Abstract
This study investigated the relationship between nutrient levels, source of fecal contamination, and pathogenic Leptospira in Puerto Rico’s northern coast and San Juan Bay Estuary (SJBE) aquatic ecosystems. Microbial source tracking (MST) was also used to investigate the connections between sources of feces contamination and the presence of Leptospira. Eighty-seven water samples were collected during the June (n=44) and August (n=43) in 2020. To quantify phosphorus and nitrogen concentrations, standard USEPA protocols were utilized, specifically Methods 365.4 for total and dissolved phosphorus, 351.2 for total Kjeldahl nitrogen and ammonium, and 353.2 for nitrate. Lipl32 gene-specific quantitative polymerase chain reaction (qPCR) was used to detect the presence of Leptospira. Human (HF183), canine (BacCan-UCD), and equine (HoF597) MST assays were utilized to trace the origins of fecal contamination. Forty one percent of the locations exceeded Puerto Rico’s authorized total phosphorus limit of 160 g L−1, while 34% exceeded the total nitrogen limit of 1700 g L−1. Nearly half of the streams examined are affected by eutrophication. The MST analysis identified human and canine feces as the most prevalent contaminants, affecting approximately 50% of the sites. In addition, Leptospira was detected in 32% of the June samples. There was a significant correlation (r = 0.79) between the incidence of pathogenic Leptospira and the human bacterial marker (HF183). This study illuminates the central role of anthropogenic inputs in nutrient enrichment and pathogen proliferation in Puerto Rico’s aquatic ecosystems.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
The northern coastal zones of Puerto Rico, including the San Juan Bay and Rio Grande de Loiza (below dam) watersheds, are increasingly becoming focal points of environmental concern due to wastewater originating from human activity. This runoff, which consists of effluents from sewage treatment, septic tanks, and animal husbandry, enters coastal lagoons and estuaries, posing critical threats to these vital ecosystems. The San Juan Bay Estuary (SJBE), a significant ecological center located within these watersheds, is essential to the integrity of the coastline, especially its mangrove forests (Fretwell et al., 1996). The estuary, which serves as the drainage for the densely populated San Juan-Caguas-Guaynabo metropolitan area, home to an estimated 2.7 million people, faces the difficulty of large volumes of refuse runoff. This situation is exacerbated during the wet season, when the region receives between 1500 and 1700 mm of precipitation annually (Brandeis et al., 2014; Fretwell et al., 1996), with the heaviest precipitation occurring between August and September during hurricane season (Colón & Schaffner, 2021).
Particularly during these rain events, San Juan has been identified as a significant source of fecal pollution and nutrient discharge, raising significant public health and environmental concerns (Holman et al., 2014; Sutter & Sosa Pascual, 2018; Walker et al., 2013).
Among these threats, eutrophication, predominantly caused by excessive nitrogen (N) and phosphorus (P) from urban and fecal sources, is the most damaging. The toxic algal outbreaks that result pose a threat to marine ecosystems, with the potential for significant ecological consequences (Conley et al., 2009; Sutter & Sosa Pascual, 2018; USEPA, 2012). Attributed to accelerated population growth and agricultural expansion within these watersheds, (Mesa et al., 2015; Ngatia et al., 2019).
The Puerto Rico Environmental Quality Board regulates the water quality in these watersheds and has established total P and total N nutrient criteria of 0.160 mg L−1 and 1.70 mg L−1, respectively (USEPA, 2019). Ngatia et al. (2019) report that most island streams and rivers exhibit alarmingly high N and P concentrations, far exceeding the established thresholds. This unsettling circumstance highlights the need for stricter pollution management measures and extensive research to comprehend the full scope of these environmental challenges.
Environments influenced by fecal runoff, especially during heavy rainfall, are potential reservoirs of harmful pathogens, such as Leptospira, the causative agent of leptospirosis. This aerobic, gram-negative bacterium is arranged in a unique spiral morphology and is currently classified into 35 species within four subclades. Notably, pathogenic Leptospira species like L. interrogans, L. borgpetersenii, L. kirschneri, L. noguchii, and L. weilii, all belonging to the P1 subclade, pose significant health risks as they are often expelled into the global environment through the urine of infected hosts (Pappas et al., 2008; Vincent et al., 2019).
This disease accounts for approximately 60,000 yearly fatalities (Bierque et al., 2020). Humans and other animals become susceptible to infection upon exposure to contaminated water, often filled with bacterium-laden urine (Pappas et al., 2008; Rawlins et al., 2014). Within Puerto Rico, the estimated cases of Leptospirosis ranged from 15 to 100 in 2010, with 114 reported cases during 2014 to 2015 (Briskin et al., 2019). However, it is crucial to note that these figures likely underestimate the actual disease burden, given that many cases of Leptospirosis are asymptomatic and often go unreported (Briskin et al., 2019).
Parallel to fecal waste nutrient contamination, environmental levels of pathogens such as Leptospira tend to spike during flooding or heavy rainfall in tropical islands. These periods of increased environmental prevalence usually precede human infection outbreaks (Barragan et al., 2017; USEPA, 2012). Truitt et al. (2020) discovered pathogenic Leptospira in a number of rural rivers in Puerto Rico during the wet season but found no meaningful correlation between physicochemical parameters (such as pH, dissolved oxygen (DO), and temperature), concentrations of fecal indicator bacteria (Escherichia. coli), and the presence of pathogenic Leptospira at the sampling sites. This was attributed to the fact that Leptospira is transmitted through urine that co-occur with feces but may not always contain these pathogens (Pratt & Rajeev, 2018; Truitt et al., 2020). A limitation of this study was the need for more evaluation regarding the potential sources of these pathogenic bacteria in the environment (Truitt et al., 2020).
A diverse array of animal hosts, including dogs, pigs, cattle, rodents, mongooses, bats, and frogs, can carry these bacteria and thus contribute significantly to disease transmission (Levett et al., 2001; Pratt & Rajeev, 2018). However, crucial information about the potential sources of pathogenic Leptospira and fecal nutrients still needs to be discovered, leaving a substantial gap in our understanding of these environmental health issues. The development of polymerase chain reaction (PCR) methodologies and microbial source tracking (MST) approaches have introduced a novel capability to detect Leptospira in environmental samples rapidly and to infer their potential sources (Bridgemohan et al., 2020; Rawlins et al., 2014; Riediger et al., 2016). The successful use of a hydrolysis probe assay targeting the Lipl32 gene, which encodes a 32-kDa membrane protein unique to pathogenic Leptospira species in the P1 subclade, represents a recent advancement in this context (Rawlins et al., 2014; Stoddard et al., 2009; Truitt et al., 2020).
Microbial source tracking methodologies can link the presence of pathogens or fecal contamination in aquatic systems to a particular source (Bridgemohan et al., 2020; González-Fernández et al., 2021). The central tenet of MST is that certain animals harbor species-specific fecal bacteria to its host, such as within the genus Bacteroidales, which can be detected using PCR (Bachoon et al., 2010; Bridgemohan et al., 2020). There have been reports of the successful use of MST to identify sources of fecal pollution and pathogenic bacteria in Puerto Rico’s marine and freshwater systems (Bachoon et al., 2010; Wade et al., 2015).
Given the extant information about the San Juan Bay and Rio Grande de Loiza watersheds, we hypothesized that humans, horses, dogs, and rodents were the most likely sources of nutrient contamination and Leptospira (Benavidez et al., 2019). For this investigation, an MST assay for rodent fecal bacteria was unavailable. However, we propose the integration of MST, nutrient level assessments, and PCR detection of Leptospira to determine the source of surface water contamination. This study aims to determine the relationship between nutrient concentrations (N and P) with fecal pollution sources, the presence of pathogenic Leptospira, and the potential sources of these contaminants in the SJBE and Rio Grande de Loiza watersheds during June and August of 2020. Once the sources of contamination are identified, appropriate mitigation measures can be implemented to address these environmental challenges.
Materials and methods
Sampling sites
To represent urban and agricultural regions within the San Juan Bay Estuary (SBJE) and Rio Grande de Loiza watersheds, sampling stations were strategically chosen. The investigation included a minimum of two stations per subbasin in 2020 (see Appendix Table 3 for details). Following established protocols (Wilde et al., 1998), 87 water samples were collected using sterile plastic receptacles in June (which had weekly rainfall cumulative average of 17.53 mm to 24.38 mm) and August (dry period) with a weekly cumulative average of 4.06 mm to 2.03 mm.
Nutrient concentration evaluation
Chemical analyses included dissolved (DP) and total reactive (TP) P (EPA method 365.4), total Kjeldahl nitrogen (TKN), dissolved ammonium (NH4+) (EPA method 351.2), and nitrate (NO3−) (EPA method 353.2). Total nitrogen (TN) was obtained from the sum of (NO3−) and TKN. Samples for (NO3−), (NH4+), and DP were passed through a 0.45-μm pore size Gelman-Acrodisc filter (Waltham, MA, USA).
DNA extraction
Each sample (100 mL) was filtered through a nitrocellulose membrane filter (Type GS, Millipore, Billerica, MA, USA) with a 0.45-μm pore size (Millipore, Billerica, MA, USA) and then stored at −20 °C before being transported to Georgia College and State University. The DNA was extracted from the filters using the QIAGEN PowerSoil Kit (Qiagen, Hilden, Germany) in accordance with a modified version of the procedure described by Bachoon et al. (2010). The modification involved separating the bead solution from the beads and placing it in a 15-mL centrifuge tube containing the filter. Solutions S1 and IRS were added to the tube and vortexed vigorously for 15 min. The solution was removed from the centrifuge tube and placed in the bead tube. The manufacturer’s protocol was followed from this point. After determining the DNA concentration of each sample using a Nanodrop ND-1000 Spectrophotometer (Wilmington, DE), the samples were stored at −20 °C.
Quantitative polymerase chain assays of Leptospira interrogans
Leptospira interrogans was quantified by qPCR using a 242-bp segment of the Lipl32 gene, as previously described (Rawlins et al., 2014; Truitt et al., 2020), by running samples in duplicate using QuantiTect Probe PCR kit (Qiagen) with the Bio-Rad CFX96 (Hercules, CA, USA), as described by Truitt et al. (2020). Each reaction had a final volume of 20 μl using ~10ng of extracted DNA, 500 nM of each primer (Table 1), and 200 nM of the probe. The thermal conditions were an initial 95°C for 15 min, followed by 40 cycles at 95°C for 10 s and 64°C for 30 s. Leptospira interrogans serovar Pomona was used as the positive control, E. coli strain K-12 as a negative control, and no-template controls were performed in each assay. The positive samples were then retested in triplicate. The number of genome copies was quantified using standard curves of L. interrogans serovar Pomona gene copies (Truitt et al., 2020). DNA extracted from L. interrogans serovar Pomona was used as the positive control; E. coli strain K-12 as a negative control; and tubes lacking DNA as no-template controls. Based on the genome size of L. interrogans (4.659 Mb), genome equivalents were calculated as described (Levett et al., 2005). The lower limit of the assay was set at 5 genome copies of L. interrogans determined using serial dilutions of positive control DNA. Deoxyribonucleic acid extracts were evaluated for the presence of PCR inhibitors by evaluating shifts in Cq-values between a sample and its 10-fold diluent (Dick et al., 2010; Truitt et al., 2020). Statistical analyses were performed with the R® program.
Microbial source tracking (MST)
Human and animal fecal bacteria were detected using quantitative polymerase chain reaction (qPCR) assays on the Bio-Rad CFX 9600 Real-Time Detection system (Hercules, California, USA). Hydrolysis probe-based assays utilizing the QuantiTect Probe PCR (Qiagen) reagents were utilized for human and canine samples, whereas the SsoFast Evagreen (Bio-Rad) dye-based assay was utilized for equine samples. Based on previously reported sensitivity and selectivity metrics, assay primers were selected and optimized to prevent non-specific cross-reactions with fecal DNA from local horse, cattle, dog, and human sources. This was done by testing each fecal host MST assay against the panel of host fecal DNA extracts (Table 1).
Polymerase chain reaction assays were conducted with 1 μL of sample DNA (approximately 10 ng μL−1) and a ten-fold diluted DNA sample with annealing temperatures and primer/probe sequences for each source marker gene as listed in Table 1. Standard curves using host-specific fecal DNA for each qPCR (controls) were linear with coefficients of determination (r2) of 0.99 and efficiencies of over 90%. Negative controls without DNA, which were run with each reaction, always exceeded the cycle threshold at a mean CT limit of the assay which was set at 5 genome copies of L. interrogans of 39.58. Each hydrolysis probe (human and dog) was labeled at the 5’ end with the reporter dye 6-FAM (6-carboxy-fluorescein) and at the 3’ end with the quencher dye TAMRA (6-carboxytetramethyl-rhodamine). For human (HF183-1), the qPCR assay used a modified protocol of Green et al. (2014), with Bacteroides dorei DSM 17855 (DSMZ) and human sewage used as a positive control and E. coli strain B from Sigma® D48890-1UN and horse and dog fecal DNA extracts as a negative control. The 20 μl assay contained 250 nM of each primer, 0.2 mg of bovine serum albumin (Sigma), and 80 nM 6-FAM™ labeled Hydrolysis® probe. Water samples were assessed for possible PCR inhibition by amending with positive control bacteria DNA as described by Bachoon et al. (2010). Changes of less than two CT value were observed, which indicates that the extracted DNA did not contain impurities that significantly inhibited the MST assay (Bachoon et al., 2010). In addition, ten-fold dilutions of sample DNA extracts were compared to the undiluted sample DNA extract, and a change in CT shift of < 3 CT indicated no significant PCR inhibition (Dick et al., 2010). The samples were run at 95 °C for 10 min, 40 cycles at 95 °C for 10 s, and at the required annealing/extension temperature (Table 1). The qPCR detection limits were determined from the lowest concentration of target DNA that could be detected at 95% confidence and gene copies ranged from human 8, dog 10, and horse 25 gene copies. Data was statistically analyzed using InfoStat 2012 (Di Rienzo et al., 2012). Data from all the experiments were analyzed using a one-way ANOVA as a completely randomized design. Significant differences among treatment means were determined using Fisher’s protected LSD at p = 0.05. Spearman correlation was also used to analyze data with significant correlation coefficients with a value greater than 0.7. Statistical analysis of the relationship between TP and TN with the marker for human fecal contamination was performed using an odds ratio.
Results
Nutrients concentration levels
Thirty-six of the eighty-seven water samples (41%) collected in 2020 had TP concentration that exceeded 160 μg L−1 which is the regulatory limit for Puerto Rico (USEPA, 2019) (Appendix Table 4). The highest levels of TP were observed after periods of rain during the June sampling events in the Quebrada Angela subbasin (3210 μg L−1) and in the Josefina subbasin (1860 μg L−1, 1,574 μg L−1). These same sites exhibited elevated levels of DP ranging from ~1500 to 2800 μg L−1, while the other sites ranged from ~2 to 955 μg L−1 (Appendix Table 4). Statistical analysis of the relationship between TP and the marker for human fecal contamination indicates that the probability of a water sample being positive for the human fecal bacterial marker when TP exceeds the regulatory limit of 160 μg L−1 is 16 times greater than when TP is less than 40 μg L−1 (p < 0.000).
Thirty of the eighty-seven water samples (34%) exceeded the regulatory limit of 1700 μg L−1 of TN (Appendix Table 4) (USEPA, 2019). It should be noted that TN in five water samples was > 4800 μg L−1, and the sites that these samples were collected from include Juan Méndez, San Antón, Quebrada Angela, and Bocaforma. More specifically, the greatest concentrations of TN were measured at one site in Quebrada Angela during the June sampling event (34,013 μg L−1) followed by two sites in the Bocaforma (5683 μg L−1, 5,962 μg L−1) subbasin in August. The lowest concentrations of TN were measured in Río Canóvanas during the June sampling event (116.56 μg L−1, 108.43 μg L−1) and the August sampling event (81.10 μg L−1). Statistical analysis of the relationship between TN and the marker for human fecal contamination indicates that the probability of a water sample being positive for the human marker when TN exceeds the regulatory limit of 1700 μg L−1 is 16 times greater than when TN is less than 40 μg L−1 (p = 0.001). Nitrate in water samples during both sampling events ranged from 0.84 μg L−1 in Río Canovanillas to 1339.59 μg L−1 in Rio Piedras Sur. Fourteen of the water samples collected contained NO3− concentrations > 1000 μg L−1, and most of these samples were collected during the dry period in August (Appendix Table 4).
Leptospira detection and MST
DNA amplified from water samples with a cycle threshold (Ct) < 40 were considered positive when evaluating qPCR assay results for both L. interrogans and MST markers. The qPCR assay for L. interrogans was considered successful with an efficiency of over 95% with an r2 = 0.987 or greater, and the detection limit of the assay was set at 5 genome copies of L. interrogans. The results from the Lipl32 gene qPCR assay indicated that pathogenic Leptospira was present only in June samples after rainfall and was undetectable in the dryer month of August (p < 0.0001) (Table 2). During June, L. interrogans were detected in 14 of the 44 water samples, with an average gene copy of ~ 400 ± 199 Leptospirosis per 100 mL sample. Bocaforma had the highest level of L. interrogans with approximately 737 genome copies per 100ml of water. Most of the samples that contained L. interrogans were from the western regions, Rio Piedras, Juan Mendez, and Dona Ana, that experienced between 17.53 and 24.38 mm of rain over 7 days (Table 2).
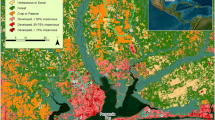
Water samples were assayed for the presence of human, dog, and horse fecal bacteria using the MST markers listed in Table 1. The BacCan MST assay indicated that dogs (26/44) were the most common source of fecal pollution in June. The next most prevalent source of fecal contamination in June was by humans (20/44), and horse fecal pollution was present in only four sites. During the August sampling, human fecal pollution was detected in 22 sites, dog fecal waste was detected in 15 sites, and horse fecal bacteria were detected in four sites (Fig 1, Table 2). Spearmen’s correlation indicated a significant positive relationship (r =0.89) between the sites with human fecal contamination and the sites with pathogenic Leptospira. These sites were mostly located on the western part of the SJBE (Fig. 1). There was a moderate correlation (r = 0.58) between dog fecal contamination and the sites with pathogenic Leptospira which could be influenced by the cross-reaction of the BacCan assay (Schriewer et al., 2013), and horse fecal pollution did not have a significant co-relationship (r = 0.14) to the presence of L. interrogans in the water samples.
Discussion
Nutrient concentration in SJBE and MST
The primary objective of this study was to assess nutrient concentrations in the waters flowing into the San Juan Bay Estuary (SJBE) and to employ microbial source tracking (MST) to identify potential fecal sources contributing to elevated nitrogen (N) and phosphorus (P) concentrations. Prior research conducted in Puerto Rico by Larsen and Webb (2009) demonstrates a startling ten-fold increase in N and P concentrations compared to presettlement values, which is primarily attributable to human landscape modification.
The SJBE in Puerto Rico, which is characterized by extensive urban and residential development, is susceptible to increased nutrient loading from a variety of sources, such as sewage discharges, animal waste, fertilizers, fossil fuels, and industrial wastewater (Conley et al., 2009; Galloway et al., 2008). This nutrient enrichment, particularly of N and P, is well documented to have detrimental effects on the integrity of estuarine ecosystems, thereby compromising their healthy functioning (Correll, 1999; Ngatia et al., 2019). To contribute to a broader understanding of human-induced nutrient loading and its implications for environmental health and resilience, the purpose of this study is to conduct a comprehensive evaluation of these crucial aspects.
Thirty-six out of the eighty-seven samples exceeded 160 μg L−1 of TP indicating that nearly half of the water sampled was in hypereutrophic conditions (Carlson, 1977). The areas that had the greatest levels of TP, Josephina (1860 μg L−1, 1574 μg L1) and Quebrada Angela (3210 μg L−1), were also impacted by human (Fig. 1) and dog fecal pollution (Table 2, Appendix Table 3). In addition, the greatest concentration of TN measured in this study (34,013 μg L−1) came from water collected at the Quebrada Angela location after periods of rainfall in June. Molecular markers for dog and human fecal pollution were also detected in these waters during the wet period in June (Table 2), while only the human fecal pollution was detected during the dry sampling event (data not shown). These results are not surprising since Quebrada Angela is located near urban areas with a high human population density. It should be noted that the concentration of NO3− in all water samples was below the USEPA regulatory limit of 10,000 μg L−1 (Appendix Table 4).
Dogs have been previously identified as significant sources of fecal pollution in coastal areas (Martin & Gruber, 2005; Wright et al., 2009), and the BacCan MST assay in our study indicates that dogs were the most common source of fecal pollution in June (26 sites) and the second most common source in August (15 sites). However, this assay is very sensitive but is reported to sometime exhibit cross-reactions with non-canine species (Boehm et al., 2013; Schriewer et al., 2013). However, the population of dogs in an area should be considered when developing management practices to limit the input of N and P in surface waters.
Horses are common in the sampled watersheds due to recreational and personal use. The molecular marker of horse fecal bacteria indicating horse fecal contamination was only present in four sites in each month; however, molecular markers for human and/or dog fecal contamination were also present in six of the eight horse-positive samples (Table 2). This was not surprising because humans, dogs, and horses are usually present in the same areas of the island. Horses typically produce a larger volume of waste per animal unit compared to humans and dogs, and their activity makes soil more susceptible to nutrient loss. For example, horse activity has been shown to decrease soil aggregate stability (Beever & Herrick, 2006) and increase bare ground in riparian buffers (Boyd et al., 2017) both of which increase the chance of surface runoff and subsequent nutrient loading. Furthermore, elevated nutrient levels have been previously linked to horse fecal pollution from uncontrolled horse manure runoff (Parvage et al., 2015; Skelly, 2015). Even though horse activity has been shown to increase nutrient loading in other watersheds, our MST results indicate that concentrations of N and P in surface waters of Puerto Rico are not significantly impacted by the presence of horses alone. Instead, our MST in Figure 1 suggests that fecal pollution in the affected watersheds is due to human population densities. Therefore, nutrient pollution in these watersheds is the result of high human population density, and dogs and horses can be considered complementary sources that are derived from human settlement.
Leptospira detection in wet and dry season
The concurrent occurrence of fecal waste and urine contamination in environmental samples implies that surface water containing high nitrogen and phosphorous from fecal runoff may also contain higher concentrations of Leptospira that is carried in the urine of infected host (Bierque et al., 2020; Fouts et al., 2016). Another aim of this study was to determine the relationship among nutrient levels, MST of human, horse and dog fecal contamination, and qPCR detection of pathogenic Leptospira in SJBE. Water samples were screened using the Lipl32 gene qPCR assay that was optimized for environmental samples (Riediger et al., 2016; Truitt et al., 2020). During June sampling event, L. interrogans were detected in approximately 32% of the sites with an average gene copy number of 400 leptospires per 100 ml. (Table 2). This low concentration of L. interrogans may still pose a public health risk since reports suggest that a low dose of L. interrogans is infectious to humans and these bacteria can remain infectious for up to 20 months in water (Adesiyun et al., 2011; Hochedez et al., 2013). In addition, the DNA extraction protocol used recovers approximately 42% of bacterial DNA which implies that the actual concentration of leptospires in the water could be greater than the detected value (Morrison et al., 2008). These trends in the detection of L. interrogans are comparable to those in another Caribbean Island (St. Kitts) and rural Puerto Rico (Rawlins et al., 2014; Truitt et al., 2020). It was not surprising that L. interrogans were detected following rainfall in June because most studies have reported an increase in the detection of L. interrogans in freshwater environments following rain events (Keenum et al., 2021; Riediger et al., 2016; Truitt et al., 2020).
The molecular source tracking of humans, horses, and dogs was used to link the presence of L. interrogans in sampling sites to a possible source of fecal pollution in watersheds that flow into the SJBE. Unfortunately, an MST assay for rodents, which are a major reservoir of Leptospira, was not available during this study (Casanovas-Massana et al., 2018). Urban development in the San Juan region will attract rodents, which are the most common animal to shed L. interrogans in their urine (Casanovas-Massana et al., 2018). Molecular source tracking for human fecal contamination using the HF183-1 marker (Green et al., 2014) indicated a significant correlation between human fecal contamination and pathogenic L. interrogans (r = 0.79) during the June sampling event. There was also a positive correlation (r = 0.70) between dog fecal contamination and L. interrogans in the June samples which is likely due to the large population of stray dogs in Puerto Rico (The Sato Project, 2021) that could be potential host for L. interrogans and easily transmit it to waterways. In contrast, there was no meaningful relationship between horse fecal pollution and L. interrogans in the sampled watersheds (r = 0.02).
Conclusions
-
The SJBE and Rio Grande de Loiza watersheds are impacted by elevated levels of TN and TP due to human activity.
-
Leptospira was only present in the June samples and was linked to human and dog fecal pollution.
-
The combined use of nutrient measurements, MST, and the qPCR assay for the Lipl32 gene and the source of the pathogenic Leptospira and nutrient contamination in environmental samples can be determined.
-
Further studies need to be conducted to examine other seasonal changes in the area regarding L. interrogans presence.
-
There is a need for mitigation efforts towards public health concerns and environmental impacts regarding nutrient concentrations and fecal pollution in SJBE.
Data availability
The data set used is included in this manuscript and is available from the corresponding author on reasonable request.
References
Adesiyun, A. A., Baboolal, S., Suepaul, S., Dookeran, S., & Stewart-Johnson, A. (2011). Human Leptospirosis in the Caribbean, 1997-2005: Characteristics and serotyping of clinical samples from 14 countries. Pan American Journal of Public Health, 29, 350–357.
Bachoon, D. S., Miller, C. M., Green, C. P., & Otero, E. (2010). Comparison of four polymerase chain reaction methods for the rapid detection of human fecal pollution in marine and inland waters. International Journal of Microbiology, 2010, 1–7. https://doi.org/10.1155/2010/595692
Barragan, V., Olivas, S., Keim, P., & Pearson, T. (2017). Critical knowledge gaps in our understanding of environmental cycling and transmission of Leptospira spp. Applied and Environmental Microbiology, 83. https://doi.org/10.1128/AEM.01190-17
Beever, E. A., & Herrick, J. E. (2006). Effects of feral horses in great basin landscapes on soils and ants: Direct and indirect mechanisms. Journal of Arid Environments, 66, 96–112. https://doi.org/10.1016/j.jaridenv.2005.11.006
Benavidez, K. M., Guerra, T., Torres, M., Rodriguez, D., Veech, J. A., Hahn, D., Miller, R. J., Soltero, F. V., Ramírez, A. E. P., León, A. P., & de Castro-Arellano, I. (2019). The prevalence of Leptospira among invasive small mammals on Puerto Rican cattle farms. PLOS Neglected Tropical Diseases, 13, e0007236. https://doi.org/10.1371/journal.pntd.0007236
Bierque, E., Soupé-Gilbert, M.-E., Thibeaux, R., Girault, D., Guentas, L., & Goarant, C. (2020). Leptospira interrogans retains direct virulence after long starvation in water. Current Microbiology, 77, 3035–3043. https://doi.org/10.1007/s00284-020-02128-7
Boehm, A. B., van De Werfhorst, L. C., Griffith, J. F., Holden, P. A., Jay, J. A., Shanks, O. C., Wang, D., & Weisberg, S. B. (2013). Performance of forty-one microbial source tracking methods: A twenty-seven lab evaluation study. Water Research, 47(18), 6812–6828.
Boyd, C. S., Davies, K. W., & Collins, G. H. (2017). Impacts of feral horse use on herbaceous riparian vegetation within a sagebrush steppe ecosystem. Rangeland Ecology & Management, 70, 411–417. https://doi.org/10.1016/j.rama.2017.02.001
Brandeis, T., Escobedo, Francisco J., Staudhammer, Christina L., Nowak, David J., Zipperer WC. (2014). San Juan bay estuary watershed urban forest inventory. Gen. Tech. Rep. SRS-GTR-190. Asheville, NC: USDA-Forest Service, Southern Research Station. 44 p. https://doi.org/10.2737/SRS-GTR-190
Bridgemohan, R. S. H., Bachoon, D. S., Wang, Y., Bridgemohan, P., Mutiti, C., & Ramsubhag, A. (2020). Identifying the primary sources of fecal contamination along the beaches and rivers of Trinidad. Journal of Water and Health, 18, 229–238. https://doi.org/10.2166/wh.2020.047
Briskin, E. A., Casanovas-Massana, A., Ryff, K. R., Morales-Estrada, S., Hamond, C., Perez-Rodriguez, N. M., Benavidez, K. M., Weinberger, D. M., Castro-Arellano, I., Wunder, E. A., Sharp, T. M., Rivera-Garcia, B., & Ko, A. I. (2019). Seroprevalence, risk factors, and rodent reservoirs of Leptospirosis in an urban community of Puerto Rico, (2015). The Journal of Infectious Diseases, 220, 1489–1497. https://doi.org/10.1093/infdis/jiz339
Carlson, R. E. (1977). A Trophic state index for lakes. Limnology and Oceanography, 22(2), 361–369.
Casanovas-Massana, A., Costa, F., Riediger, I. N., Cunha, M., Oliveira, D., de Mota, D. C., Sousa, E., Querino, V. A., Nery, N., Reis, M. G., Wunder, E. A., Diggle, P. J., & Ko, A. I. (2018). Spatial and temporal dynamics of pathogenic Leptospira in surface waters from the urban slum environment. Water Research, 130, 176–184. https://doi.org/10.1016/j.watres.2017.11.068
Colón, Y. M. S., & Schaffner, F. C. (2021). A case study of the effects of management interventions on phosphorus dynamics at a coastal, eutrophic, Caribbean lagoon (Laguna Cartagena, Puerto Rico). Water, 13, 449. https://doi.org/10.3390/w13040449
Conley, D. J., Paerl, H. W., Howarth, R. W., Boesch, D. F., Seitzinger, S. P., Havens, K. E., Lancelot, C., & Likens, G. E. (2009). Controlling eutrophication: Nitrogen and phosphorus. Science, 323, 1014–1015. https://doi.org/10.1126/science.1167755
Correll, D. L. (1999). Phosphorus: A rate limiting nutrient in surface waters. Poultry Science, 78, 674–682. https://doi.org/10.1093/ps/78.5.674
Di Rienzo, J. A., Casanoves, F., Balzarini, M. G., Gonzalez, L., Tablada, M., & Robledo, C. W. (2012). InfoStat. versión 2011. In Grupo InfoStat, FCA. Universidad Nacional de Córdoba http://www.infostat.com.ar
Dick, L. K., Bernhard, A. E., Brodeur, T. J., Domingo, J. W. S., Simpson, J. M., Walters, S. P., & Field, K. G. (2005). Host distributions of uncultivated fecal Bacteroidales bacteria reveal genetic markers for fecal source identification. Applied and Environmental Microbiology, 71, 3184–3191. https://doi.org/10.1128/AEM.71.6.3184-3191.2005
Dick, L. K., Stelzer, E. A., Bertke, E. E., Fong, D. L., & Stoeckel, D. M. (2010). Relative decay of Bacteroidales microbial source tracking markers and cultivated Escherichia coli in freshwater microcosms. Applied and Environmental Microbiology, 76, 3255–3262. https://doi.org/10.1128/AEM.02636-09
Fouts, D. E., Matthias, M. A., Adhikarla, H., Adler, B., Amorim-Santos, L., Berg, D. E., Bulach, D., Buschiazzo, A., Chang, Y.-F., Galloway, R. L., Haake, D. A., Haft, D. H., Hartskeerl, R., Ko, A. I., Levett, P. N., Matsunaga, J., Mechaly, A. E., Monk, J. M., Nascimento, A. L. T., et al. (2016). What makes a bacterial species pathogenic: Comparative genomic analysis of the genus Leptospira. PLOS Neglected Tropical Diseases, 10, e0004403. https://doi.org/10.1371/journal.pntd.0004403
Fretwell, J. D., Williams, J., & Redman, P. (1996). National water summary on wetland resources. US Government Printing Office. https://doi.org/10.3133/wsp2425
Galloway, J. N., Townsend, A. R., Erisman, J. W., Bekunda, M., Cai, Z., Freney, J. R., Martinelli, L. A., Seitzinger, S. P., & Sutton, M. A. (2008). Transformation of the nitrogen cycle: Recent trends, questions, and potential solutions. Science, 320, 889–892. https://doi.org/10.1126/science.1136674
González-Fernández, A., Symonds, E. M., Gallard-Gongora, J. F., Mull, B., Lukasik, J. O., Navarro, P. R., Aguilar, A. B., Peraud, J., Brown, M. L., Alvarado, D. M., Breitbart, M., Cairns, M. R., & Harwood, V. J. (2021). Relationships among microbial indicators of fecal pollution, microbial source tracking markers, and pathogens in Costa Rican coastal waters. Water Research, 188, 116507. https://doi.org/10.1016/j.watres.2020.116507
Green, H. C., Haugland, R. A., Varma, M., Millen, H. T., Borchardt, M. A., Field, K. G., Walters, W. A., Knight, R., Sivaganesan, M., Kelty, C. A., & Shanks, O. C. (2014). Improved HF183 quantitative real-time PCR assay for characterization of human fecal pollution in ambient surface water samples. Applied and Environmental Microbiology, 80, 3086–3094. https://doi.org/10.1128/AEM.04137-13
Hochedez, P., Escher, M., Decoussy, H., Pasgrimaud, L., Martinez, R., Rosine, J., Théodose, R., Bourhy, P., Picardeau, M., Olive, C., Ledrans, M., & Cabié, A. (2013). Outbreak of Leptospirosis among canyoning participants, Martinique, 2011. Eurosurveillance, 18. https://doi.org/10.2807/ese.18.18.20472-en
Holman, C. B., Bachoon, D. S., Otero, E., & Ramsubhag, A. (2014). Detection of Helicobacter pylori in the coastal waters of Georgia, Puerto Rico, and Trinidad. Marine Pollution Bulletin, 79, 354–358. https://doi.org/10.1016/j.marpolbul.2013.11.021
Keenum, I., Medina, M. C., Garner, E., Pieper, K. J., Blair, M. F., Milligan, E., Pruden, A., Ramirez-Toro, G., & Rhoads, W. J. (2021). Source-to-tap assessment of microbiological water quality in small rural drinking water systems in Puerto Rico six months after hurricane Maria. Environmental Science & Technology, 55, 3775–3785. https://doi.org/10.1021/acs.est.0c08814
Kildare, B. J., Leutenegger, C. M., McSwain, B. S., Bambic, D. G., Rajal, V. B., & Wuertz, S. (2007). 16S rRNA-based Assays for quantitative detection of universal, human-, cow-, and dog-specific fecal Bacteroidales: A Bayesian approach. Water Research, 41, 3701–3715. https://doi.org/10.1016/j.watres.2007.06.037
Larsen, M. C., & Webb, R. M. T. (2009). Potential effects of runoff, fluvial sediment, and nutrient discharges on the coral reefs of Puerto Rico. Journal of Coastal Research, 251, 189–208. https://doi.org/10.2112/07-0920.1
Levett, P. N., Branch, S. L., Whittington, C. U., Edwards, C. N., & Paxton, H. (2001). Two methods for rapid serological diagnosis of acute Leptospirosis. Clinical Diagnostic Laboratory Immunology, 8, 349–351. https://doi.org/10.1128/CDLI.8.2.349-351.2001
Levett, P. N., Morey, R. E., Galloway, R. L., Turner, D. E., Steigerwalt, A. G., & Mayer, L. W. (2005). Detection of pathogenic leptospires by real-time quantitative PCR. Journal of Medical Microbiology, 54, 45–49. https://doi.org/10.1099/jmm.0.45860-0
Martin, A., & Gruber, S. (2005). Amplification of indicator bacteria in organic debris on southern California beaches. Weston Solutions. StormCon® 2005 July 18-21, 2005 JW Marriott Grande Lakes Orlando, FL, USA
Mesa, L., Mayora, G., Saigo, M., & Giri, F. (2015). Nutrient dynamics in wetlands of the middle Paraná river subjected to rotational cattle management. Wetlands, 35, 1117–1125. https://doi.org/10.1007/s13157-015-0699-2
Morrison, C., Bachoon, D., & Gates, K. (2008). Quantification of enterococci and Bifidobacteria in Georgia estuaries using conventional and molecular methods. Water Research, 42, 4001–4009.
Ngatia, L., Grace, J., Moriasi, D., & Taylor, R. (2019). Nitrogen and phosphorus eutrophication in marine ecosystems. Monitoring of Marine Pollution, 1, 1–17. https://doi.org/10.5772/intechopen.81869
Pappas, G., Papadimitriou, P., Siozopoulou, V., Christou, L., & Akritidis, N. (2008). The globalization of Leptospirosis: Worldwide incidence trends. International Journal of Infectious Diseases, 12, 351–357. https://doi.org/10.1016/j.ijid.2007.09.011
Parvage, M. M., Ulén, B., & Kirchmann, H. (2015). Are horse paddocks threatening water quality through excess loading of nutrients? Journal of Environmental Management, 147, 306–313. https://doi.org/10.1016/j.jenvman.2014.09.019
Pratt, N., & Rajeev, S. (2018). Leptospira seroprevalence in animals in the Caribbean region: A systematic review. Acta Tropica, 182, 34–42. https://doi.org/10.1016/j.actatropica.2018.02.011
Rawlins, J., Portanova, A., Zuckerman, I., Loftis, A., Ceccato, P., Willingham, A., & Verma, A. (2014). Molecular detection of Leptospiral DNA in environmental water on St. Kitts. International Journal of Environmental Research and Public Health, 11, 7953–7960. https://doi.org/10.3390/ijerph110807953
Riediger, I. N., Hoffmaster, A. R., Casanovas-Massana, A., Biondo, A. W., Ko, A. I., & Stoddard, R. A. (2016). An optimized method for quantification of pathogenic Leptospira in environmental water samples. PLOS ONE, 11, e0160523. https://doi.org/10.1371/journal.pone.0160523
Schriewer, A., Goodwin, K. D., Sinigalliano, C. D., Cox, A. M., Wanless, D., Bartkowiak, J., et al. (2013). Performance evaluation of canine-associated Bacteroidales assays in a multi-laboratory comparison study. Water Research, 47(18), 6909–6920.
Skelly, C. (2015). One horse or a hundred: Manure and water don’t mix. Michigan State University Extension. October 23, 2015.
Stoddard, R. A., Gee, J. E., Wilkins, P. P., McCaustland, K., & Hoffmaster, A. R. (2009). Detection of pathogenic Leptospira spp. through TaqMan polymerase chain reaction targeting the Lipl32 gene. Diagnostic Microbiology and Infectious Disease, 64, 247–255. https://doi.org/10.1016/j.diagmicrobio.2009.03.014
Sutter, J.D., & Sosa Pascual, O. (2018). Deaths from bacterial disease in Puerto Rico spiked after Maria. Advisory Board, Daily Briefing. July 10, 2018. The Sato Project, 2021. https://thesatoproject.org/. Accessed May 04, 2021.
Truitt, Z. G., Poon-Kwong, B., Bachoon, D. S., & Otero, E. (2020). Seasonal shifts in the presence of pathogenic Leptospires, Escherichia coli, and physicochemical properties in coastal rivers and streams of Puerto Rico. Journal of Environmental Quality, 49, 1264–1272. https://doi.org/10.1002/jeq2.20091
USEPA, (2012). Recreational water quality criteria (No. 820- F-12–058). United States Environmental Protection Agency.
USEPA. (2019). Water quality standards regulations. Puerto Rico.
Vincent, A. T., Schiettekatte, O., Goarant, C., Neela, V. K., Bernet, E., Thibeaux, R., Ismail, N., Khalid, M. K. N. M., Amran, F., Masuzawa, T., Nakao, R., Korba, A. A., Bourhy, P., Veyrier, F. J., & Picardeau, M. (2019). Revisiting the taxonomy and evolution of pathogenicity of the genus Leptospira through the prism of genomics. PLOS Neglected Tropical Diseases, 13, e0007270. https://doi.org/10.1371/journal.pntd.0007270
Wade, C., Otero, E., Poon-Kwong, B., Rozier, R., & Bachoon, D. (2015). Detection of human-derived fecal contamination in Puerto Rico using Carbamazepine, HF183 Bacteroides, and fecal indicator bacteria. Marine Pollution Bulletin, 101, 872–877. https://doi.org/10.1016/j.marpolbul.2015.11.016
Walker, T. J., Bachoon, D. S., Otero, E., & Ramsubhag, A. (2013). Detection of verotoxin producing Escherichia coli in marine environments of the Caribbean. Marine Pollution Bulletin, 76, 406–410. https://doi.org/10.1016/j.marpolbul.2013.08.019
Wilde, F.D., D.B. Radtke, J. Gibs, &. Iwatsubo, R.T. (1998). National field manual for the collection of water-quality data. Techniques of Water-Resources Investigations, Book 9, Handbooks for water-resources investigations. United States Geological Survey, .
Wright, M. E., Solo-Gabriele, H. M., Elmir, S., & Fleming, L. E. (2009). Microbial load from animal feces at a recreational beach. Marine Pollution Bulletin, 58, 1649–1656. https://doi.org/10.1016/j.marpolbul.2009.07.003
Acknowledgements
We thank the researchers at the University of Puerto Rico for collecting and shipping the water filter samples.
Funding
This project was funded by the Corporation for the Conservation of the San Juan Bay Estuary,C72-250-03/UPRM-AES, and Georgia Collage and State University. Additional funding was provided by Georgia College and State University, Milledgeville, GA 31061, USA.
Author information
Authors and Affiliations
Contributions
Everyone contributed to the final version of this manuscript. D.S.B. and T.C. analyzed the data, conducted the PCR analysis, and wrote the main manuscript text. G.A.M. collected the samples and prepared the maps and figures and assisted with the manuscript editing and statistics. C.D.B. assisted with the proof reading, statistics, nutrient data analysis, and discussion. W.C.D. assisted with the sample preparation, PCR analysis, and tables.
Corresponding author
Ethics declarations
Ethics approval
All authors have read, understood, and have complied as applicable with the statement on “Ethical responsibilities of Authors” as found in the Instructions for Authors and are aware that with minor exceptions, no changes can be made to authorship once the paper is submitted. This is an observational study. The Research Ethics Committee has confirmed that no ethical approval is required.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Appendix
Appendix
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Chapman, T., Bachoon, D.S., Martinez, G.A. et al. Tracking the sources of Leptospira and nutrient flows in two urban watersheds of Puerto Rico. Environ Monit Assess 195, 1318 (2023). https://doi.org/10.1007/s10661-023-11948-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10661-023-11948-6