Abstract
Despite the number of studies on antibiotic-resistant enterococci from Serbian clinical settings, there are no data about environmental contamination with these bacteria. Thus, this study investigated the prevalence of antibiotic-resistant enterococci in Belgrade, Serbia. Enterococcus species collected from ten surface water sites, including a lake, two major river systems, and springs, were tested. Among enterococci, we found single (21.7 %), double (17.4 %), and multiple antibiotic resistance patterns (56.3 %). Vancomycin-resistant strains were not found, indicating that their abundance in Belgrade is tightly linked to clinical settings. The multiple drug-resistant strains Enterococcus faecalis, Enterococcus faecium, and Enterococcus mundtii were frequently detected in the lake during the swimming season and in the rivers near industrial zones. We confirmed the presence of ermB, ermC, ant(6)-Ia, tetM, and tetL and mutations in gyrA genes. The phylogenetic analysis of 16S rRNA gene of E. faecium isolates that harbor esp gene classified them into two groups based on high-bootstraps scores in the tree analysis. Pulsed-field gel electrophoresis analysis of antibiotic-resistant enterococci revealed genomic similarity ranging from 75 to 100 %. This study indicates the importance of anthropogenic impact to the spread of antibiotic-resistant enterococci in environmental waters of Belgrade, Serbia.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Enterococci are Gram-positive bacteria primarily associated with indigenous human and animal gastrointestinal flora (Leroy et al. 2003). Due to their ability to survive adverse conditions, enterococci are widely distributed in nature (Franz et al. 2003). Once released into the environment through human and animal feces, enterococci are able to inhabit different ecological niches such as soil, surface water, and various plants (Giraffa 2003). For this reason, they have received widespread interest as useful indicators of fecal pollution in aquatic ecosystems (Davis et al. 2005).
Their exceptional metabolic adaptability allows them to fulfill a variety of roles as commensals and opportunistic pathogens (Giménez–Pereira 2005). Under certain circumstances, they are able to cause a variety of infections in humans and are now recognized as a major cause of clinical infections with limited therapeutic options due to their great ability to acquire resistance to the most clinically relevant antimicrobial agents (Teixeira and Merquior 2013). As commensals of the human gastrointestinal tract, enterococci are routinely exposed to a variety of antibiotics due to medical treatments. The antibiotic resistance of enterococci plays an important role in the ecological dynamics during and after antibiotic therapy (Kristich et al. 2014). The widespread release of antibiotics into the environment as a result of their use in human and animal medicine has led to an increased spread of resistant enterococci in soil, water, and food (Cabello 2006).
Enterococci are not indigenous to water and their prevalence in water sources depends on environmental factors including the geographical area, potential sources and types of sewage, and the presence of industrial zones and agricultural sites (Sidhu et al. 2014). Detailed studies of individual locations are necessary in order to understand the distribution of Enterococcus species and provide useful information about the dissemination of resistance genes into surface water (Di Cesare et al. 2014; Niederhausern et al. 2013).
Contaminated surface water can become a reservoir of virulent and antibiotic-resistant strains of fecal bacteria, including enterococci species (Di Cesare et al. 2012), thus contributing to the dissemination of resistance genes into surface water by horizontal gene transfer mechanisms (Di Cesare et al. 2014).
Serbia was, for decades, among the countries where the use, over-use, misuse, and abuse of antibiotics were high due to poor regulation, until recently (http://www.who.int/drugresistance/documents/situationanalysis/en/). However, the years of lax restriction and enforcement together with the fact that antibiotics were sold in pharmacies to anyone who could afford them could and probably did lead to a higher incidence of antibiotic-resistant strains, including enterococci. Vancomycin-resistant enterococci (VRE) are designated as pathogens of great importance in Serbian hospitals (Mioljevic et al. 2013; Stosovic et al. 2004). Although most VRE infections are nosocomial, environmental contamination with VRE is of special interest since enterococci are ubiquitously detected in aquatic environments where they are able to survive long period of time (Nishiyama et al. 2015). Accordingly, we analyzed surface water samples from the Belgrade area in which hospitals VRE were previously described (Mioljevic et al. 2013; Stosovic et al. 2004) in order to determine whether the environment pool of multiresistant enterococci and VRE.
The analyzed waters included three springs (Brace Jerkovic, Hajducka Cesma, and Sakinac) where the human impact is minimal, three rivers (the Topcider River and the major rivers Danube and Sava) and a lake (Ada Ciganlija Lake) where anthropogenic influence is of particular importance, especially during the swimming season. Many of these waters are used to supply water for domestic, recreational, industrial, and agricultural uses. To evaluate the incidence and the prevalence of antibiotic-resistant enterococci from three different types of surface water in the Belgrade area that may represent a transmission route for resistant genes to healthy organisms, both culture-dependent and molecular tools were used.
Material and methods
Localities and sampling strategy
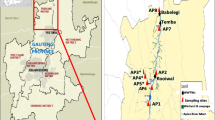
Sampling from surface waters in the area of Belgrade, Serbia was performed from July to October 2013. Water samples were collected as follows: four samples from the localities situated across the major river systems in Belgrade [the Danube-Dorcol (DDE) and Sava (RSE) rivers, the Topcider River (TRE), and the Danube River-Ada Huja (AHE)], three samples from the lake at Ada Ciganlija [before (AJE), during (AAE) and after (ASE) the swimming season], and three samples from springs [Brace Jerkovic (BJE), Hajducka Cesma (CHE), and Sakinac (SACE)] (Fig 1). The Danube River-Ada Huja (AHE) is located in an industrial zone of Belgrade City. Ada Ciganlija Lake is the most famous beach and recreation centre in Belgrade with up to 150,000 visitors daily during the swimming season. The springs are distant from the urban area, surrounded by vegetation.
Locations of the sampling sites in the Belgrade area. The TRE, RSE, DDE, and AHE river water systems are indicated by circles; CHE, BJE, and SACE springs are indicated by triangles; and water samples from the lake, AJE (before swimming season), AAE (swimming season), ASE (after swimming season), are indicated by squares. The map was taken from https://www.google.rs/maps/@44.7989863,20.4441833,12637m/data=!3m1!1e3?hl=sr
Isolation and identification of enterococci
Isolation of enterococci from water samples was performed using a membrane filtration technique. Water samples (500 mL) were collected in sterile bottles approximately 50 cm below the water’s surface, and concentrated using 0.45 μm membrane filters (Sarstedt, USA). Enrichment of samples was performed as described previously with minor modifications (Messer and Dufour 1997). The filters with precipitate were incubated in sterile Petri plates with M17 medium supplemented with 0.5 % (w/v) glucose (GM17), sodium azide (0.15 g/L), and cycloheximide (10 μL/mL). The samples were grown up to 7 days at 37 °C. Bacteria grown on filter surface were collected and serial dilutions (10−1 to 10−7) of each sample were prepared and 100 μL of each dilution was plated on GM17 agar plates and incubated for 48 h at 37 °C in aerobic conditions. Upon cultivation, the colonies with typical enterococcal morphology were selected and isolates were identified to the genus level by Gram staining, catalase testing, and black zone formation on bile esculin agar (Himedia, Mumbai, India). Species identification was carried out by polymerase chain reaction (PCR) using primers for amplification 16S rRNA, UNI16SF (5′-GAGAGTTTGATCCTGGC 3′) and UNI16SR (5′-AGG AGGTGATCCAGCCG-3′; Jovcic et al. 2009), and for sodA gene degenerate primers d1 (5′-CCITAYICITAYGAYGCIYTIGARCC-3′) and d2 (5′-ARRTARTAIGCRTGYTCCCAIACRTC-3′; Poyart et al. 2000). The PCR amplicons obtained were purified using kit (Thermo Scientific, Lithuania) and sequenced (Macrogen, Amsterdam, the Netherlands). The BLAST algorithm was used to determine the most related sequence relatives in the NCBI nucleotide sequence database (http://www.ncbi.nlm.nih.gov/ BLAST).
Phylogenetic analysis
The phylogenetic inferences were generated by MEGA version 6.0. (Tamura et al. 2013). The multiple DNA sequence alignments were performed using Vector NTI program that uses the ClustalW algorithm with default parameters. The construction of an Enterococcus faecium16S rRNA phylogenetic tree was conducted by the maximum-likelihood (ML) method using a Jones–Taylor–Thornton (JTT) model. The confidence levels of ML trees were obtained by bootstrapping of 1,000 replicates. Eighteen E. faecium strains, harbouring the esp gene, were included in this analysis.
Antimicrobial susceptibility
Antibiotic susceptibility was tested for 16 antibiotics of interest in animal and human medicine (microgram per disk): penicillin (PEN,10), vancomycin (VAN, 30), teicoplanin (TEI, 30), ampicillin (AMP, 10), streptomycin (STR, 300), gentamicin (GEN, 120), chloramphenicol (CHL, 30), tetracycline (TET, 30), erythromycin (ERY, 15), quinupristin–dalfopristin (QD, 15), ciprofloxacin, (CIP, 5), levofloxacin (LVX, 5), nitrofurantoin (FUR, 300), rifampicin (RFM, 5), linezolid (LIN, 30), and bacitracin (BAC, 130), by the disk diffusion method (Clinical and Laboratory Standards Institute 2013). Antibiotic disks were obtained from BioRad, Mernes-la-Coquette, France. The diameters of antibiotics inhibition zones were measured and recorded as susceptible (S), intermediate (I), or resistant (R).
Enterococci isolates showing intermediate or resistant phenotypes were submitted to further tests. Determination of the minimal inhibitory concentration (MIC) was performed by microdilution tests in Mueller Hinton broth (Oxoid, Hampshire, UK). Appropriate cell culture was added in wells of the microtiter plate containing increasing concentrations of antibiotics dissolved in 180 μL MH medium. Cell density was monitored by OD600 measurements after 24 h of incubation at 37 °C in a microtiter plate reader (Multiscan RC, Labsystems, UK). The lowest concentration of antibiotic at which no growth of bacteria was detected was taken as the minimum inhibitory concentration. Experiments were done in triplicate.
PCR detection of resistance genes
The presence of antibiotic resistance genes was tested by PCR in all enterococcal isolates which showed resistance to the tested antibiotics. The primer sequences of the target genes, the expected amplicon sizes, and annealing temperatures (Th) are given in Table 1. PCR conditions were as previously described (references are given in Table 1). The PCR reaction mixture (50 μL) contained: 5 ng of bacterial DNA, 1× reaction buffer (Kapabiosystems, USA), 200 μM of each dNTPs, 10 pmol of the primer/s, and 1 U Tag DNA polymerase (Kapabiosystems, USA, 5 U/μL). Gene sequencing and analysis was described previously. The sequences of antibiotic resistance genes were submitted to the European Nucleotide Archive (http://www.ebi.ac.uk/ena/data/view/LN611411-LN611416) under accession No. LN611411-LN611416.
Pulsed-field gel electrophoresis
To investigate genetic relatedness among resistant enterococci strains, pulsed-field gel electrophoresis (PFGE) was carried out as previously described (Kojic et al. 2005). Briefly, PFGE with SmaI-digested genomic DNA was performed for 18 h at 300 V at 9 °C using a 2015 Pulsafor unit apparatus (LKB Instruments, Bromma, Sweden). Grouping of isolates based on the PFGE results was done using SPSS software package, version 20 (IBM Inc., Chicago, IL).
Statistical analysis
Classical ecology indexes were used to obtain the richness (S), the biodiversity (H′), and the dominance (D) of the species studied:
-
Species richness (S): this is the simplest measurement of diversity, being defined as the number of the species found in a defined area.
-
Shannon–Wiener index (H′): used to obtain the general biodiversity:
$$ H^{\prime } = - {\varSigma}^S{p}_ilo{g}_2(pi) $$where S is the number of species and p i is the proportion of thesample belonging to its species. Simpson’s index (D), which gives a strong weighting to the dominant species:
$$ D = {\varSigma}^S{\left({p}_i\right)}^2 $$where S is the number of species and p i is the proportion of the sample belonging to its species.
Results and discussion
Isolation and identification of Enterococcus spp.
Several studies have focused on enterococci isolates from surface water (Sidhu et al. 2014; Michael et al. 2013) and from the environment to achieve a greater understanding of the origin, spread, and persistence of antibiotic-resistant enterococci. However, data about antibiotic-resistant enterococci from Balkan Peninsula surface waters are still unavailable. To our knowledge, this is the first study carried out on water from the Belgrade area, despite that this area includes major rivers, namely the Danube River and the Sava River.
In order to examine the presence of enterococcal species and the distribution of antibiotic resistance genes among them, a total of 124 lactic acid bacteria were isolated from nine out of ten localities samples of surface water in the Belgrade area (Fig. 1). A total of 91 enterococcal isolates were chosen for further investigation based on growing on selective media, Gram staining, and negative results obtained after catalase tests.
After the sequencing of sodA genes and gene for the 16S rRNA, we found that isolated enterococci belong to ten distinct species with the following distribution: E. faecium (38.5 %), Enterococcus faecalis (20.9 %), Enterococcus durans (16.5 %), Enterococcus mundtii (6.6 %), Enterococcus hirae (6.6 %), Enterococcus thailandicus/sanquinicola/lactis (5.5 %), Enterococcus rivorum (2.2 %), Enterococcus casseliflavus (1.1 %), Enterococcus lactis (1.1 %), and Enterococcus gallinarum (1.1 %). Specimen tags, locations, and enterococci isolates are indicated in Table 2.
The analysis of species distribution indicated that E. faecium and E. faecalis were predominant in all types of surface water except the springs (Fig. 2). E. hirae was found in the rivers, the Topcider (66.7 %) and the Danube River-Dorcol (16.7 %), as well as the Danube River-Ada Huja (21.4 %). The species E. lactis (3.1 %) and E. casseliflavus (3.1 %) were present only in Ada Ciganlija Lake during the swimming season, while E. thailandicus/sanquinicola/lactis (26.3 %) and E. rivorum (10.5 %) were found only in Ada Ciganlija Lake after swimming season. Additionally, E. durans was present before (100 %) and after (15.8 %) swimming season at Ada Ciganlija Lake. The species E. mundtii was isolated from Brace Jerkovic spring (100 %), the Sava River (33.3 %), and Ada Ciganlija Lake (12.5 %) during swimming season. The species E. gallinarum was found only in the spring Hajducka Cesma (100 %). Regarding the enterococci richness (S), ten different species were isolated in all water samples (Fig. 2). The general index of biodiversity (H′) and the concentration of dominance (D) were calculated on the basis of the number of identified species by 16S rRNA sequencing. The highest diversity of enterococci was found in Ada Ciganlija Lake after swimming season (H′ = 2.14) where no fewer than five different enterococci species were identified (E. faecalis, E. faecium, E. durans, E. rivorum, and E.thailandicus/sanquinicola/lactis), and no dominance of any of species was scored (D = 0.25). Also, the highest biodiversity of enterococci (H1′ = 1.50), and the lowest concentration of dominance (D = 0.47) was found in Ada Ciganlija Lake during swimming season. On the contrary, the Ada Ciganlija Lake before swimming season and both springs (Hajducka Cesma and Brace Jerkovic) exhibited the lowest Shannon’s index (H2′ = 0) and the highest dominance index (D = 1). No enterococci were isolated from Sakinac spring (Fig. 2).
Relative abundance (%) of enterococci species among distinct locations in the Belgrade area. Diversity indices of species richness (S), Shannon–Wiener index (H′) indicating general biodiversity, and Simpson's index (D) for evaluating dominance are presented for enterococci for each locations of the sampling sites. Sites were marked as follows: 1 Ada Ciganlija Lake before swimming season, 2 Ada Ciganlija Lake during swimming season, 3 Ada Ciganlija Lake after swimming season, 4 Spring Hajducka Cesma, 5 Spring Brace Jerkovic, 6 Topcider River, 7 Sava River, 8 Danube River-Dorcol, and 9 Danube River-Ada Huja
This species distribution in the Belgrade area surface waters, at all locations, was in agreement with previously reported results (Moore et al. 2008; Švec and Sedlaček 1999) where the prevalence of E. faecalis and E. faecium species was evident, especially in waters where the influence of humans was the most significant. In addition, it was observed that in rivers, enterococci were less prevalent than in Ada Ciganlija Lake. Even though higher number of enterococci species was scored in rivers near industrial zone, the Danube River-Ada Huja (14) and the Danube River-Dorcol (6), than in the Topcider River near Kosutnjak forest (3) and in the Sava River (3). Finally, the springs had the lowest number of species scored: Brace Jerkovic (1), Hajducka Cesma (1), and Sakinac spring (0) that could be related to the lowest anthropogenic impact. Most importantly, in the water from Brace Jerkovic spring, only E. mundtii species, usually found on plants (Aarestrup and Hasman 2004), was isolated, indicating that another possible source of enterococci in water could be soil and vegetation (Sidhu et al. 2014). Interestingly, the number of isolates is consistent with the distribution of species; the highest diversity (H′ = 2.14) was observed in the recreational lake while the lowest diversity (H′ = 0) was found in the springs.
Phylogenetic analysis of E. faecium species harboring esp gene
The presence of enterococci is considered an indicator of faecal contamination of environmental water sources, reflecting the human health risk of drinking and recreational waters (WHO 2004). The increased number of enterococci during and after the swimming season could be attributed to anthropogenic influences. The enterococcal surface protein gene, esp, is a major putative pathogenicity marker in clinical isolates of E. faecium and E. faecalis. This gene can be exchanged between enterococcal strains by conjugation (Oancea et al. 2004). E. faecalis Esp has been implicated as a contributing factor in colonization and persistence of infection within the urinary tract (Shankar et al. 1999, 2001). An esp homologue has been identified in E. faecium (Eaton and Gasson 2001). Since previous studies reported that the esp gene may be a useful marker for Enterococcus originating from human feces (Kim et al. 2010; Scott et al. 2005), we evaluated the presence of esp gene in E. faecium strains isolated from surface water (Table 2). Interestingly, the results revealed that 51.42 % of tested E. faecium strains, originating from Ada Ciganlija Lake during and after swimming season, the Danube River-Dorcol and the Danube River-Ada Huja, were positive for the esp gene thus indicating that human faecal contamination is one of the major causes of pollution.
The diversity among 18 E. faecium strains, harboring the esp gene, was studied using 16S rRNA gene sequences. Maximum likelihood (ML) 16S rRNA tree (Fig. 3) classified E. faecium isolates into two distinct groups, A and B. The strains in both groups were clustered at high bootstrap (BT) value (100 %). The strains of group A includes nine strains isolated from the major river systems in Belgrade, the Danube River-Dorcol (DDE), the Danube River-Ada Huja (AHE), and one strain, ASE2-5, from the Ada Ciganlija Lake, after the swimming season (Fig. 3). Group B was a cluster of eight E. faecium strains isolated from the Ada Ciganlija Lake (AAE) during the swimming season.
Phylogenetic inferences of 16S rRNA gene among E. faecium strains. A phylogenetic tree of 16S rRNA gene was constructed with the maximum likelihood (ML) method using a Jones–Taylor–Thornton (JTT) model distance matrix. The percentages of 1,000 bootstrap samplings supporting the clusters are indicated at bifurcations (only bootstrap results above 50 % are reported). The isolates from the major river systems Danube River–Dorcol (DDE) and the Danube River–Ada Huja (AHE) are represented by black and white squares, respectively. The isolates from Ada Ciganlija Lake after the swimming season and during the swimming season are represented by white and black triangles, respectively.
Although all studied E. faecium strains originating from human feces, ML phylogenetic analysis suggested the possibility that they come from various sources of pollution, since they are clearly separated into two distinct groups. Human fecal pollution of rivers from which were isolated strains from a group A may occur from failing septic systems, leachate from dumps and landfills, or from improper disposal of sewage. Also, water from rain or snowmelt can transfer pathogens released by poorly maintained septic systems and discharge them into a sewage system or directly to a lake, stream or river. In contrast to the river pollution, the source of Ada Ciganlija Lake pollution, which is a public beach whose water quality is checked regularly, probably could be attributed to direct anthropogenic influences during and after the swimming season.
Antibiotic susceptibility testing
Apart from being indicator organisms, enterococci are important nosocomial pathogens resistant to many commonly used antibiotics. Several studies have shown a strong and direct relationship between enterococci and an increased number of swimmers suffering from gastrointestinal diseases (Boehm and Soller 2011).
An antibiotic susceptibility test, using a disk diffusion method, enabled rapid screening of resistance to 16 antibiotics among a total of 91 enterococcal isolates. The prevalence of antibiotic resistance is shown in Table 3.
The highest percentage of multiple resistance was found among the isolates of E. faecalis, E. faecium, and E. mundtii, which were resistant to 13 distinct antibiotics, in total (Table 3). It was found that all enterococcal isolates were sensitive to teicoplanin, while the majority of the isolates (over 98 %) were sensitive to tetracycline, and vancomycin. As mentioned, the isolates were susceptible to chloramphenicol. The isolates of E. faecalis (72 %) sampled from Ada Ciganlija Lake after swimming season had intermediate resistance to vancomycin. All isolates of E. thailandicus/sanquinicola/lactis and E. rivorum species exhibited a resistance to seven antibiotics (Bac, Cip, Ery, Gen, Lvx, QD, and Rfa). The isolates of E. casseliflavus, E. gallinarum, E. hirae, and E. lactis showed resistance to almost the same group of antibiotics (see Table 3). Interestingly, E. durans were resistant only to rifampicin and intermediately resistant to ciprofloxacin, nitrofurantoin, gentamicin, and quinupristin–dalfopristin (Table 3).
Determination of minimum inhibitory concentration (MIC)
A total of 67 enterococci were chosen, based on the preliminary results obtained by the disk diffusion assay, to determine their MIC values by microdilution assay. All strains that were found to be resistant to penicillin, ampicillin, linezolid, nitrofurantoin, vancomycin, and chloramphenicol in the disc diffusion assay were sensitive to those antibiotics in the microdilution assay. In contrast, the results of the microdilution assay were in accordance with the disk diffusion assay for enterococci resistant to bacitracin, rifampicin, levofloxacin, erythromycin and tetracycline.
Among aforementioned isolates, we found strains with single, double, and multiple antibiotic resistance patterns. The isolates resistant to one antibiotic were present at 21.7 %, on two antibiotics 17.4 %, three 4.3 %, four 13 %, five 17.4 %, six 13 %, seven 4.3 %, or eight 4.3 %. E. thailandicus/sanquinicola/lactis and E. durans species isolated from Ada Ciganlija Lake after the swimming season harbored resistance to bacitracin–rifampicin–erythromycin–levofloxacin, which represented the core of the most frequent multiple drug resistance pattern. The E. faecalis isolates collected from the Danube River-Dorcol showed resistance to all tested antibiotics (Table 4). These isolates exhibited a high-level resistant phenotype to streptomycin (≥2,000 μg/mL), gentamycin (≤2,000 μg/mL), bacitracin (≥256 μg/mL), erythromycin (256 μg/mL), tetracycline (256 μg/mL), levofloxacin (64 μg/mL), rifampicin (16 μg/mL), and ciprofloxacin (16 μg/mL). Also, the E. faecium strains isolated from the Danube River-Ada Huja, the Sava River, and the Danube River-Dorcol near locations belonging to industrial zones and E. faecalis, E. faecium, and E. mundtii isolated from Ada Ciganlija Lake during the swimming season,when the number of visitors is the highest, harbor resistance to between five and eight antibiotics (Table 4). These results indicate the high anthropogenic impact on the spread of drug-resistant strains. Single resistance was observed for erythromycin-resistant E. faecium isolated from Ada Ciganlija Lake after the swimming season and E. hirae isolated from the Topcider River and the Danube River-Ada Huja (Table 4). E. durans isolated from Ada Ciganlija Lake before the swimming season and E. hirae isolated from the Danube River-Dorcol carried single resistance to rifampicin (Table 4). In addition, E. lactis was sensitive to all tested antibiotics in the microdilution assay.
Detection of resistance genes by PCR
The 67 antibiotic-resistant enterococcal isolates were screened for the presence of the resistance genes most frequently reported in enterococci as follows: erythromycin resistance (ermA, ermB, ermC, mef, and msr), tetracycline resistance (tetM, tetL, and RPP), aminoglycoside resistance (ant(6)-Ia and acc(6')-aph(2')), and beta-lactam resistance (blaZ). Also, point mutations associated with fluoroquinolone resistance (within gyrA and parC) and beta-lactams (pbp5) were analyzed.
Genetic determiners for erythromycin (84 %) were the most prevalent, followed by those for streptomycin (10 %) and tetracycline (3 %), while the least prevalent was for ciprofloxacin (1.5 %). However, among all of the tested enterococci, the resistance genes for gentamicin, penicillin, and ampicillin were not found, though widespread common resistance to gentamicin and, less common, to ampicillin and penicillin was observed. These data might indicate the presence of other genes involved in resistance or the existence of intrinsic resistance.
Resistance to erythromycin was determined by the presence of erm genes. Regarding ERY-resistant isolates (n = 56), the frequencies of ermB and ermC genes were 100 and 76.8 %, respectively, while the presence of ermA was not detected. Interestingly, the presence of ermB and ermC genes was observed among all ten distinct species found in all types of surface waters. It is assumed that the high frequency of ermC genes is consequence of its plasmid localization and it is possible that bacterial conjugation enabled its horizontal spreading. Also it is important to emphasize that antibiotics like tylosin, lincomycin, and neomycin that are the prime antibiotics that are commonly used in animal husbandry also lead to dissemination of ermA, B, and C genes (Thumu and Halami 2012). It is known that ermA is the most common among clinical enetrococci isolates (Sutcliffe et al. 1996); therefore, the fact that ermA was not found among the enterococci isolated from water was not a surprise. Only three isolates, E. faecalis DDE3, E. faecium DDE4, and E. thailandicus/sanquinicola/lactis ASE2-1, harbored both ermB and ermC genes with a high-level-resistant phenotype to erythromycin (256 ug/ml). Our findings are in correlation with those observed by other authors (Sadowy and Luczkiewicz 2014). Nevertheless, there are enterococci isolated from different water types where erm genes were not found (Di Cesare et al. 2012). The presence of two erythromycin-effluxes pump genes, mef and msr, were not detected in the analyzed enterococci strains.
The ant(6)-Ia gene responsible for streptomycin resistance (n = 9) was found in enterococci strains isolated from the rivers and lake of the Belgrade area, with a frequency of 77.8 %. Gene ant(6)-Ia has also been recorded among enterococci isolated from water (González et al. 2009) and clinical isolates (Padmasini et al. 2014) as well as enterococci isolated from soil (Abriouel et al. 2008).
Although the number of gentamicin-resistant strains was significant (n = 61; MIC ≤ 2,000), the acc(6')-aph(2') gene was not detected in the analyzed strains, which is in correlation with published data. High-level of gentamicin resistance in enterococci (MIC ≥ 2,000) is usually associated with the presence of the acc(6')-aph(2') gene (Chow et al. 1998).
Among TET-resistant enterococci (n = 2) both isolates carried tetM and tetL genes. Tetracycline resistance is prevalently mediated by tetL and tetK, which code for the active efflux of the drug across the cell membrane, and tetM, a ribosomal protection gene mostly carried by Tn916-like elements. The strains E. faecalis DDE3 and E. faecium AHE1-2 harbor two tet genes, tetM and tetL, which are responsible for high-level tetracycline resistance, 256 and 32 ug/mL, respectively. These results are in agreement with other authors (Rathnayake et al. 2012) that showed that tetM and tetL genes were predominantly identified in tetracycline-resistant isolates. In addition, the clinical isolates exhibited similar tetracycline resistance to enterococci isolates from water (Jia et al. 2014).
Among ampicillin-resistant isolates (n = 7), neither blaZ gene, nor point mutations in pbp5 gene were detected. However, mutations in other penicillin-binding proteins (PBPs) could alter their affinity to ampicillin and lead to resistant phenotype.
In total, 4 of 67 tested enterococci were resistant to ciprofloxacin. Among 4 ciprofloxacin-resistant E. faecalis strains, only one strain, E. faecalis DDE3, isolated from Danube River-Dorcol carried a mutation in the gyrA gene responsible for resistance to quinolone antibiotics (Jacoby 2005). This isolate had glutamic acid to tyrosine substitution at position 83. The strain E. faecalis DDE3 was the only one which grew at 16 μg/mL of ciprofloxacin. In contrast to the enterococci isolated from water, ciprofloxacin resistance is more prevalent in clinical isolates that harbor mutations in gyrA and parC genes (Rathnayake et al. 2012). In addition, a commonly found mutation in parC, an amino acid substitution at position 80, was not observed.
Genetic relatedness of antibiotic-resistant enterococci
Besides the identification of the Enterococccus species found in urban waters, we employed PFGE to further characterize enterococci isolates for potential genetic relatedness. Out of 67 analyzed antibiotic-resistant enterococcal isolates, 36 representatives of different types and from distinct locations were included in the PFGE analysis. The dendrogram produced by SPSS software demonstrated genomic similarity ranging from 75 to 100 % (Fig. 4). Two clusters with differences up to 25 % were observed. Cluster I comprises two pulsotypes (A and B), while cluster II comprises only one pulsotype (C) (Fig. 4). Pulsotype A contained 14 strains with genetic similarity from 83 % to 100 %, whereas pulsotype B contained 8 strains with genetic differences up to 14 %. Both pulsotypes A and B included the E. faecium and E. faecalis isolates sampled from the same locations (Ada Ciganlija Lake during swimming season and the Danube River-Ada Huja). In addition, pulsotype B included the isolates from the springs (Hajducka Cesma and Brace Jerkovic) as well as the multi-resistant strain E. faecalis DDE3 isolated from the Danube River-Dorcol. Pulsotype C encompassed 14 strains with genetic similarity from 84 to 100 %. Pulsotype C comprises isolates from all of the river localities and Ada Ciganlija Lake during and after swimming season where the majority of strains were E. faecium (50 %), E. faecalis (14 %), and E. hirae (14 %). Notably, the existence of significant differences among isolates of the same species in the different pulsotypes as well as the presence of the same resistance genes in distinct pulsotypes indicate that these genes are likely disseminating among Enterococccus species by horizontal gene transfer as well as clonal spread.
Among 91 Enterococcus spp. strains, multiple antibiotic resistance patterns were found in 56.3 %. They were the most frequently detected in Ada Ciganlija Lake during the swimming season and in the rivers near industrial zones. However, it is very important to indicate that environmental waters in Belgrade area, Serbia are not contaminated with VRE, thus cannot be a source of these enterococci.
In conclusion, this is the first study that has reported the distribution, as well as the occurrence of antibiotic resistance, of Enterococcus spp. isolated from urban surface waters in the Western Balkan Countries. The results of this study showed that the majority of environmental enterococcal isolates act as a reservoir of resistance genes.
Special attention should be drawn to the relationship between community-acquired enterococcal infections and recreational activities. The presence of clinical resistances and the high prevalence of reduced susceptibility to various antibiotics among environmental isolates of Enterococcus sp. are most likely attributed to the uncontrolled use of antibiotics due to poor regulations in Serbia. Finally, our findings lead to necessity for the continuous education on use of antibiotics in the public sector in order to reduce the risk of spreading of antibiotic resistance in the future.
References
Aarestrup, F. M., & Hasman, H. (2004). Susceptibility of different bacterial species isolated from food animals to copper sulphate, zinc chloride and antimicrobial substances used for disinfection. Veterinary Microbiology, 100, 83–89.
Abriouel, H., Ben Omar, N., Molinos, A. C., López, R. L., Grande, M. J., Martínez-Viedma, P., et al. (2008). Comparative analysis of genetic diversity and incidence of virulence factors and antibiotic resistance among enterococcal populations from raw fruit and vegetable foods, water and soil, and clinical samples. International Journal of Food Microbiology, 123, 38–43.
Aminov, R. I., Garrigues-Jeanjean, N., & Mackie, R. I. (2001). Molecular ecology of tetracycline resistance: development and validation of primer for detection of tetracycline resistance genes encoding ribosomal protection proteins. Applied Environmental Microbiology, 67, 22–32.
Boehm, A.B. & Soller, J.A. (2011). Risks associated with recreational waters: pathogens and fecal indicators. In R. A. Meyers (Ed.), Encyclopedia of Sustainability Science and Technology.
Cabello, F. C. (2006). Heavy use of prophylactic antibiotics in aquaculture: a growing problem for human and animal health and for the environment. Environmental Microbiology, 8, 1137–1144.
Chow, J. W., Donabedian, S. M., Clewell, D. B., Sahm, D. F., & Zervos, M. J. (1998). In vitro susceptibility and molecular analysis of gentamicin-resistant enterococci. Diagnostic Microbiology & Infectious Disease, 32, 141–146.
Clinical and Laboratory Standards Institute (CLSI) (2013). Performance standards for antimicrobial susceptibility testing; 23rd informational supplement. CLSI document M100-S23. Clinical Laboratory Standards, Institute, Wayne, PA
Davis, K., Anderson, M. A., & Yates, M. V. (2005). Distribution of indicator bacteria in Canyon Lake, California. Water Research, 39, 1277–1288.
Di Cesare, A., Vignaroli, C., Luna, G. M., Pasquaroli, S., & Biavasco, F. (2012). Antibiotic-resistant enterococci in seawater and sediments from a coastal fish farm. Microbial Drug Resistance, 18, 502–509.
Di Cesare, A., Pasquaroli, S., Vignaroli, C., Paroncini, P., Luna, G. M., Manso, E., et al. (2014). The marine environment as a reservoir of enterococci carrying resistance and virulence genes strongly associated with clinical strains. Environmental Microbiology Reports, 6, 184–190.
Eaton, J. T., & Gasson, J. M. (2001). Molecular screening of Enterococcus virulence determinants and potential for genetic exchange between food and medical isolates. Applied Environmental Microbiology, 67, 1628–1635.
Franz, C. M. A. P., Stiles, M. E., Schleifer, K. H., & Holzapfel, W. H. (2003). Enterococci in foods—a conundrum for food safety. International Journal of Food Microbiology, 88, 105–122.
Garofalo, C., Vignaroli, C., Zandri, G., Aquilanti, L., Bordoni, D., Osimani, A., et al. (2007). Direct detection of antibiotic resistance genes in specimens of chicken and pork meat. International Journal of Food Microbiology, 113, 75–83.
Gevers, D., Danielsen, M., Huys, G., & Swings, J. (2003). Molecular characterization of tet(M) genes in Lactobacillus isolates from different types of fermented dry sausage. Applied Environmental Microbiology, 69, 1270–1275.
Giménez–Pereira, M. L. (2005). Enterococci in milk products. PhD thesis, Massey University Palmerston North, New Zealand.
Giraffa, G. (2003). Functionality of enterococci in dairy products. International Journal of Food Microbiology, 88, 215–222.
González, M., Afonso, O., & Tejedor, M. T. (2009). Antimicrobial susceptibility and molecular typing of Enterococcus faecium isolated from humans, chickens and environment in Canary Islands (Spain). Revista Española de Quimioterapia, 22, 120–126.
Jacoby, G. A. (2005). Mechanisms of resistance to quinolones. Clinical Infectious Diseases, 41, S120–S126.
Jensen, L. B., Frimodt-Moller, N., & Aarestrup, F. M. (1999). Presence of erm gene classes in Gram-positive bacteria of animal and human origin in Denmark. FEMS Microbiology Letters, 170, 151–158.
Jia, W., Li, G., & Wang, W. (2014). Prevalence and antimicrobial resistance of Enterococcus species: a hospital-based study in China. International Journal of Environmental Research and Public Health, 11, 3424–3442.
Jovcic, B., Begovic, J., Lozo, J., Topisirovic, L., & Kojic, M. (2009). Dynamic of sodium dodecyl sulfate utilization and antibiotic susceptibility of strain Pseudomonas sp. ATCC19151. Archives of Biological Sciences, 61, 159–165.
Kim, S. Y., Lee, J. E., Lee, S., Lee, H. T., Hur, H. G., & Ko, G. (2010). Characterization of Enterococcus spp. from human and animal feces using 16S rRNA sequences, the esp gene, and PFGE for microbial source tracking in Korea. Environmental Science & Technology, 44, 3423–3428.
Kojic, M., Jovcic, B., Vindigni, A., Odreman, F., & Venturi, V. (2005). Novel target genes of PsrA transcriptional regulator of Pseudomonas aeruginosa. FEMS Microbiology Letters, 246, 175–181.
Kristich, C. J., Djorić, D., & Little, J. L. (2014). Vancomycin-enhanced cephalosporin susceptibility in vancomycin-resistant enterococci revealed using counter selection with dominant-negative thymidylate synthase. Antimicrobial Agents & Chemotherapy, 58, 1556–1564.
Leroy, F., Foulquie Moreno, M. R., & De Vuyst, L. (2003). Enterococcus faecium RZS C5, an interesting bacteriocin producer to be used as a co-culture in food fermentation. International Journal of Food Microbiology, 88, 235–240.
Messer, J. W., & Dufour, A. P. (1997). A rapid, specific membrane filtration procedure for enumeration of enterococci in recreational water. Applied Environmental Microbiology, 64, 678–680.
Michael, I., Rizzo, L., Mc Ardell, C. S., Manaia, C. M., Merlin, C., Schwartz, T., et al. (2013). Urban wastewater treatment plants as hotspots for the release of antibiotics in the environment: a review. Water Research, 47, 957–995.
Mioljevic, V., Markovic Denic, L. J., Vidovic, A., Jovanovic, M., Tosic, T., & Tomin, D. (2013). Risk factors for vancomycin-resistant Enterococcus colonization in hematologic patients. Vojnosanitarni Pregled, 70, 1109–1116.
Moore, D. F., Guzman, J. A., & McGee, C. (2008). Species distribution and antimicrobial resistance of enterococci isolated from surface and ocean water. Journal of Applied Microbiology, 105, 1017–1025.
Niederhausern, S. D., Bondi, M., Anacarso, I., Iseppi, R., Sabia, C., Bitonte, F., & Messi, P. (2013). Antibiotics and heavy metals resistance and other biological characters in enterococci isolated from surface water of Monte Cotugno Lake (Italy). Journal of Environmental Science & Health Part A, Environmental Science & Engineering & Toxic and Hazardous Substance Control, 48, 939–946.
Nishiyama, M., Iguchi, A., & Suzuki, Y. (2015). Identification of Enterococcus faecius and Enterococcus faecalis as vanC-type vancomycin-resistant enterococci (VRE) from sewage and river water in the provincial city of Miyazaki, Japan. Journal of Environmental Science & Health Part A, Environmental Science & Engineering & Toxic and Hazardous Substance Control, 50, 16–25.
Oancea, C., Klare, I., Witte, W., & Werner, G. (2004). Conjugative transfer of the virulence gene, esp, among isolates of Enterococcus faecium and Enterococcus faecalis. Journal of Antimicrobial Chemotherapy, 54(1), 232–235.
Padmasini, E., Padmaraj, R., & Ramesh, S. S. (2014). High level aminoglycoside resistance and distribution of aminoglycoside resistant genes among clinical isolates of Enterococcus species in Chennai, India. Scientific World Journal 1–5
Poyart, C., Quesnes, G., & Trieu-Cuot, P. (2000). Sequencing the gene encoding manganese-dependent superoxide dismutase for rapid species identification of enterococci. Journal of Clinical Microbiology, 38, 415–418.
Radhouani, H., Igrejas, G., Pinto, L., Goncalves, A., Coelho, C., Rodrigues, J., & Poeta, P. (2011). Molecular characterization of antibiotic resistance in enterococci recovered from seagulls (Larus cachinnans) representing an environmental health problem. Journal of Environmental Monitoring, 13, 2227–2233.
Rathnayake, I. U., Hargreaves, M., & Huygens, F. (2012). Antibiotic resistance and virulence traits in clinical and environmental Enterococcus faecalis and Enterococcus faecium isolates. Systematic & Applied Microbiology, 35, 326–333.
Sadowy, E., & Luczkiewicz, A. (2014). Drug-resistant and hospital-associated Enterococcus faecium from wastewater, riverine estuary and anthropogenically impacted marine catchment basin. BMC Microbiology, 14, 66.
Scott, T., Jenkins, T., Lukasik, J., & Rose, J. (2005). Potential use of a host associated molecular marker in Enterococcus faecium as an index of human fecal pollution. Environmental Science & Technology, 39, 283–287.
Shankar, V., Baghdayan, A. S., Huycke, M. M., Lindahl, G., & Gilmore, M. S. (1999). Infection-derived Enterococcus faecalis strains are enriched in esp, a gene encoding a novel surface protein. Infection & Immunity, 67(1), 193–200.
Shankar, N., Lockatell, C. V., Baghdayan, A. S., Drachenberg, C., Gilmore, M. S., & Johnson, D. E. (2001). Role of Enterococcus faecalis surface protein ESP in the pathogenesis of ascending urinary tract infection. Infection & Immunity, 69(7), 4366–4372.
Sidhu, J. P. S., Skelly, E., Hodgers, L., Ahmed, W., Li, Y., & Toze, S. (2014). Prevalence of Enterococcus species and their virulence genes in fresh water prior to and after storm events. Environmental Science & Technology, 48, 2979–2988.
Stosovic, B., Stepanovic, S., Donabedian, S., Tosic, T., & Jovanovic, M. (2004). Vancomycin-resistant Enterococcus faecalis in Serbia. Emerging Infectious Diseases, 10, 157–158.
Strommenger, B., Kettlitz, C., Werner, G., & Witte, W. (2003). Multiplex PCR assay for simultaneous detection of nine clinically relevant antibiotic resistance genes in Staphylococcus aureus. Journal of Clinical Microbiology, 41, 4089–4094.
Sutcliffe, J. E., Grebe, T., Tait-Kamradt, A., & Wondrack, L. (1996). Detection of erytromycin-resistant determinants by PCR. Antimicrobial Agents & Chemotherapy, 40, 2562–2566.
Švec, P., & Sedlaček, I. (1999). Occurrence of Enterococcus spp. in waters. Folia Microbiologica, 44, 3–10.
Tamura, K., Stecher, G., Peterson, D., Filipski, A., & Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Molecular Biology & Evolution, 30, 2725–2729.
Teixeira, L. M. & Merquior, V. L. C. (2013). “Enterococcus” molecular typing in bacterial infections. Humana Press, 17–26
Thumu, S. C. R., & Halami, P. M. (2012). Acquired resistance to macrolide–lincosamide–streptogramin antibiotics in lactic acid bacteria of food origin. Indian Journal of Microbiology, 52(4), 530–537.
WHO; (2004). Guidelines for drinking-water quality, p 515. Recommendations, 3rd ed, vol 1. World Health Organization, Geneva, Switzerland
Acknowledgments
The Ministry of Education, Science and Technological Development, Republic of Serbia (Grant No. 173019) supported this work. The authors are grateful to Nathaniel Aaron Sprinkle, a native English editor for the proofreading of the manuscript.
Conflict of interest
The authors have declared that no competing interest exists.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Veljović, K., Popović, N., Vidojević, A.T. et al. Environmental waters as a source of antibiotic-resistant Enterococcus species in Belgrade, Serbia. Environ Monit Assess 187, 599 (2015). https://doi.org/10.1007/s10661-015-4814-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10661-015-4814-x