Abstract
Cassava root rot disease (CRRD) severely affects productivity in several countries. The objective of this study was to estimate genetic parameters and to identify multiple resistance sources against pathogens associated with CRRD. The symptoms caused by Fusarium spp., Phytophthora spp., and Botryosphaeriaceae species in peel and pulp from the roots were evaluated in 277 accessions using a whole tuberous root inoculation method. The resistance data were obtained by REML/BLUP (restricted maximum likelihood/best linear unbiased predictor). The classic selection index (CI), multiplicative (MI), and sum of ranks (SRI) were used to identify the accessions with multiple resistance. For all pathogens, the environmental variance (\(\sigma_{e}^{2}\)) was the most important component. Individual heritability \(\left( {h_{g}^{2} } \right)\) was of low magnitude for resistance to most pathogens (0.16 ± 0.02—peel and 0.31 ± 0.03—pulp for Fusarium spp.; 0.26 ± 0.03—peel and 0.30 ± 0.03—pulp for Phytophthora spp.; and 0.28 ± 0.03—peel and 0.27 ± 0.03—pulp for Botryosphaereacea species). The distribution of CRRD symptoms indicated the presence of quantitative inheritance. The direct selection of the 15 more resistant accessions based on the genotypic predicted values result in high reductions of disease (>50%). However, there was a low matching rate of the most resistant accessions for each pathogen and the different parts of the tuberous roots (peel and pulp). The CI and MI were the most promising compared to the SRI to ensure high and balanced resistance for each pathogen. Understanding the genetic basis of resistance to CRRD and the identification of sources with multiple resistance may be useful in various management strategies to control the disease.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In many African countries, cassava (Manihot esculenta Crantz) is considered a subsistence crop that grows and produces relatively well in low fertility soils and in areas with water deficits and limited use of agricultural inputs (Hillocks 2014). It is the third most important source of calories in the tropics after rice and maize (FAO 2015). Besides the use as food in its fresh form, cassava starch has a wide range of uses in the food industry as well as for paper and cellulose, textiles, biodegradable polymers, pharmaceuticals, beyond its traditional use in animal feed and recently in ethanol production (Rickard et al. 1991; Hoover 2001; Suppakul et al. 2013).
In Brazil, which is the second largest producer of cassava with 11.43% of the total world production of about 204 million tonnes in 2013 (FAO 2015), cassava is cultivated throughout the country. With continental dimensions, cassava production systems in Brazil are very diverse, considering climatic aspects or the occurrence of pests. However, some diseases, such as cassava root rot, have widespread occurrence throughout the country.
According to Bandyopadhyay et al. (2006), among the major cassava biotic constraints, the root rot disease complex is less understood due to the low number of researchers who are dedicated to study this disease. The few studies on cassava root rot disease (CRRD) have showed the following: (1) generally, in several situations the cassava root system enables the plant to remain asymptomatic until harvest even with high disease severity (Msikita et al. 2005); (2) the yield losses may vary from 25% to 100% (Onyeka et al. 2005a; Bandyopadhyay et al. 2006; Oliveira et al. 2013); (3) pathogens causing CRRD can colonize the plant independent of their age; (4) the pathogens are related to different genera and can be divided into three groups based on the symptoms produced: (i) soft root rot where the main species involved are Phytophthora spp. and Pythium spp.; (ii) dry root rot, caused by Fusarium spp., Corallomycetella repens (equivalent to Nectria mauritiicola and Sphaerostilbe repens), Armillaria mellea and Sclerotium rolfsii; and (iii) black root rot, caused by Botryosphaeriaceae species such as Neoscytalidium hyalinum, Lasiodiplodia spp. and Neofusicoccum mangiferae (equivalent to Nattrassia mangiferae) (Bandyopadhyay et al. 2006; Okechukwu et al. 2009; Banito et al. 2010; Guo et al. 2012; Machado et al. 2014; Vilas Boas et al. 2017).
Integrated disease management based on planting resistant cultivars associated with soil and water management practices, crop rotation and use of healthy, high-quality planting material are economic and reliable techniques to control cassava root rot. However, the main management practice used is variety resistance (Onyeka et al. 2005a; Oliveira et al. 2013), although the genetic variability for this trait has not been fully explored in the germplasm of M. esculenta.
In the routine of breeding programs, the selection of genotypes resistant varieties to CRRD has been performed based on the expression of symptoms in field conditions. The main drawback of this approach is the occurrence of plants that escape the disease due to non-uniform distribution of the inoculum of the pathogen in the soil. An example of this was presented by Onyeka et al. (2005b) for field sites in Nigeria. Moreover, Onyeka et al. (2005c) developed standardized methodologies for CRRD inoculation, in which the results allowed the separation of resistant and susceptible genotypes quite accurately. Even with the existence of standardized assessment methods, we still need to invest in understanding and obtaining estimates of genetic parameters as well as in identifying the sources of resistance to the main pathogens associated with this disease in order to incorporate resistance in commercial varieties.
Genetic resistance to CRRD caused by L. theobromae (Botryodiplodia theobromae) has been reported in Africa (Onyeka et al. 2005a, b), while the resistance to dry root rot caused by Fusarium sp. has been demonstrated in cassava accessions from Brazil (Oliveira et al. 2013; Vilas Boas et al. 2016). However, for the development of more durable resistance, the identification of sources of resistance for multiple pathogens commonly associated with CRRD in Brazil such as dry root rot (Fusarium spp.), soft root rot caused by Phytophthora spp., and black root rot caused by Botryosphaeriaceae species should be done simultaneously, considering that pathogens can occur in the same area causing yield lost in cassava (Banito et al. 2010). The objective of this study was to estimate the genetic parameters for the main soil-borne pathogens associated with cassava root rot disease in Brazil and to carry out simultaneous selection to search for resistance in the M. esculenta germplasm.
Materials and methods
Plant material
In total, 277 germplasm accessions within the Cassava Germplasm Bank (CGB) at Embrapa Cassava and Fruits (Cruz das Almas, Bahia, Brazil) were evaluated. One elite clone (9624-09), twelve improved varieties resulting from conventional breeding procedures (BRS Amansa Burro, BRS Aramaris, BRS Caipira, BRS Dourada, BRS Gema de Ovo, BRS Kiriris, and IAC90), and three landraces widely grown in Brazil (Mani Branca, Eucalipto, and Fécula Branca) were used as controls.
Experimental design and pathogen inoculation
Field experiments were conducted in different areas in Cruz das Almas (Bahia, Brazil), located at 12°40′19′′S, 39°06′22′′W, altitude 220 m. The climate is hot and humid, tropical, Aw to Am, according to the Köppen classification. The planting was carried out using stem cuttings of 20 cm in the rainy season (March–August) using spacing of 0.90 m between rows and 0.80 between plants. The plots comprised two rows of 16 plants in total. The cropping system followed the regional recommendations (Souza et al. 2006) in well-drained soil and the absence of root rot.
The evaluations were made in the 2013 crop year. The harvest was performed manually between 10 and 12 months after planting, taking care to avoid injury to the roots. The tuberous roots were standardized for size (20–25 cm long and 4–6 cm in diameter) and shape, respecting the inherent characteristics of each accession.
For the identification of sources of resistance to CCRD, whole cassava tuberous roots were washed and disinfected with sodium hypochlorite (0.5%) and placed on sterile filter paper to dry. For inoculation, wounds (3–4-mm depth) were produced in the central region of the tuberous root using a metal punch (Ø 0.6 cm). The experimental design was a randomized block with nine repetitions for each accession, in which each experimental unit was represented by a tuberous root with three points of inoculation. Each root repetition was placed on a granite countertop without any contact between roots.
The isolates used were divided according to the symptoms produced in cassava tuberous roots and inoculated as a “mix” of the most aggressive isolates from each class (dry, soft, and black root rot). For the assessment of the reaction to dry root rot, the species F. solani, F. oxysporum, and F. lateritium were used; for the soft root rot, the isolates were from P. dreschsleri and P. melonis; and for the black root rot, the species used were from the Botryosphaeriaceae family (N. hyalinum, L. theobromae, and L. euphorbicola). The selection of the isolates was done based on the survey conducted by Vilas Boas et al. (2017), on the major cassava production regions of Brazil.
The fungi isolates grew on PDA and the oomycetes on V8 amended with β-sitosterol. The tuberous roots were inoculated with a 100 µl drop of spore suspension adjusted to 2 × 105 spores ml−1; a mock inoculation (only water) was also used based on the methodology described by Onyeka et al. (Onyeka et al. 2005a, b, c). After the inoculation, the tuberous roots were placed in trays containing moistened cotton, wrapped in plastic bags (moist chamber), and then stored at 26 ± 2 °C for 10 days.
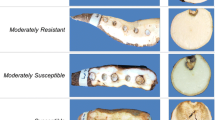
At the time of evaluation, tuberous roots were cut longitudinally and were digitally photographed for later analysis of the images with the aid of ImageTool v.3.0 software (UTHSCSA, University of Texas Health Science Center, San Antonio) in order to obtain the area colonized by the pathogen. After the spatial calibration, using a ruler, the lesioned area was manually selected by trace contouring, and area of each lesion was automatically calculated by the software. Two measures were taken: the first one was the severity of symptoms in the peel (cortex) of the tuberous roots; which is comprised of cortical parenchyma, phloem and sclerenchyma, and the second was the severity of the disease in the pulp of the tuberous roots; which is comprised by approximately 80% of xylem vessels and starch-containing cells. The images were also manually edited in order to identify the effects of cassava root rot and avoid confounding with physiological post-harvest deterioration symptoms.
Genetic parameters
The linear mixed model y = Xb + Zg + Wp + e was used (Resende 2002), where y is the data vector; b is the vector of fixed effects associated with general mean and blocks effect; g is the vector of random genetic effects; p is the vector of random effects of plots; e is the random error vector, and X, Z, and W are the incidence matrix that associates the unknown parameters b, g and p, respectively, to the y vector.
The covariance between all random effects models is given by the following:
where \(E\left[ {\begin{array}{*{20}c} y \\ g \\ p \\ e \\ \end{array} } \right] = \left[ {\begin{array}{*{20}c} {Xb} \\ 0 \\ 0 \\ 0 \\ \end{array} } \right]\) and \({\text{Var}}\left[ {\begin{array}{*{20}c} y \\ g \\ p \\ e \\ \end{array} } \right] = \left[ {\begin{array}{*{20}l} V \hfill & {ZG} \hfill & {WC} \hfill & R \hfill \\ {GZ^{\prime}} \hfill & G \hfill & 0 \hfill & 0 \hfill \\ {CW^{\prime}} \hfill & 0 \hfill & C \hfill & 0 \hfill \\ R \hfill & 0 \hfill & 0 \hfill & R \hfill \\ \end{array} } \right],\) and \(G = A\hat{\sigma }_{g}^{2}\), \(R = I\hat{\sigma }_{p}^{2}\), \(C = I\hat{\sigma }_{e}^{2}\) and \(V = ZA\widehat{\sigma }_{g}^{2} Z^{\prime} + WI\widehat{\sigma }_{p}^{2} W + I\hat{\sigma }_{e}^{2} = ZGZ^{\prime} + WCW^{\prime} + R\).
The mixed model methodology allows for the estimation of b using a generalized least squares procedure and g and p using BLUP. The system of linear equations (mixed model equations—MME) that were used to obtain the solutions of the model was
wherein \(\lambda_{1} = \frac{{\sigma_{e}^{2} }}{{\sigma_{g}^{2} }} = \frac{{1 - h^{2} - c^{2} }}{{h^{2} }}\) and \(\lambda_{2} = \frac{{\sigma_{e}^{2} }}{{\sigma_{p}^{2} }} = \frac{{1 - h^{2} - c^{2} }}{{c^{2} }}\) are the shrinkage factors of the random effects of the mixed model equations, in which \(\sigma_{g}^{2}\) is the additive genetic variance, \(\sigma_{p}^{2}\) is the variance of the effect of the plot, \(\sigma_{e}^{2}\) is the residual variance (environment + non-additive), A is the matrix of additive genetic correlation between individuals, and I is the identity matrix.
REML estimates of the variance components were obtained using the EM (expectation and maximization) algorithm according to the following expressions:
where C 22 and C 33 are derived from
which is the generalized inverse of the coefficient MME matrix, tr is the trace of a matrix, r(x) is the rank of the X matrix, N-r(x) is the number of degrees of freedom of the error, q is the number individuals, s is the number of plots, and N is the total number of observations.
The coefficients of the individual broad-sense heritability of the block were estimated according to \(\hat{h}_{g}^{2} = \frac{{\hat{\sigma }_{g}^{2} }}{{\hat{\sigma }_{g}^{2} + \hat{\sigma }_{p}^{2} + \hat{\sigma }_{e}^{2} }}\). The standard deviation of the heritability coefficients was obtained from the information matrix of the mixed model equations. The other estimates of variances and genetic parameters are given by \(\hat{\sigma }_{f}^{2} = \hat{\sigma }_{g}^{2} + \hat{\sigma }_{p}^{2} + \hat{\sigma }_{e}^{2}\) (phenotypic variance individual) and \(\hat{h}_{m}^{2} = \frac{{\hat{\sigma }_{g}^{2} }}{{\hat{\sigma }_{g}^{2} + \frac{{\hat{\sigma }_{e}^{2} }}{b}}}\) (heritability adjusted in average of accessions), where b is the number of blocks. The accuracy of the selection of clones was given by \(Ac = \sqrt {h_{m}^{2} }\). The estimation of variance components and non-biased prediction effects were estimated by REML using the Selegen software (Resende 2007).
Selection for multiple resistance
To select cassava accessions with multiple resistance to the pathogens causing CRRD, the classic selection index (CI), multiplicative index (MI), and sum of the ranks (SRI) were used based on the genotypic values from REML/BLUP according to the following:
-CI = ((p × FsppPeel) × (GV × FsppPeel)) + ((p × FsppPulp) × (GV × FsppPulp)) + ((p × PsppPeel) × (GV × PsppPeel)) + ((p × PsppPulp) × (GV × PsppPulp)) + ((p × BsppPeel) × (GV × BsppPeel)) + ((p × BsppPulp) × (GV × BsppPulp))
-MI = (GV × FsppPeel) × (GV × FsppPulp) × (GV × PsppPeel) × (GV × PsppPulp) × (GV × BsppPeel) × (GV × BsppPulp)
-SRI = (r × GV × FsppPeel) + (r × GV × FsppPulp) + (r × GV × PsppPeel) + (r × GV × PsppPulp) + (r × GV × BsppPeel) + (r × GV × BsppPulp);
where p is the weight established for the pathogen (one for all); GV is the predicted genotypic value; r is the rank of the accession for each pathogen; FsppPeel and FsppPulp refers to the severity of Fusarium spp., in the peel and pulp, respectively; PsppPeel and PsppPulp refers to the severity of Phytophthora spp. in the peel and pulp, respectively; and BsppPeel and BsppPulp refers to the severity of Botryosphaeriaceae species, peel and pulp, respectively.
In addition, the genetic gains using the selection indices and based on direct selection for each disease group (dry, soft, and black rot) were obtained. The estimated genetic gains were obtained by averaging the GV of the selected accessions.
Results and discussion
Estimates of genetic parameters
The components of variance, heritability estimates, coefficient of variation, and accuracy are shown in Table 1. For all pathogens causing root rot, environmental variance (\(\sigma_{e}^{2}\)) was a strong component of the phenotypic variance, in which the variation was 40.5% (BsppPulp) to 78.8% (FsppPulp). Except for Phytophthora spp., the \(\sigma_{e}^{2}\) varied considerably in the severity of the peel and pulp, i.e., 68.7 and 78.8% for FsppPeel and FsppPulp, respectively, and 71.3 and 40.5% for BsppPeel and BsppPulp, respectively. Similar results were observed for other diseases in plants such as the soybean rust, in which 53% of the phenotypic variation was due to environmental effects (Kiryowa et al. 2008).
The main influence of the environment on expression of the root rot symptoms resulted in low magnitudes for individual heritability coefficients (\(h_{g}^{2}\)) for most of the pathogens group. The \(h_{g}^{2}\) was very similar for resistance to the Phytophthora spp. (0.26 and 0.30 in the peel and pulp, respectively) and Botryosphaeriaceae species (0.28 and 0.27 in the peel and pulp, respectively). In contrast, the \(h_{g}^{2}\) for resistance of Fusarium spp. was twice for the peel compared to the pulp (0.31 and 0.16, respectively), indicating high environmental effect for the expression of the symptoms when the pathogen reached the pulp of the tuberous roots. The standard deviations associated with each \(h_{g}^{2}\) estimate ranged from 9.6 to 13.3% (FsppPeel and FsppPulp, respectively) and are therefore considered of low magnitude; this ensures a good prediction of genetic values according to Resende (2002).
Although \(h_{g}^{2}\) was of low magnitude for most pathogens causing root rot, the \(h_{g}^{2}\) comprises mainly the additive and dominance variances between the selection units that are effectively used for the selection of the best plants. In the particular case of cassava, whose propagation is predominantly asexual (stakes), breeders can select elite accessions and transmit all the genetic variance for the following plantations through cloning.
The heritability for the average of accessions (\(h_{m}^{2}\)) was virtually the same as \(h_{g}^{2}\) for the Fusarium spp and Phytophthora spp. (peel and pulp) and Botryosphaeriaceae species (peel). In contrast, the \(h_{m}^{2}\) for the Botryosphaeriaceae species in the pulp showed a significant increase compared with \(h_{g}^{2}\) (0.40). The main observed difference between \(h_{g}^{2}\) and \(h_{m}^{2}\) is due to lower phenotypic variation in the estimate of heritability due to the reduction of the residual variance and absence of the plot variance.
The success of breeding programs of any species depends on the availability of information on the genetic variability to be improved as well as the availability of estimates of genetic parameters for the traits of interest. Specifically for cassava root rot disease, studies on genetic parameters associated with genetic resistance are scarce and limited to a few pathogens, for example, those associated with soft root rot (Alvarez et al. 2002; Llano et al. 2004). However, genetic parameters for other pathogen group are currently unknown. On the other hand, genetic parameters such as heritability are very important to breeders, as they determine to what extend important traits will be transferred from parents to their progeny. Using simulated data, Viana (2007) showed proportionality between the trait heritability and genetic gain in maize depended on the trait, the improved level of the population, and selection strategy adopted. Therefore, genetic parameters help the breeder to define the optimal selection methods to be used.
High values of the coefficient of determination due to the common environments of the experimental unit (\(\sigma_{b}^{2}\)) indicated greater environmental variability between plots only for the Botryosphaeriaceae species in the pulp of the tuberous roots. For the remaining pathogens, the values ranged from 0.00 to 0.04 (Table 1) where coefficients smaller than 0.10 indicate good accuracy of the experiments, according to Resende (2002).
The accuracy values or correlation between the true and predicted genotypic values were greater than 0.50 for most pathogens, except for Fusarium spp. in the pulp (0.40). Accuracies greater than 0.50 are considered of medium magnitude, although Resende and Duarte (2007) recommend the implementation of selection in the early and intermediate stages of breeding program only for traits with accuracy above 0.7.
High genotypic coefficients of variation (CVg) were observed for all pathogens (ranging from 42.28 to 54.10% to Fusarium spp. in the peel and pulp, respectively), indicating the presence of high genotypic variability between accessions to cassava root rot. This genetic variability is a prerequisite for carrying out the selection of the best accessions. In contrast, the residual variation coefficient (CVe) whose variation was 55.28–93.46% (Table 1) was quite high mainly for Fusarium spp. and Phytophthora spp.
CVe above 50% were observed for shoot weight and weight of non-commercial roots in sweet potato (Ipomoea batatas L.) (Borges et al. 2010) and for shoot weight, yield per plant, and yield per area in cassava (Aina et al. 2007). According to Borges et al. (2010), large coefficients of variation are common when evaluating traits underground, in which the control of the environmental is difficult.
Selection for multiple pathogens related to cassava root rot
The existence of accessions that are resistant to a particular pathogen that causes CRRD and that are susceptible to other pathogens creates difficulties for the development of new varieties once it requires specific breeding programs for each pathogen. However, this is difficult for breeding programs in terms of restrictions in the number of populations to be evaluated annually, the limited availability of planting material to conduct separate evaluations, as well as financial and human costs. Therefore, the priority in the selection of parentals for cassava breeding programs should be the identification of accessions with favorable genotypic performance against various pathogens causing cassava root rot. Thus, the application of selection indices is an attractive alternative for the simultaneous selection of multiple traits.
The classic selection index (CI) selected accessions with reduced the root rot symptoms caused by Fusarium spp in −45.24 and −46.08% in the peel and pulp, respectively (Table 2). These values are only 9.88 and 17.55% lower than the direct gains with the selection of the most resistant accessions to Fusarium spp, in the peel and pulp, respectively. In contrast, when using the CI, the reductions in −28.13 and −21.12% in the peel and pulp, respectively, for Phytophthora spp. were quite low compared to the −50.43 and −56.83% in the direct selection to this pathogen. The reductions in the severity caused by the Botryosphaeriaceae species were −41.26 and −38.57%, in the peel and pulp, respectively, which represents a reduction of 17.74 and 36.85% compared to direct selection for this pathogen.
By using the multiplicative index (MI), the reductions in the severity of Fusarium spp were lower than the CI, with an average of −38.33 and −40.41%, in the peel and pulp, respectively, which represents a reduction of 23.65 and 27.70% in relation to the direct selection of the most resistant accessions to both tuberous root parts (Table 3). In contrast, the higher gains for resistance to Phytophthora spp. were observed using MI, i.e., −28.78 and −39.90% reduction in the symptoms of CRRD in the peel and pulp, respectively. However, these reductions are still lower compared to the direct gain of more resistant accessions in the peel (42.93%) and pulp (29.80%), respectively. In relation to resistance of the Botryosphaeriaceae species, the symptom reductions in the peel and pulp were −33.51 and −48.79%, respectively. Nevertheless, these reductions are 33.19 and 20.12% lower in comparison with the direct gain in the selection of the most resistant accessions to Botryosphaeriaceae species individually, considering the severity in the peel and pulp, respectively.
The use of the sum of ranks index (SRI) presented the smaller gain in terms of reducing the severity of CRRD pathogens (Table 4). The reduction of severity caused by Fusarium spp. was −7.68 and −22.44% in the peel and pulp, respectively. Therefore, this index was lower than the direct selection in 84.70 and 59.85% for symptoms in the peel and pulp, respectively. For resistance to Phytophthora spp., the symptoms decreased in the peel but was very low (−4.68%), whereas for the pulp there was an increase in the severity of Phytophthora spp. (1.70%). Similar to those observed for other pathogens, the reduction in severity caused by the Botryosphaeriaceae species was quite low with use of SRI (−5.70 and −6.70%, respectively, in the peel and pulp of tuberous roots). This represents a reduction of 88.64 and 89.04% in the peel and pulp of the tuberous roots compared to the direct gain.
By using the REML/BLUP method to obtain gains in agronomic and nutritional traits in progenies of Brachiaria humidicola, Figueiredo et al. (2012) carried out direct selection based on two selection indices. The first one considered simultaneous gains only for agronomic traits, and the second one used 70 and 30% weights for agronomic and nutritional traits, respectively. Based on the selection of the 10 best progenies these authors also showed that the gains were always higher for direct selection compared to the use of selection indices, although it was possible to select high performance progenies based on the selection indices. Moreover, these authors stated that the REML/BLUP offers the advantage of obtaining the gains for selection directly by BLUP, once these are the predicted genotypic values that have already been adjusted for fixed environmental effects.
Even without the use of selection indices, the identification of sources of resistance to multiple pathogens in cassava has been evaluated in the literature. In Nigeria, Fokunang et al. (2000) evaluated 35 cassava accessions for incidence and severity caused by anthracnose (CAD), bacterial blight (CBB), and African mosaic virus (ACMV). Under natural conditions of pathogen infection, they demonstrated that these diseases presented significant negative correlations with yield traits such as the number and weight of tuberous roots and dry matter content.
Table 5 shows the matching rate of selected accessions by direct selection for each pathogen individually, using the CI, MI, and SRI indices. In general, the selection indices were poor matches. The highest matching rate was observed between MI × IC (0.60) and between MI × direct selection for Fusarium spp. in the peel (0.47), and IC × direct selection for Fusarium spp. in the peel (0.47). Low matching rate of the best genotypes selected by using different selection indices and REML/BLUP method was observed by Freitas et al. (2013) in full-sib progeny of popcorn for yield and popping expansion. According to those authors, even with the strong negative association between yield and popping expansion, the REML/BLUP method was more efficient in selecting genotypes with high performance and promising estimated genetic gain compared with the selection indices.
No common accessions were observed among the SRI and the other selection indices. A similar situation was also observed when considering the matching rate between the selected accessions based on the direct selection for different pathogens causing root rot. Surprisingly, within a specific pathogen, the matching rate between the most resistant accessions considering the severity in the peel and pulp was also low, reinforcing the hypothesis of different mechanisms of resistance in both cassava tuberous root structures.
In rubber tree, Jayasuriya (2004) reported that the main defense mechanisms against pathogens causing root rot (Phellinus noxius and Rigidoporus lignosus) are related to cellular hypertrophy and hyperplasia, cambium activity stimulation, lignification, and suberification of certain cell walls. Although such studies are not available for the pathogens causing root rot in cassava, resistance to other diseases such as anthracnose, bacterial blight, and African mosaic virus have been associated with metabolic changes of phenols, increase in peroxidase activity, phytoalexin action, anthocyanins, and cyanogenic glycosides (Fokunang et al. 2000). In addition, the processes related to cell lignification may also restrict the development of pathogenic agents by increasing the mechanical strength of the cell wall, reducing the degradation of the cell wall by extracellular enzymes, restricting the diffusion of pathotoxins and nutrients, and also inhibiting the growth of the pathogens by the action of toxic lignin precursors (Fokunang et al. 2000).
In cassava, the content of cyanogenic compounds may play an important role in the plant-pathogen interaction, since cyanide is toxic to many microorganisms (Fokunang et al. 2000). Therefore, with the differences in the behavior of accessions for the severity of CRRD in the peel and pulp of the tuberous roots, it is possible to speculate the effect of the cyanogenic content in the development of pathogens, whereas higher levels of these toxic compounds is found in peel of the tuberous roots in comparison with the pulp (Sornyotha et al. 2007). Thus, considering the differential resistance to Fusarium spp., Phytophthora spp., and the Botryosphaeriaceae species in the peel and pulp of tuberous roots, further studies on the plant-pathogen interaction should be conducted to elucidate the process of penetration, colonization, and degradation of different cassava tuberous roots tissues in accessions with different levels of genetic resistance.
Perspectives for breeding
Resistance to cassava root rot is complex considering the environmental interactions, the presence of different pathogenic strains of a same species as well as differential expression of the symptoms in different parts of the plant, similar to what was observed in the peel and pulp of cassava tuberous roots. In contrast, quantitative analysis of CRRD can aid the study of the resistance against various pathogens and identify areas of research that can add information to this pathosystem considering physiological and molecular aspects.
According to Falconer and Mackay (1996), the heritability of certain traits is not constant and may be changed depending on the breeding methods adopted. However, the low estimate of heritability for the most pathogens causing CRRD in detached tuberous roots indicates that effective selection methods should be used to ensure higher genetic gains in the early breeding generations.
Furthermore, from the point of view of selection for breeding purposes, the low matching rate of cassava accessions for resistance to different pathogens causing root rot, and especially the low matching rate of the different parts of the tuberous roots (peel and pulp), indicates the presence of different resistance genes controlling this disease. More research is needed on the methods of assessment the root rot resistance as a prerequisite to more detailed breeding and inheritance studies in cassava.
The distribution of the root rot symptoms caused by Fusarium spp, Phytophthora spp. and Botryosphaeriaceae species indicates the presence of quantitative inheritance, in which the resistance is the result of many genes with a small effect. In this case, breeding programs should adopt strategies to combine and monitor the presence of the different genes associated with resistance, which is not always easy by using conventional methods due to the existence of epistatic effects. In contrast, the use of cassava resistant accessions to root rot provides the opportunity to identify and map molecular markers linked to the resistance genes. Molecular mapping can open the opportunity to confirm the presence of multiple genes in certain genotypes. The availability of several types of molecular markers, along with strategies of molecular-assisted selection, can help the development of new cassava varieties with more durable resistance to root rot, similar to what has been done in other species (Ashkani et al. 2012).
Continuous evaluations of cassava germplasm for resistance to the main pathogens related to root rot as well as understanding the genetic basis of resistance should be used for implementation of integrated management strategies, identification and introduction of multiple resistance to ensure the crop sustainability (especially in areas affected by soil pathogens), and increase the crop yield potential. Although, some reports have shown that field resistance to cassava root rot disease presented a strong correlation with laboratory methods (Onyeka et al. 2005c), future comparison between genotypes response in the field and after artificial inoculation will be necessary to check the agreement (accuracy) between estimated cassava root rot severity and true values (field resistance), especially regarding the genetic parameters and ranking of the accessions with multiple resistance to cassava root rot.
References
Aina OO, Dixon AGO, Akinrinde EA (2007) Genetic variability in cassava as it influences storage root yield in Nigeria. J Biol Sci 7:765–770
Alvarez E, Loke J, Rivera S, Llano G (2002) Genética de la resistencia a pudrición causada por Phytophthora tropicalis en dos poblaciones segregantes de yuca (Manihot esculenta Crantz). Fitopatol Colomb 26:61–66
Ashkani S, Rafii MY, Rusli I, Sariah M, Abdullah SNA, Harun AR, Latif MA (2012) SSRs for marker-assisted selection for blast resistance in rice (Oryza sativa L.). Plant Mol Biol Rep 30:79–86
Bandyopadhyay R, Mwangi M, Aigbe SO, Leslie JF (2006) Fusarium species from the cassava root rot complex in West Africa. Phytopathology 96:673–676
Banito A, Kpémoua KE, Bissang B, Wydra K (2010) Assessment of cassava root and stem rots in ecozones of Togo and evaluation of the pathogen virulence. Pak J Bot 42:2059–2068
Borges V, Ferreira PV, Soares L, Santos GM, Santos AMM (2010) Seleção de clones de batata-doce pelo procedimento REML/BLUP. Acta Sci Agron 32:643–649
Falconer DS, Mackay TFC (1996) Introduction to quantitative genetics, 4th edn. Pearson Education Limited, London
FAO (2015) FAOSTAT database. FAO, Rome. http://faostat.fao.org/. Accessed 26 Jan 2015
Figueiredo UJ, Nunes JAR, Valle CB (2012) Estimation of genetic parameters and selection of Brachiaria humidicola progenies using a selection index. Crop Breed Appl Biotechnol 12:237–244
Fokunang CN, Akem CN, Dixon AGO, Ikotun T (2000) Evaluation of a cassava germplasm collection for reaction to three major diseases and the effect on yield. Genet Resour Crop Evol 47:63–71
Freitas ILJ, Amaral Junior AT, Viana AP, Pena GF, Cabral OS, Vittorazzi C, Silva TRC (2013) Ganho genético avaliado com índices de seleção e com REML/Blup em milho pipoca. Pesq Agropec Bras 48:1464–1471
Guo H, Li CP, Shi T, Fan CJ, Huang GX (2012) First report of Phytophthora palmivora causing root rot of cassava in China. Plant Dis 96:1072
Hillocks RJ (2014) Addressing the yield gap in Sub-Saharan Africa. Outlook Agric 43:85–90
Hoover H (2001) Composition, molecular structure, and physicochemical properties of tuber and root starches: a review. Carbohydr Polym 45:253–267
Jayasuriya KE (2004) Factors affecting disease tolerance of rubber tree and research needs for developing disease tolerant genotypes for the sustainability of rubber industry. Bull Rubber Inst Sri Lanka 45:1–10
Kiryowa M, Tukamuhabwa P, Adipala E (2008) Genetic analysis of resistance to soybean rust disease. Afr Crop Sci J 16:211–217
Llano GA, Álvarez E, Muñoz JE, Fregene M (2004) Identificación de genes análogos de resistencia a enfermedades en yuca (Manihot esculenta Crantz) y su relación con la resistencia a tres especies de Phytophthora. Acta Agron 53:15–24
Machado AR, Pinho DB, Oliveira SAS, Pereira OL (2014) New occurrences of Botryosphaeriaceae causing black root rot of cassava in Brazil. Trop Plant Pathol 39:464-470.
Msikita W, Bissang B, James BD, Baimey H, Wilkinson HT, Ahounou M, Fagbemisi R (2005) Prevalence and severity of Nattrassia mangiferae root and stem rot pathogen of cassava in Benin. Plant Dis 89:12–16
Okechukwu RU, Dixon AGO, Akoroda MO, Mwangi M, Bandyopadhyarr R (2009) Root rot resistance in new cassava varieties introduced to farmers in Nigeria. Exp Agric 45:15–24
Oliveira SAS, Hohenfeld CS, Santos VS, Haddad F, Oliveira EJ (2013) Resistance to Fusarium dry root rot disease in cassava accessions. Pesq Agropec Bras 48:1414–1417
Onyeka TJ, Dixon AGO, Ekpo EJA (2005a) Identification of levels of resistance to cassava root rot disease (Botryodiplodia theobromae) in African landraces and improved germplasm using in vitro inoculation method. Euphytica 145:281–288
Onyeka TJ, Dixon AGO, Ekpo EJA (2005b) Field evaluation of root rot disease and relationship between disease severity and yield in cassava. Exp Agric 41:357–363
Onyeka TJ, Dixon AGO, Ekpo EJA (2005c) Assessment of laboratory methods for evaluating cassava genotypes for resistance to root rot disease. Mycopathologia 159:461–467
Resende MDV (2002a) Genética biométrica e estatística no melhoramento de plantas perenes. Embrapa Informação Tecnológica, Brasília
Resende MDV (2007) Selegen-REML/BLUP: sistema estatístico e seleção genética computadorizada via modelos lineares mistos. Embrapa Florestas
Resende MDV, Duarte JB (2007) Precisão e controle da qualidade de experimentos para avaliação de cultivares. Pesq Agropec Trop 37:182–194
Rickard JE, Asaoke M, Blanshard JMV (1991) The physico-chemical properties of cassava starch. Trop Sci 31:189–207
Sornyotha S, Kyu KL, Ratanakhanokchai K (2007) Purification and detection of linamarin from cassava root cortex by high performance liquid chromatography. Food Chem 104:1750–1754
Souza LS, Farias AR, Mattos PLP, Fukuda WMG (2006) Aspectos socioeconômicos e agronômicos da mandioca. Embrapa Mandioca e Fruticultura Tropical, Cruz das Almas
Suppakul P, Chalernsook B, Ratisuthawat B, Prapasitthi S, Munchukangwan N (2013) Empirical modeling of moisture sorption characteristics and mechanical and barrier properties of cassava flour film and their relation to plasticizing-antiplasticizing effects. LWT Food Sci Technol 50:290–297
Viana JMS (2007) Breeding strategies for recurrent selection of maize. Pesq Agropec Bras 42:1383–1391
Vilas Boas SAV, Hohenfeld CS, Oliveira SAS, Santos VS, Oliveira EJ (2016) Sources of resistance to cassava root rot caused by Fusarium spp.: a genotypic approach. Euphytica 209:237–251
Vilas Boas SAV, Oliveira SAS, Bragança CAD, Ramos JB, Oliveira EJ (2017) Survey of fungi associated with cassava root rot from different producing regions in Brazil. Sci Agricola 74:60–67
Acknowledgements
The authors thank the Fapesb, CAPES and CNPq for the financial assistance and scholarship support.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
de Oliveira, E.J., de Oliveira, S.A.S., Boas, S.A.V. et al. Selection of cassava accessions with multiple resistance to pathogens associated with root rot disease. Euphytica 213, 185 (2017). https://doi.org/10.1007/s10681-017-1973-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10681-017-1973-6