Abstract
Fire blight, caused by Erwinia amylovora, is a destructive bacterial disease that severely hampers apple production. To conduct Quantitative Trait Locus (QTL) studies for breeding resistant apple cultivars, phenotyping of large genetic mapping populations of apples for fire blight resistance is essential. This, however, necessitates precise, quantitative data spanning multiple years, locations, and pathogen strains. It can be time-consuming and resource-intensive to keep QTL mapping populations for apples in the field and greenhouse. This creates a bottleneck for identifying novel QTL for fire blight resistance or developing resistant cultivars. To address this challenge, we present an image-based method for rapid and accurate phenotyping fire blight resistance using apple leaf discs. This leaf disc assay demonstrates significant (p < 0.05) percent disease area (PDA) differences in fire blight inoculations among eight apple genotypes with well-known resistance levels. Furthermore, the image-based leaf disc assay consistently shows a 40–70% difference in PDA between resistant and susceptible checks. We also report high within and across trial broad sense heritability values ranging from 0.86 – 0.97. We demonstrate the use of K-means clustering and best linear unbiased estimators to combine multiple trials. This assay offers an efficient alternative to traditional fire blight screening methods, potentially improving our understanding of the host response, and accelerating the development of resistant apple cultivars.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Modern crop breeding and genetic mapping studies rely on precise phenotyping of complex traits, including disease resistance. However, accurate measures of disease severity require skilled evaluators and repeated experiments across multiple years, locations, and pathogen strains/isolates (Chiang et al., 2016). Visual assessment rating scales are the most used method for evaluating plant disease severity (Bock et al., 2022). These discrete rating scales are limited by a low throughput, high labor cost, rater biases and represent an imperfect measurement of the continuous underlying phenotypic variation (Bock et al., 2010; Chiang et al., 2016). As a result, there has been a growing interest in image-based phenotyping methods that use sensors and cameras to accurately evaluate plant traits (Li et al., 2014). Image-based analysis of disease symptoms can improve the standardization of rating scales, increase throughput, and lower the cost of data acquisition (Bock et al., 2008, Mutka and Bart, 2015). Additionally, wavelengths outside the visible spectrum, such as ultra-violet, near infrared, and short-wave infrared, can measure traits beyond human capabilities (Bock et al., 2010; Li et al., 2014). These methods are particularly valuable for the decomposition of complex traits and can have a positive impact on crop genetics research (Mutka and Bart, 2015; Mir et al., 2019).
Disease resistance is often a complex trait controlled by many additive minor effect loci with a complicated genetic architecture (Brachi et al. 2011). Therefore, discrete disease rating scales can lead to a loss of variation that makes it difficult to detect minor and moderate effect resistance Quantitative Trait Locus (QTL) (Mir et al., 2019). Though major effect QTL can be easily observed visually, loci with a minor to moderate effect size require precise quantitative data (Desnoues et al., 2018). The loci often have low heritability as their effects are highly influenced by the environment, pathogen strain, and experimental factors (Desnoues et al., 2018). Biased and inaccurate phenotype data further compound these challenges and can result in the advancement of inferior breeding lines, insufficient experimental power, or the identification of ghost QTL (Wallin et al., 2021). Employing image-based phenotyping methods that enhance throughput compared to visual counterparts, increases the number of samples collected and improve the statistical power needed to accurately identify QTL for breeding (Bock et al., 2010). Moreover, the evaluation of more time points and pathogen strains can lead to the selective advancement of lines with more robust disease resistance (Cowger & Brown, 2019). This can be particularly impactful for crops where generating large population sizes is challenging.
Breeding for disease resistance in long-cycle woody perennial crops like apples is a slow and difficult process. The priority of many apple breeding programs is host resistance to the most destructive bacterial disease, fire blight, caused by Erwinia amylovora (Khan & Korban, 2022; Peil et al., 2021). However, the development of apple populations for breeding or QTL mapping is time-consuming and requires considerable resources (Khan & Korban, 2022; Peil et al., 2021). This is partly due to the high number of replicates needed to accurately measure fire blight resistance (Kostick et al., 2021). The screening of fire blight resistance in apples is performed via controlled single-strain inoculations of either grafted plants in the greenhouse or established orchard trees (Desnoues et al., 2018). Manual quantitative measurements of black or brown necrotic tissue are used to estimate disease severity (Desnoues et al., 2018; Kostick et al., 2019). The inoculation of young shoots of greenhouse-grown plants is the most cost-effective and space-efficient method. This method has been effective in identifying several important fire blight resistance QTL (Durel et al., 2009; Emeriewen et al., 2021; Khan et al., 2006; Peil et al., 2008). However, it is limited to single E. amylovora strain infections of vegetative tissue and may not accurately reflect the resistance level expected in the field (Peil et al., 2019). Field inoculations, performed by either shoot inoculation or blossom spray, provide the most biologically relevant results (Peil et al., 2019). However, field inoculations are expensive, require considerable maintenance of mapping populations, and are subject to variable environmental conditions (Kostick et al., 2021). In both cases, inoculations take several weeks to conduct and can only be performed once or twice a year on the same plants (Emeriewen et al., 2021). Any more would risk long-term damage to important germplasm. These current fire blight inoculation methods are laborious and slow down identification of novel QTLs.
Leaf disc assays have been proposed to screen large populations of crops for genetic mapping of disease resistance, and to overcome the challenges of phenotyping. An image-based leaf disc assay offers a highly standardized method to efficiently evaluate many genotypes for genetic mapping studies (Zendler et al., 2021). These rapid and non-destructive disease screening methods have been successfully applied to many crops, including grapes, cucumber, and potato (Leonards-Schippers et al., 1994; Longzhou et al., 2008; Zendler et al., 2021). Previous studies explored alternative fire blight assays using whole detached apple leaves (Donovan, 1991, Martínez-Bilbao et al., 2009). Martínez-Bilbao et al. (2009) found a moderate correlation (0.56) between greenhouse-inoculated plants and inoculated detached leaves of the same genotypes. These inoculations on whole leaves had limitations. These include an arbitrary visual rating system and maintaining detached leaves for 5–6 days, which can introduce confounding abiotic factors. Additionally, both studies found disease severity scores were affected by the age of the leaf, indicating the influence of ontogenic resistance (Panter & Jones, 2002). Recently, Zendler et al., 2021 demonstrated the potential of coupling a grape downy mildew leaf disc assay with an automated image analysis pipeline to screen a genetic mapping population.
In this study, we aimed to develop a non-destructive and high-throughput image-based leaf disc assay for rapidly phenotyping apple genotypes for fire blight resistance. We performed this assay, on eight genotypes known to have varying degrees of fire blight resistance, across three repeated trials. The goals of this experiment were to 1) determine if the leaf disc assay and image-analysis pipeline could reliably discriminate high, moderate, and low levels of fire blight resistance and 2) combine multiple fire blight resistance screening trials to improve the power of the assay.
Materials and methods
Plant material and leaf collection
We selected a panel of apple genotypes comprising both wild Malus accessions and cultivars with known fire blight resistance levels. The panel included eight genotypes representing a range of fire blight resistance classes, namely highly susceptible (HS), moderately resistant (M), resistant (R) and highly resistant (HR). The HS class comprised M. domestica cv. 'Gala' (PI392303) (Harshman et al., 2017; Kostick et al., 2019). The M class included M. domestica cv. 'Cox's Orange Pippin' (PI588853) and M. domestica cv. 'Enterprise' (PI590210), while the R class consisted of M. sieversii 'KAZ 95 18–14' (PI657054) and M. hybrid 'KAZ 96 01–03' (PI657085) (Khan et al., 2006; Khan et al., 2007; Harshman et al., 2017; van de Weg et al., 2018; Kostick et al., 2019). The HR class comprised M. hybrid 'KAZ 96 08-01P-37' (PI657115), M. floribunda 'Floribunda 821' (PI589827), and M. × robusta 'Robusta 5' (PI588825) (Peil et al. 2007; Durel et al., 2009; Harshman et al., 2017).
Leaves were collected from healthy, actively growing apple trees from the McCarthy Research Farm, at the United States Department of Agriculture—Plant Genetic Resources Unit (USDA-PGRU), Geneva, NY. Leaf collection was conducted between June and July 2023. Only young leaves with a length of 3 to 5 cm and a slightly glossy cuticle were selected for the assay. The leaves were detached at the petiole, placed in labeled plastic Ziploc bags, and transported in a cooler with an ice pack. The total collection and transport time was approximately one hour. Prior to use, the leaves were cleaned with a 1% bleach solution by submerging for about 20 s and rinsed with deionized water. Leaves were left to dry on a clean paper towel for five minutes. All equipment and surfaces were sterilized using 70% ethanol. Using an 8 mm diameter leather punch, 1 to 2 leaf discs were cut from each leaf (Fig. 1a). Leaf discs were immediately plated in a clear acrylic imaging tray on 1% water agar solution grouped by genotype using a grid template to separate each disc into 2 cm2 cells (Fig. 1a).
Graphical representation of the major steps of the leaf disc assay to phenotype fire blight resistance in apples. A Apple leaves on the upper portion of the panel demonstrate the age of the leaf and number of leaf discs taken from each sample. The bottom portion shows the leaf discs plated on a water agar tray to be sprayed with a Erwinia amylovora liquid inoculum. B This panel shows the flatbed scanner used for image data collection and an image of the leaf disc matrix that is used for image analysis. C The steps of segmenting each leaf disc in the matrix is shown in the upper portion of the panel. Two representative leaf discs with a high and low amount of disease severity are shown on the bottom portion of the panel. To the right of each disc is the pseudo-colored classified mask (red = necrosis, yellow = pre-symptomatic, green = healthy)
Image data collection
Custom acrylic scanner trays (TAP Plastics; San Leano, CA) specifically designed for the Epson 12000XL flatbed scanner (Epson; Los Alamitos, CA) were utilized for the assay and imaging. The dimensions of the tray are 40 cm × 30 cm × 2 cm. Each tray was thoroughly sterilized with a 70% ethanol solution before use. A 300 ml 1% water agar solution was prepared in advance and poured evenly into the tray. Any bubbles formed during the pouring process were removed with a pipette tip to ensure that the leaf discs were not obscured during imaging.
Erwinia amylovora cultures were obtained from glycerol stock stored at -80 °C. The plates were streaked 16 h prior to inoculation on lysogeny broth (LB) media and incubated at 28 °C. The bacterial colonies were washed from the plates and prepared as a liquid bacterial suspension in sterile type I water at a concentration of 1 × 109 CFU/ml. The fire blight inoculum was prepared as a liquid suspension using the aggressive Canadian strain of E. amlyovora, Ea4001. The fire blight inoculum (25 ml) was evenly sprayed across all samples using an 88 ml manual spray bottle (Good to Go, Navajo Inc; Denver, CO) over the entire tray from approximately 12 cm away. The tray was covered with plastic cling wrap to maintain high humidity and incubated in the dark for 48 h at 25 °C.
The trays were imaged at 48 h post inoculation (hpi) using an Epson 12000XL flatbed scanner and Epson Scan 2 app (Fig. 1b). The main settings were adjusted to document source as transparency unit, document type as color positive film, image type as 48-bit color, resolution at contrast to 50%, and saturation to 100%, while all other parameters were set to default.
Image analysis
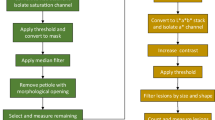
To manually preprocess the images, we used Fiji version 2.3.1 (Schindelin et al., 2012). We cropped the image edges to remove the tray and used the paint tool to eliminate any extraneous objects (dirt, leaf debris, double-stacked discs). The resulting images were cropped to isolate each column containing a single genotype, and the.jpeg file names were annotated with the date, experiment, and genotype.
For image analysis, a custom python script was developed to automate the phenotyping of the inoculated leaf discs using the OpenCV and PlantCV python libraries as the foundation (Gehan et al. 2017). The code is available on GitHub at the following link: https://github.com/KhanLab-AppleDiseases/Fire-Blight-Leaf-Disc-Assay. Additional documentation can be found at https://plantcv.readthedocs.io/en/latest/naive_bayes_classifier/. The script converts the input image of the leaf discs to grayscale using the lightness channel and applies a threshold filter to remove background noise. From the grayscale images, binary masks are applied to find the edge of the discs and segment the objects. Each disc was auto cropped to create a single image of each disc for classification (Fig. 1c). Pixels in each single-disc image were classified using a naïve Bayes classifier, which was trained with a text file containing red, blue, and green (RGB) pixel data using Fiji Pixel Inspector. The file consists of 20 pixels of training data for each of the four classes: healthy (H), pre-symptomatic (P), necrotic (N), and white background (WB). The classifier uses a probability density function to bin pixels into the appropriate category. The resulting image mask recolors the leaf disc image so that H pixels appear green, P pixels appear yellow, N pixels appear red, and WB pixels appear white (Fig. S1). The trait analyzed in our study is the percent disease area (PDA), which is calculated as the percentage of N pixels to the total area of the leaf disc (N + P + H). Before generating a.csv dataset with the image names, replicate number, and disease severity scores, the output images were checked to ensure that no unrelated objects in the tray were analyzed.
Statistical analysis
All statistics were performed in R (version 4.0.1; R Core Team, 2021). One-way analysis of variance (ANOVA) followed by Tukey–Kramer multiple comparison test was performed to compare the PDA of multiple groups (Fig. 2). To check the normality of residuals QQ plots and Shapiro-Wilks tests were used. To test the homogeneity of variances across genotypes and trials, Levene’s Test was applied with the leveneTest() function from the R package ‘Car’ (Fox et al. 2019). To satisfy the assumptions of normality of residuals and homogeneity of variances, a BoxCox transformation was applied to the PDA data with a lambda value of 0.3. Broad sense heritability (H2) was calculated using Eq. 1 (Calenge et al., 2005; Kruijer et al., 2014).
Horizontal bar plots showing the mean percent diseased area values for fire blight severity in eight apple genotypes (standard error bars are indicated). Each color represents a different repeated trial (A-orange; B-blue; C-green; All Trials-grey). Each genotype name is labeled with the abbreviated fire blight resistance class in parentheses (HS = highly susceptible; M = Moderate; R = resistant; HR = highly resistant). Tukey–Kramer groups are shown with letters at the end of each bar representing statistical significance (p < 0.05) within a group. Plant introduction (PI) number is the unique identifier from United States Department of Agriculture—Germplasm Resources Information Network database
The terms are defined as genetic variance (\({\upsigma }_{G}^{2}\)), environmental variance (\({\upsigma }_{E}^{2}\)), and average number of replicates (\({n}_{rep}\)). Variance components were estimated by calculating the mean square (MS) error of genotype and residual error from ANOVA, where \({\upsigma }_{G}^{2}\) = MS(genotype) – MS(residual error) / \({n}_{rep}\) and \({\upsigma }_{E}^{2}\) = MS(residual error) (Kruijer et al., 2014).
Multi-trial PDA best linear unbiased estimators (BLUEs) of the response variable (\({Y}_{ijk}\)) were estimated using Eq. 2 with a mixed linear model fit by restricted maximum likelihood (REML) using the lme4 package (Bates et al. 2015). In this context, BLUEs are used as a more accurate estimation of the phenotype by incorporating the effects of experimental factors to account for multiple sources of variation. The response variable, \({Y}_{ijk}\), is the PDA value for the ith genotype, jth trial, and kth replicate.
The model terms include grand mean (\(\mu\)), trial treatment effect (\({T}_{j}\)), genotype (\({G}_{i}\)) effect, genotype by trial interaction (\({GT}_{ij}\)), and residual error (\({\varepsilon }_{ijk}\)). The model term \({G}_{i}\) was fitted as a fixed effect, while all other terms were fitted as random effects. Trial was defined as an independent repeated experiment each performed on a single tray. The BLUE values were used to rank genotypes and compare resistance levels among checks.
For K-mean clustering, the data was formatted to have each genotype as a row name and the PDA value of each trial as a separate column. The randomized seed was set to 1 and the PDA values were divided by 100 to get scaled values from 0 to 1. The Kmeans() function from the R package ‘Stats’ (R Core Team, 2022) was used to calculate the K centers and cluster values. A custom R script was used to loop through center values 1 to 6 to find the best fitting number of K by calculating the total within-cluster sum of squares (WSS) values. The best fitting K value was determined by plotting K by WSS and finding the value K associated with the inflection point in the curve. We used the Kmeans() function to calculate the cluster positions for the leaf disc assay data with the best fitting K value and a random starting set of centroids (nstart) as 25. We plotted the clusters with the fviz_cluster() function and ellipses from the ‘factoextra’ R package (Kassambara & Mundt, 2017). We assessed the disease rating of each individual by examining the cluster membership in relation to the susceptible and resistant checks.
Results
Phenotypic quality and repeatability
There was a wide variation in PDA values across the three trials ranging from 0.5% to 95%. The K-means clusters reflected the results of the Tukey–Kramer means separation which identified four groups consistent with the original HS, M, R, and HR resistance classes (Fig. 2, Fig. S1 ), Within trials these groups were inconsistent with only the HS and HR classes having consistent mean differences. The groups with the greatest difference in PDA mean across all trials were between the highly susceptible check ‘Gala’ in the ‘a’ group and the highly resistant check ‘Robusta 5’ in the ‘d’ group (Fig. 2). The difference in PDA values between ‘Gala’ and ‘Robusta 5’ within trials A, B, and C were 38%, 76%, 43%, respectively (Fig. 2). Across all trials the PDA difference between ‘Gala’ and ‘Robusta 5’ was 53% (Fig. 2). The ‘b’ group was comprised of all genotypes in the M and R classes (Fig. 2). These included ‘Enterprise’, ‘Cox’s Orange Pippin’, M. sieversii PI657054, and M. hybrid PI657085. The ‘c’ group was formed from the HR genotypes ‘Floribunda 821’ and M. hybrid PI657115 (Fig. 2).
As a measure of repeatability, the broad sense heritability (H2) was calculated within and across trials. The within-trial broad-sense heritability of trials A, B, and C were 0.93, 0.95, and 0.86, respectively. The across-trial broad-sense heritability was 0.97. This shows the environmental/experimental effects are minimal, and the data is useful for predicting genetic effects. The Pearson correlation coefficient for trial A × B was 0.7, A × C was 0.8, and B × C was 0.86. The pairwise Pearson correlations showed each trial was significantly (p < 0.05) correlated to each other. Across an average replicate number of 14 discs per genotype, the standard deviation in PDA values was 17 with a maximum of 31 (Table S1). The various metrics of repeatability and data quality indicate that the phenotype data generated from the leaf disc assay method is reliable.
Multi-trial genotype classification
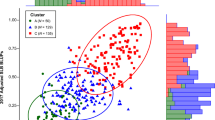
The K-mean cluster analysis combining trials A-C was able to capture 96.9% of the phenotypic variation across three repeated trials. The first two dimensions captured 86.4% and 10.5% of the variation in the data, respectively (Fig. 3). Three clusters formed based on the mean PDA differences (Fig. 3). Cluster 1 was formed with only the genotype ‘Gala’ representing a highly susceptible group with a mean PDA of 57.9%. Cluster 2 contained each genotype that was classified as M or R including M. sieversii PI657054, M. hybrid PI657085, Cox’s Orange Pippin, and Enterprise. The mean PDA for that cluster was 32.6%. Cluster 3 was formed with all HR genotypes, M. hybrid PI657115, Floribunda 821, and Robusta 5. The mean PDA for cluster 3 is 13.5%. These classes follow closely with the Tukey–Kramer results where four significantly (p < 0.05) distinct groups were identified that followed the rank orders of the resistance classes (Fig. 2). The only difference is that with the K-mean clustering the M and R classes were combined into a single group. Each genotype was ranked based on the scaled PDA BLUE value (Fig. 4). The ranked values largely matched the results from the prior analyses (Fig. S1) . The pre-assigned classes were ordered correctly based on expected level of fire blight resistance (Fig. 4). The genotypes in the HS group and the HR group were ranked on both extreme ends of the distribution ranging from approximately -4 to + 1. Genotypes from the M and R class have highly similar values around -1 ranked accurately in the middle of the distribution. ‘Floribunda 821’ and M. hybrid PI657115 are both in the HR class and have similar PDA BLUE values between -2 and -3.
Apple genotypes screened for fire blight resistance with the leaf disc assay grouped into three clusters via K-mean clustering. The K centers and grouping of genotypes is a based on the percent disease area values of the three repeated trials. The data was plotted on a biplot where the x and y axis are the first and second dimension, respectively. Next to each axis name, in parentheses, is the percent variance explained by that dimension. Each genotype name is labeled with the resistance class in parentheses (HS = highly susceptible; M = Moderate; R = resistant; HR = highly resistant). Cluster 1 (n = 4) is blue with circle points, cluster 2 (n = 3) is green with triangle points, and cluster 3 (n = 1) is red with square points. Plant introduction (PI) number is the unique identifier from United States Department of Agriculture—Germplasm Resources Information Network database
Horizontal bar plots with the percent disease area best linear unbiased estimator (BLUE) value for fire blight severity for eight apple genotypes. Each genotype name has the abbreviated fire blight resistance class in parentheses (HS = highly susceptible; M = Moderate; R = resistant; HR = highly resistant). Plant introduction (PI) number is the unique identifier from United States Department of Agriculture—Germplasm Resources Information Network database
Discussion
We have developed an image-based leaf disc assay that can greatly reduce the time and labor required for fire blight resistance phenotyping in apples for genetic research and breeding. The analytical pipeline of the assay utilizes standardized, high-resolution RGB images to estimate the percentage leaf disc area covered by fire blight lesions.
Leaf disc assays have been popular in studying host–pathogen interactions but have not been widely used for genetic mapping and breeding (Kortekamp, 2006), except for some fungal diseases (Leonards-Schippers et al., 1994; Longzhou et al., 2008; Zendler et al., 2021). Detached leaf assays have been developed for evaluating resistance to bacterial diseases in apple, pear, and cherry (Bedford et al., 2003; Moragrega et al., 2003; Martínez-Bilbao et al., 2009). However, all these studies primarily used subjective visual rating scales to quantify disease severity (Bock et al., 2022). Martínez-Bilbao et al. (2009) found a moderate correlation (0.56) between fire blight inoculations of shoots and detached leaves of the same cultivars when using a visual rating scale, likely due to the bias and subjectivity in severity scoring.
We have shown that this newly developed assay had biological relevance, low turn-around time and requires minimal technical expertise to perform. We were able to correctly classify (P < 0.05) eight genotypes with previously described resistance to their established classes, based on the percent area of fire blight symptoms measured using this assay. The long turn-around time of detached leaf assays for disease response evaluation can potentially introduce confounding abiotic factors (Donovan, 1991, Martínez-Bilbao et al., 2009). Detached leaf assays are time-sensitive since the leaf rapidly starts degrading upon detachment and reactive oxygen species (ROS) begin accumulating (Wang et al., 2012). Given that ROS production is common to both apple leaf senescence and the Erwinia amylovora host defense response, cross-tolerance plays a role in the assay results (Kharadi et al., 2021; Perez & Brown, 2014). Wang et al. (2012) found significant (P < 0.05) chlorophyll loss and H2O2 accumulation in detached apple leaves beginning after 4 days for the cultivar M. domestica cv. ‘Hanfu’. Although we did not observe abiotic stress on the leaf discs, we cannot rule out the impact of stress from detaching and injuring leaves, as well as physiological differences of the leaves used, on the assay. The PDA value is based only on pixels classified as necrotic, directly capturing the hypersensitive response caused by effector-triggered immunity of the host, while excluding pre-symptomatic pixels that are not specific to E. amylovora symptoms (Kharadi et al., 2021). The necrotic symptoms observed include rapidly growing orange-brown necrotic lesions that initially colonize in the midrib and produce ooze (Kharadi et al., 2021). The high broad-sense heritability (0.86–0.97) indicates that the experimental/environmental factors (i.e., field collection of leaves) contribute minimal variation to the PDA. These broad-sense heritability estimates are comparable to greenhouse screenings of fire blight QTL mapping populations (0.71–0.96) and are higher than fire blight field inoculations (0.44–0.75) (Calenge et al., 2005; Desnoues et al., 2018; Durel et al., 2009; Khan et al., 2006; Kostick et al., 2021).
Our results also showed that non-hierarchical cluster analysis using K-means clustering was successful in capturing 96.9% of the phenotypic variance in the first two dimensions, with clusters largely corresponding to anticipated resistance categories, including HS, M, and R classes combined, and HR. Kostick et al. (2019) utilized a similar K-means clustering approach to classify fire blight resistance of 94 apple cultivars, resulting in three clusters that categorized cultivars into susceptible, intermediate, and resistant classes. ‘Gala’ and ‘Enterprise’ were categorized as HS and M-HR, respectively, matching our classifications from the leaf disc assay. However, Kostick et al. (2019) classified 'Cox's Orange Pippin' as moderately susceptible, while it was clustered in the M-R group in our study. This may be due to environmental differences between these studies that directly impact this resistance locus. It is also important to note that PDA values are not absolute, but rather relative metrics meant to be ranked with HS and HR checks in every experiment. Our results and conclusions were based on only eight genotypes, and screening more genotypes would help to better understand the accuracy and sensitivity of this assay. For genotypes with unknown resistance levels, it is recommended to include checks and control genotypes similar to those used by Zendler et al. (2021). BLUEs of the genotype effects were able to correctly rank genotypes based on their expected levels of fire blight resistance across multiple trials. A mixed model approach is useful for combining data from multiple trials to estimate genotype effects while accounting for environmental/experimental variation between trials (Bernardo, 2020). BLUEs are widely used for estimating breeding values of diverse populations screened for disease resistance (Abdelraheem et al., 2020). This method is also well-suited for unbalanced data, which is a likely case for leaf disc assays where the leaves available for each genotype may vary. For studies that plan to incorporate multiple strains, severity metrics, and time-points, using a mixed model approach will improve the integration of this data to estimate genotypic effects more accurately (Bernardo, 2020).
However, we believe that the leaf-disc assay can be further improved by focusing on two key steps: increasing the training data sets and exploring additional features of the data. Currently, the study utilizes a training data set with only 20 examples of pixels from each category. Increasing the size of this data set would enhance the detection accuracy of fire blight symptoms. Additionally, incorporating more time points and analysis of longitudinal data may lead to a better identification of moderate fire blight symptoms. We strongly encourage feedback and the sharing of experiences through the ‘issues’ or ‘pull request’ features of the GitHub repository provided. This collaborative approach can lead to refinements in the performance of this assay. Moreover, the pipeline presented in this study can serve as a user-friendly foundation for rapid fire blight resistance phenotyping in other challenging crops.
Conclusion
The image-based leaf disc assay is an effective method to phenotype fire blight resistance in multiple genetic mapping populations with reduced time and labor. Additionally, it requires minimal technical expertise and familiarity with fire blight, allowing for precise quantitative data to be obtained through the 48-h inoculation and image analysis protocols. Our research further demonstrates that combining trial data from multiple experiments using multi-trial BLUEs and K-means clustering can account for random experimental variation and improve classification of fire blight resistant apple genotypes. Therefore, this assay presents a valuable alternative for phenotype fire blight in apples for breeding and mapping of fire blight resistance QTL.
References
Abdelraheem, A., Elassbli, H., Zhu, Y., Kuraparthy, V., Hinze, L., Stelly, D., Wedegaertner, T., & Zhang, J. (2020). A genome-wide association study uncovers consistent quantitative trait loci for resistance to Verticillium wilt and Fusarium wilt race 4 in the US Upland cotton. Theoretical and Applied Genetics, 133(2), 563–577. https://doi.org/10.1007/S00122-019-03487-X/FIGURES/3
Bates, D., Maechler, M., Bolker, B., Walker, S., Christensen, R. H. B., Singmann, H., Dai, B., Scheipl, F., Grothendieck, G. & Green, P. (2015). Package ‘lme4’: linear mixed-effects models using Eigen and S4 (Version 1.1-7). R Foundation for Statistical Computing. Available online at https://cran.r-project.org/web/packages/lme4/lme4.pdf. Assessed on 07 April 2023.
Bedford, K. E., Sholberg, P. L., & Kappel, F. (2003). Use of a detached leaf bioassay for screening sweet cherry cultivars for bacterial canker resistance. Acta Horticulturae, 622, 365–368. https://doi.org/10.17660/ACTAHORTIC.2003.622.37
Bernardo, R. (2020). Reinventing quantitative genetics for plant breeding: something old, something new, something borrowed, something BLUE. Heredity, 1256. 125:375–385. https://www.nature.com/articles/s41437-020-0312-1
Bock, C. H., Parker, P. E., Cook, A. Z., and Gottwald, T. R. 2008. Visual rating and the use of image analysis for assessing different symptoms of citrus canker on grapefruit leaves. Plant Disease, 92:530–541. https://pubmed.ncbi.nlm.nih.gov/30769647/
Bock, C. H., Poole, G. H., Parker, P. E., and Gottwald, T. R. (2010). Plant Disease Severity Estimated Visually, by Digital Photography and Image Analysis, and by Hyperspectral Imaging. Critical Reviews in Plant Sciences, 29(2), 59–107. https://www.tandfonline.com/doi/abs/https://doi.org/10.1080/07352681003617285
Bock, C. H., Chiang, K. S., and Del Ponte, E. M. (2022). Plant disease severity estimated visually: a century of research, best practices, and opportunities for improving methods and practices to maximize accuracy. Tropical Plant Pathology, 47:25–42. https://springerlink.bibliotecabuap.elogim.com/article/https://doi.org/10.1007/s40858-021-00439-z
Brachi, B., Morris, G. P., Borevitz, J. O. (2011). Genome-wide association studies in plants: the missing heritability is in the field. Genome Biology, 12, 232. https://doi.org/10.1186/gb-2011-12-10-232
Calenge, F., Drouet, D., Denancé, C., Van De Weg, W. E., Brisset, M. N., Paulin, J. P., et al. (2005). Identification of a major QTL together with several minor additive or epistatic QTLs for resistance to fire blight in apple in two related progenies. Theoretical Applied Genetics, 111:128–135. https://springerlink.bibliotecabuap.elogim.com/article/https://doi.org/10.1007/s00122-005-2002-z
Chiang, K. S., Bock, C. H., Lee, I. H., El Jarroudi, M., and Delfosse, P. (2016). Plant disease severity assessment-how rater Bias, assessment method, and experimental design affect hypothesis testing and resource use efficiency. Phytopathology, 106,1451–1464. https://apsjournals.apsnet.org/doi/https://doi.org/10.1094/PHYTO-12-15-0315-R
Cowger, C., and Brown, J. K. M. (2019). Durability of Quantitative Resistance in Crops: Greater Than We Know? Annual Review Phytopathology, 57, 253–277. https://pubmed.ncbi.nlm.nih.gov/31206351/
Desnoues, E., Norelli, J. L., Aldwinckle, H. S., Wisniewski, M. E., Evans, K. M., Malnoy, M., et al. (2018). Identification of Novel Strain-Specific and Environment-Dependent Minor QTLs Linked to Fire Blight Resistance in Apples. Plant Molecular Biology Report, 36, 247–256. https://springerlink.bibliotecabuap.elogim.com/article/https://doi.org/10.1007/s11105-018-1076-0
Donovan, A. (1991). Screening for fire blight resistance in apple (Malus pumila) using excised leaf assays from in vitro and in vivo grown material. Annual Applied Biology, 119, 59–68 https://onlinelibrary.wiley.com/doi/full/https://doi.org/10.1111/j.1744-7348.1991.tb04843.x
Durel, C. E., Denancé, C., and Brisset, M. N. (2009). Two distinct major QTL for resistance to fire blight co-localize on linkage group 12 in apple genotypes “Evereste” and Malus floribunda clone 821. Genome, 52,139–147 https://cdnsciencepub.com/doi/https://doi.org/10.1139/G08-111
Emeriewen, O. F., Richter, K., Flachowsky, H., Malnoy, M., and Peil, A. (2021). Genetic Analysis and Fine Mapping of the Fire Blight Resistance Locus of Malus × arnoldiana on Linkage Group 12 Reveal First Candidate Genes. Frontiers in Plant Science, 12, 663. https://www.frontiersin.org/articles/https://doi.org/10.3389/fpls.2021.667133/full
Fox, J. (2007). The car package. R Foundation for Statistical Computing. https://rdocumentation.org/packages/car/versions/3.1-1
Fox, J., Weisberg, S., Price, B., Adler, D., Bates, D., Baud-Bovy, G. & Bolker, B. (2019). car: Companion to Applied Regression. R package version 3.0-2. Website. https://CRAN.R-project.org/package=car. Accessed 24 April 2023.
Harshman, J. M., Evans, K. M., Allen, H., Potts, R., Flamenco, J., Aldwinckle, H. S., et al. (2017). Fire blight resistance in wild accessions of Malus sieversii. Plant Disease, 101, 1738–1745. https://apsjournals.apsnet.org/doi/https://doi.org/10.1094/PDIS-01-17-0077-RE
Kassambara A, Mundt F. (2017). Package ‘factoextra’. Extract and visualize the results of multivariate data analyses. http://www.sthda.com/english/rpkgs/factoextra.
Khan, A., and Korban, S. S. (2022). Breeding and genetics of disease resistance in temperate fruit trees: challenges and new opportunities. Theoretical Applied Genetics, 13511, 135:3961–3985. https://springerlink.bibliotecabuap.elogim.com/article/https://doi.org/10.1007/s00122-022-04093-0
Khan, M. A., Duffy, B., Gessler, C., and Patocchi, A. (2006). QTL mapping of fire blight resistance in apple. Molecular Breeding, 17, 299–306. https://springerlink.bibliotecabuap.elogim.com/article/https://doi.org/10.1007/s11032-006-9000-y
Khan, M. A., Durel, C. E., Duffy, B., Drouet, D., Kellerhals, M., Gessler, C., et al. (2007). Development of molecular markers linked to the “Fiesta” linkage group 7 major QTL for fire blight resistance and their application for marker-assisted selection. Genome, 50, 568–577. https://cdnsciencepub.com/doi/https://doi.org/10.1139/G07-033
Kharadi, R. R., Schachterle, J. K., Yuan, X., Castiblanco, L. F., Peng, J., Slack, S. M., Zeng, Q., & Sundin, G. W. (2021). Genetic Dissection of the Erwinia amylovora Disease Cycle. Annual Review Phytopathology, 191–212. https://doi.org/10.1146/ANNUREV-PHYTO-020620-095540
Kortekamp, A. (2006). Expression analysis of defense-related genes in grapevine leaves after inoculation with a host and a non-host pathogen. Plant Physiology and Biochemistry, 44(1), 58–67. https://doi.org/10.1016/J.PLAPHY.2006.01.008
Kostick, S. A., Norelli, J. L., and Evans, K. M. (2019). Novel metrics to classify fire blight resistance of 94 apple cultivars. Plant Pathology, 68, 985–996. https://onlinelibrary.wiley.com/doi/full/https://doi.org/10.1111/ppa.13012
Kostick, S. A., Norelli, J. L., Teh, S. L., and Evans, K. M. (2021). Quantitative variation and heritability estimates of fire blight resistance in a pedigree-connected apple germplasm set. J. Plant Pathology, 103, 65–75. https://springerlink.bibliotecabuap.elogim.com/article/https://doi.org/10.1007/s42161-020-00543-0
Kruijer, W., Boer, M. P., Malosetti, M., Flood, P. J., Engel, B., Kooke, R., Keurentjes, J. J. B., & van Eeuwijk, F. A. (2014). Marker-based estimation of heritability in immortal populations. Genetics, 199(2), 379–398. https://doi.org/10.1534/GENETICS.114.167916/-/DC1
Leonards-Schippers, C., Gieffers, W., Schafer-Pregl, R., Ritter, E., Knapp, S. J., Salamini, F., et al. (1994). Quantitative resistance to Phytophthora infestans in potato: a case study for QTL mapping in an allogamous plant species. Genetics, 137, 67–77. https://academic.oup.com/genetics/article/137/1/67/6012520
Li, L., Zhang, Q., and Huang, D. (2014). A review of imaging techniques for plant phenotyping. Sensors (Switzerland), 14, 20078–20111. https://www.mdpi.com/1424-8220/14/11/20078
Longzhou, L., Xiaojun, Y., Run, C., Junsong, P., Huanle, H., Lihua, Y., Yuan, G., & Lihuang, Z. (2008). Quantitative trait loci for resistance to powdery mildew in cucumber under seedling spray inoculation and leaf disc infection. Journal of Phytopathology, 156(11–12), 691–697. https://doi.org/10.1111/j.1439-0434.2008.01427.x
Martínez-Bilbao, A., Ortiz-Barredo, A., Montesinos, E., and Murillo, J. S. (2009). Evaluation of a Cider Apple Germplasm Collection of Local Cultivars from Spain for Resistance to Fire Blight (Erwinia amylovora) Using a Combination of Inoculation Assays on Leaves and Shoots. Horticulture Science, 44, 1223–1227 https://journals.ashs.org/hortsci/view/journals/hortsci/44/5/article-p1223.xml
Mir, R. R., Reynolds, M., Pinto, F., Khan, M. A., and Bhat, M. A. (2019). High-throughput phenotyping for crop improvement in the genomics era. Plant Science, 282, 60–72. https://pubmed.ncbi.nlm.nih.gov/31003612/
Moragrega, C., Llorente, I., Manceau, C., & Montesinos, E. (2003). Susceptibility of European pear cultivars to Pseudomonas syringae pv. syringae using immature fruit and detached leaf assays. European Journal of Plant Pathology, 109(4), 319–326. https://doi.org/10.1023/A:1023574219069/METRICS
Mutka, A. M., and Bart, R. S. (2015). Image-based phenotyping of plant disease symptoms. Front. Plant Science, 5, 734. https://www.frontiersin.org/articles/https://doi.org/10.3389/fpls.2014.00734/full
Panter, S. N., & Jones, D. A. (2002). Age-related resistance to plant pathogens. Advances in Botanical Research, 38. https://doi.org/10.1016/s0065-2296(02)38032-7
Peil, A., Garcia-Libreros, T., Richter, K., Trognitz, F. C., Trognitz, B., Hanke, M. V., & Flachowsky, H. (2007). Strong evidence for a fire blight resistance gene of Malus robusta located on linkage group 3. Plant Breeding, 126(5), 470–475. https://doi.org/10.1111/j.1439-0523.2007.01408.x
Peil, A., Hanke, M. V., Flachowsky, H., Richter, K., Garcia-Libreros, T., Celton, J. M., et al. (2008). Confirmation of the fire blight QTL of Malus × robusta 5 on linkage group 3. Acta Horticulturae, 793, 297–304. https://www.actahort.org/books/793/793_44.htm
Peil, A., Hübert, C., Wensing, A., Horner, M., Emeriewen, O. F., Richter, K., et al. (2019). Mapping of fire blight resistance in Malus × robusta 5 flowers following artificial inoculation. BMC Plant Biology, 19, 1–12. https://bmcplantbiol.biomedcentral.com/articles/https://doi.org/10.1186/s12870-019-2154-7
Peil, A., Emeriewen, O. F., Khan, A., Kostick, S., and Malnoy, M. (2021). Status of fire blight resistance breeding in Malus. Journal of Plant Pathology, 103, 3–12. https://springerlink.bibliotecabuap.elogim.com/article/https://doi.org/10.1007/s42161-020-00581-8
Perez, L. B., & Brown, P. J. (2014). The role of ROS signaling in cross-tolerance: From model to crop. Frontiers in Plant Science, (DEC), 1–6. https://doi.org/10.3389/FPLS.2014.00754/BIBTEX
R Core Team. (2021). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.r-project.org/
R Core Team. (2022). R: A language and environment for statistical computing. R Foundation for Statistical Computing. https://www.R-project.org/
Schindelin, J., Arganda-Carreras, I., Frise, E., Kaynig, V., Longair, M., Pietzsch, T., et al. (2012). Fiji: an open-source platform for biological-image analysis. Nature Methods, 9, 676–682 https://www.nature.com/articles/nmeth.2019
Wallin, J., Bogdan, M., Szulc, P. A., Doerge, R. W., and Siegmund, D. O. (2021). Ghost QTL and hotspots in experimental crosses: novel approach for modeling polygenic effects. Genetics. https://academic.oup.com/genetics/article/217/3/iyaa041/6067404
Wang, P., Yin, L., Liang, D., Li, C., Ma, F., & Yue, Z. (2012). Delayed senescence of apple leaves by exogenous melatonin treatment: Toward regulating the ascorbate–glutathione cycle. Journal of Pineal Research, 53(1), 11–20. https://doi.org/10.1111/J.1600-079X.2011.00966.X
van de Weg, E., Di Guardo, M., Jänsch, M., Socquet-Juglard, D., Costa, F., Baumgartner, I., et al. (2018). Epistatic fire blight resistance QTL alleles in the apple cultivar ‘Enterprise’ and selection X-6398 discovered and characterized through pedigee-informed analysis. Molecular Breeding, 38, 1–18. https://springerlink.bibliotecabuap.elogim.com/article/https://doi.org/10.1007/s11032-017-0755-0
Zendler, D., Malagol, N., Schwandner, A., Töpfer, R., Hausmann, L., and Zyprian, E. (2021). High-Throughput Phenotyping of Leaf Discs Infected with Grapevine Downy Mildew Using Shallow Convolutional Neural Networks. Agronomy, 11, 1768 https://www.mdpi.com/2073-4395/11/9/1768/htm
Acknowledgements
This research was funded by the New York State Department of Agriculture & Markets, Apple Research & Development Program (ARDP), grant number: CM04068AQ, Khan. We would like to acknowledge the USDA (United States Department of Agriculture) Plant Genetic Resources Unit (PGRU) in Geneva, New York, for maintaining the US national Malus collection, where all plant material was collected.
Author information
Authors and Affiliations
Contributions
A.K., conceptualized, designed, and managed the project. R.T., and D.H., done the experiments, analysis and interpretation. R.T., drafted the manuscript. R.T., and D.H., K.R., and A.K., revised, and finalized the manuscript. All authors read and approved the final version.
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing interests.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Tegtmeier, R., Hickok, D., Robbins, K. et al. An image-analysis based leaf disc assay for the rapid evaluation of genetic resistance to fire blight in apples. Eur J Plant Pathol 168, 249–259 (2024). https://doi.org/10.1007/s10658-023-02750-8
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10658-023-02750-8