY, Meng XY, Zeng X, Wang ZH (2015b) Enhanced salinity tolerance in transgenic maize plants expressing a BADH gene from Atriplex micrantha. Euphytica 206:775–783. https://doi.org/10.1007/s10681-015-1515-z
Ellis J, Jones D (1998) Structure and function of proteins controlling strain-specific pathogen resistance in plants. Curr Opin Plant Biol 1:288–293. https://doi.org/10.1016/1369-5266(88)80048-7
Ellis J, Dodds P, Pryor T (2000) Structure, function and evolution of plant disease resistance genes. Curr Opin Plant Biol 4:278–284. https://doi.org/10.1016/S1369-5266(00)00080-7
Flor HH (1942) Inheritance of pathogenicity in Melampsora lini. Phytopathology 32:653–669
Gao Z, Chung EH, Eitas TK, Dangl JL (2011) Plant intracellular innate immune receptor resistance to Pseudomonas syringae pv. maculicola 1 (RPM1) is activated at, and functions on, the plasma membrane. Proc Natl Acad Sci USA 108:7619–7624. https://doi.org/10.1073/pnas.1104410108
Gillissen B, Bergemann J, Sandmann C, Schroeer B, Bölker M, Kahmann R (1992) A two-component regulatory system for self/non-self-recognition in Ustilago maydis. Cell 68:647–657. https://doi.org/10.1016/0092-8674(92)90141-X
Hammond-Kosack KE, Jones JDG (1997) Plant disease resistance genes. Annu Rev Plant Physiol Plant Mol Biol 48:575–607. https://doi.org/10.1146/annurev.arplant.48.1.575
Höfgen R, Willmitzer L (1988) Storage of competent cells for Agrobacterium transformation. Nucl Acids Res 16(20):9877
Holt BF, Boyes DC, Ellerström M, Siefers N, Wiig A, Kauffman S, Dangl JL (2002) An evolutionarily conserved mediator of plant disease resistance gene function is required for normal Arabidopsis development. Dev Cell 2(6):807–817. https://doi.org/10.1016/S1534-5807(02)00174-0
Jia RZ, Ming R, Zhu YJ (2013) Genome-wide analysis of nucleotide-binding site (NBS) disease resistance (R) genes in sacred lotus (Nelumbo nucifera Gaertn.) reveals their transition role during early evolution of land plants. Trop Plant Biol 6:98–116. https://doi.org/10.1007/s12042-013-9122-4
Johal GS, Briggs SP (1992) Reductase activity encoded by the HM1 disease resistance gene in maize. Science 258(5084):985–987. https://doi.org/10.1126/science.1359642
Kanazin V, Marek LF, Shoemarker RC (1996) Resistance gene analogs are conserved and clustered in soybean. Proc Natl Acad Sci USA 93:11746–11750. https://doi.org/10.1073/pnas.93.21.11746
Lamb CJ, Lawton MA, Dron M, Dixon RA (1989) Signals and transduction mechanisms for activation of plant defenses against microbial attack. Cell 56:215–224. https://doi.org/10.1016/0092-8674(89)90894-5
Lee SK, Song MY, Seo YS, Kim HK, Ko S, Cao PJ, Suh JP, Yi G, Roh JH, Lee S, An G, Hahn TR, Wang GL, Ronald P, Jeon JS (2009) Rice Pi5-mediated resistance to Magnaporthe oryzae requires the presence of two coiled-coil-nucleotide-binding-leucine-rich repeat genes. Genetics 181:1627–1638. https://doi.org/10.1534/genetics.108.099226
Leister D, Ballvora A, Salamini F, Gebhardt C (1996) A PCR-based approach for isolating pathogen resistance genes from potato with potential for wide application in plants. Nat Genet 14:421–429. https://doi.org/10.1038/ng1296-421
Leister D, Kurth J, Laurie DA, Yano M, Sasaki T, Devos K, Graner A, Schulze-Lefert P (1998) Rapid reorganization of resistance gene homologues in cereal genomes. Proc Natl Acad Sci USA 95:370–375
Li XH, Wang ZH, Gao SR, Shi HL, Zhang SH, George MLC, Li MS, Xie CX (2008) Analysis of QTL for resistance to head smut (Sporisorium reiliana) in maize. Field Crops Res 106:148–155
Martinez C, Roux C, Dargent R (1999) Biotrophic development of Sporisorium reilianum f. sp. zeae in vegetative shoot apex of maize. Biochem Cell Biol 89:247–253. https://doi.org/10.1094/PHYTO.1999.89.3.247
Meyers BC, Dickerman AW, Michelmore RW, Sivaramakrishnan S, Sobral BW, Young ND (1999) Plant disease resistance genes encode members of an ancient and diverse protein family within the nucleotide-binding superfamily. Plant J 20:317–332. https://doi.org/10.1046/j.1365-313X.1999.00606.x
Murray MG, Thompson WF (1980) Rapid isolation of high molecular weight plant DNA. Nucl Acids Res 8:4321–4326
Penuela S, Danesh D, Young ND (2002) Targeted isolation, sequence analysis, and physical mapping of nonTIR NBS–LRR genes in soybean. Theor Appl Genet 104:261–272. https://doi.org/10.1007/s00122-001-0785-0
Rivkin MI, Vallejos CE, McClean PE (1999) Disease-resistance related sequences in common bean. Genome 42:41–47. https://doi.org/10.1139/g98-097
Saraste M, Sibbald PR, Wittinghofer A (1990) The P-loop–a common motif in ATP- and GTP-binding proteins. Trends Biochem Sci 15:430–434. https://doi.org/10.1016/0968-0004(90)90281-F
Schirawski J, Heinze B, Wagenknecht M, Kahmann R (2005) Mating type loci of Sporisorium reilianum: novel pattern with three a and multiple b specificities. Eukaryot Cell 4:1317–1327. https://doi.org/10.1128/EC.4.8.1317-1327.2005
Shen KA, Meyers BC, Islam-Faridi MN, Chin DB, Stelley DM, Michelmore RW (1998) Resistance gene candidates identified by PCR with degenerate oligo nucleotide primers map to clusters of resistance genes in lettuce. Mol Plant Microbe Interact 11:815–823. https://doi.org/10.1094/MPMI.1998.11.8.815
Tameling WI, Elzinga SD, Darmin PS, Vossen JH, Takken FL, Haring MA, Cornelissen BJ (2002) The tomato R gene products I-2 and Mi-1 are functional ATP binding proteins with ATPase activity. Plant Cell 14(11):2929–2939
Tameling WI, Vossen JH, Albrecht M, Lengauer T, Berden JA, Haring MA, Cornelissen BJ, Takken FL (2006) Mutations in the NB-ARC domain of I-2 that impair ATP hydrolysis cause auto activation. Plant Physiol 140:1233–1245
Tao Y, Yuan F, Leister RT, Ausubel FM, Katagiri F (2000) Mutational analysis of the Arabidopsis nucleotide binding site-leucine-rich repeat resistance gene RPS2. Plant Cell 12:2541–2554. https://doi.org/10.1105/tpc.12.12.2541
Walker JE, Saraste M, Runswick MJ, Gay NJ (1982) Distantly related sequences in the α- and β-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J 1:945–954
Wei HL, Li W, Sun XW, Zhu SJ, Zhu J (2013) Systematic analysis and comparison of nucleotide-binding site disease resistance genes in a diploid cotton Gossypium raimondii. PLoS ONE 8(8):e68435. https://doi.org/10.1111/j.1742-4658.2012.08621.x
Weng JF, Liu XJ, Wang ZH, Wang JJ, Zhang L, Hao ZF, Xie CX, Li MS, Zhang DG, Bai L, Liu CL, Zhang SH, Li XH (2012) Molecular mapping of the major resistance quantitative trait locus qHS2.09 with simple sequence repeat and single nucleotide polymorphism markers in maize. Phytopathology 102(7):692–699. https://doi.org/10.1094/PHYTO-12-11-0330
Williamson VM (1999) Plant nematode resistance genes. Curr Opin Plant Biol 2:327–331. https://doi.org/10.1016/S1369-5266(99)80057-0
Yu YG, Buss GR, Maroof MA (1996) Isolation of a superfamily of candidate disease resistance genes in soybean based on a conserved nucleotide-binding site. Proc Natl Acad Sci USA 93:11751–11756. https://doi.org/10.1073/pnas.93.21.11751
Yuan B, Zhai C, Wang W, Zeng X, Xu X, Hu H, Lin F, Wang L, Pan Q (2011) The Pik-p resistance to Magnaporthe oryzae in rice is mediated by a pair of closely linked CC–NBS–LRR genes. Theor Appl Genet 122:1017–1028. https://doi.org/10.1007/s00122-010-1506-3
Zhao BY, Lin XH, Poland J, Trick H, Leach JE, Hulbert SH (2005) A maize resistance gene functions against bacterial streak disease in rice. Proc Natl Acad Sci USA 102(43):15383–15388. https://doi.org/10.1073/pnas.0503023102
Zhou B, Qu SH, Liu GF, Dolan M, Sakai H, Lu G, Bellizzi M, Wang GL (2006) The eight amino-acid differences within three leucine-rich repeats between Pi2 and Piz-t resistance proteins determine the resistance specificity to Magnaporthe grisea. Mol Plant Microbe Interact 19:1216–1228. https://doi.org/10.1094/MPMI-19-1216
Zuo WL, Chao Q, Zhang N, Ye JR, Tan GQ, Li BL, Xing YX, Zhang BQ, Liu HJ, Fengler KA, Zhao J, Zhao XR, Chen YS, Lai JS, Yan JB, Xu ML (2015) A maize wall-associated kinase confers quantitative resistance to head smut. Nat Genet 47(2):151–157. https://doi.org/10.1038/ng.3170
Acknowledgements
This work was supported financially by the National Natural Science Foundation of China (No. 31371634) and Genetically Modified Organisms Breeding Major Projects of China (2016ZX08003003). We would like to thank Prof. Schirawski for providing the S. reilianum strains SRZ1 and SRZ2.
Author information
Authors and Affiliations
Corresponding author
Additional information
Hong Di and Tao Yu contributed equally to this research.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Di, H., Yu, T., Deng, Y. et al. Complementary DNA (cDNA) cloning and functional verification of resistance to head smut disease (Sphacelotheca reiliana) of an NBS–LRR gene ZmNL in maize (Zea mays). Euphytica 213, 288 (2017). https://doi.org/10.1007/s10681-017-2083-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10681-017-2083-1