Abstract
To improve biocontrol approaches in Saudi Arabia, it is necessary to collect and screen suitable native Trichoderma strains. In this study, the biocontrol potential of 20 native Trichoderma asperellum strains was assessed using dual culture and antibiosis assays against Fusarium oxysporum f. sp. radicis-cucumerinum (Forc), the causal agent of Fusarium root and stem rot (FRSR) in cucumber plants. We identified two T. asperellum strains (TAS23 and TAS27) with the highest in vitro antagonistic capacity against Forc. The compatibility between the two strains was identified in vitro. We found that treating cucumber plants with these antagonistic strains separately was effective in delaying the occurrence of FRSR in greenhouse trials. However, treatment with strain mixture TASMix (TAS23 + TAS27) had a synergistic effect and resulted in the highest reduction (P < 0.05) in disease incidence and severity index by 51% and 59.6%, respectively. The decrease in growth due to pathogen-induced stress was significantly less in the TASMix-treated plants than in those treated with individual strains. Real-time PCR assay revealed that the reduction of FRSR in plants treated with TASMix was accompanied by a significant reduction in Forc populations in cucumber stems and rhizosphere. The results of this study suggest that TASMix-controlled FRSR is achieved by reducing reactive oxygen species accumulation, limiting cellular damage, and increasing the activities of antioxidant enzymes in cucumber roots. In summary, a synergistic approach with the application of a mixture of native Trichoderma strains seems promising for managing FRSR in cucumber under organic farming conditions in semi-arid regions.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cucumber (Cucumis sativus L.) is an important vegetable worldwide, including Saudi Arabia; however, its production is greatly hampered by Fusarium oxysporum f. sp. radicis-cucumerinum D.J. Vakalounakis (Forc) (Lievens et al., 2007). This necrotrophic soil-borne pathogen causes Fusarium root and stem rot of cucumber (FRSR), which often appears as an epidemic (Rose et al., 2003). In Saudi Arabia, FRSR has become more widespread in commercial cucumber-greenhouses around Al Kharj area of Riyadh region, and is now severely reducing crop production in susceptible cultivars, with disease incidence ranging from 45 to 60% (El_komy et al., 2021). The common symptoms of FRSR include wilting, yellowing, crown and root rot, and stem disintegration, which can result in severe yield loss and even the death of the plant, especially when grown under high temperatures (El_komy et al., 2021). Forc can survive as chlamydospores for several years in the soil or embedded in plant debris, which makes it difficult to control (Pavlou & Vakalounakis, 2005). Farmers generally apply fungicide-based strategies (Pavlou & Vakalounakis, 2005; Rose et al., 2003) to prevent this disease. However, this method is costly and has hazardous effects on the environment and farmers’ health. Furthermore, there is concern regarding the possibility of the emergence of new fungicide-resistant strains of pathogens (Pavlou & Vakalounakis, 2005). Therefore, alternative measures are urgently required to control this disease in an eco-friendly and economical way.
In this regard, the incorporation of biocontrol agents belonging to the genus Trichoderma (Ascomycota, Hypocreales) within an integrated control system offers a safer and ecologically acceptable alternative to chemical control, especially in organic agriculture (Aleandri et al., 2015; El_komy et al., 2015). Trichoderma spp. are soil inhabitants and plant symbionts that are relatively easy to isolate and grow quickly in the soil and plant rhizosphere (Pimentel et al. 2020; Hermosa et al., 2014). Isolates of several Trichoderma species have been reported to effectively reduce soilborne diseases in greenhouses as well as field crop production (Pimentel et al. 2020; Rojo et al., 2007; Abo-Elyousr et al., 2009; El_komy et al., 2016). They act as biocontrol agents through competition for space and/or nutrients; production of cell wall degrading enzymes and antifungal, diffusible, and volatile metabolites; and mycoparasitism (Howell, 2003; Benítez et al., 2004). In addition to these properties, certain Trichoderma species are able to trigger host defenses against some plant pathogens and improve plant growth by greatly boosting their biological arsenal, thereby acting as biofertilizers and/or antagonists (biopesticides) to pathogens (Howell, 2003; Benítez et al., 2004; Pimentel et al. 2020). Furthermore, Trichoderma can promote plant-beneficial microbial communities and decrease pathogen attack in general (Tandon et al., 2018).
Trichoderma strains are important resources for many commercial biopesticides, but their successful application in commercial agriculture is largely affected by environmental conditions (Yu et al., 2021). The environment affects the colonization, survival, and efficiency of biocontrol agents (Guetsky et al., 2002; Khan et al., 2020). Saudi Arabia is one of the most arid regions in the world and is characterized by harsh environmental conditions, including a lack of soil moisture and poor fertility, represented by a low content of organic matter as well as high calcium carbonate content and salinity (Khan et al., 2020). Therefore, collecting and screening suitable native antagonistic Trichoderma strains would improve biocontrol management strategy in such arid regions as these strains are naturally adapted to harsh arid ecosystems and are expected to exhibit better performance than exotic strains in the rhizosphere soil (Aleandri et al., 2015; El_Komy et al., 2020; Yu et al., 2021).
The majority of biocontrol approaches rely on the application of a single biocontrol agent for disease suppression; however, the effect of a single agent may not be stable under diverse soil environments in different pathosystems (Cong et al., 2019; El_komy et al., 2020). In recent years, the strategy of mixing several antagonistic strains into a single inoculum that could theoretically adapt more readily to a wide range of environmental conditions has been studied (Cong et al., 2019; El_komy et al., 2020; Guetsky et al., 2002; Liu et al., 2018). It suggests a stronger antagonistic potential, attributed to the synergistic action of multiple biocontrol agents as compared to that of a single strain (Chemeltorit et al., 2017; Guetsky et al., 2002; Mendoza García et al., 2003). This combination of beneficial properties is interesting and attractive for plant protection. In previous studies, multiple strain mixtures of biocontrol agents have been shown to be highly effective in controlling soil-borne pathogens (Guetsky et al., 2002; Abo-Elyousr et al., 2009; Singh & Singh, 2012; Huang 2020; Jangir et al., 2019; Karuppiah et al., 2019), these include mixtures of fungi (Aleandri et al., 2015; Cong et al., 2019), bacteria (El_komy et al., 2020), and bacteria and fungi (Chemeltorit et al., 2017). However, not all biocontrol mixtures lead to increased control efficacy (Jambhulkar et al., 2018). In some cases, lower disease incidences have been reported (Janousek et al., 2009) while in the others, no reduction in disease incidence has been observed (Martinez-Medina et al., 2004).
In this study, we hypothesized that the effectiveness of an antagonistic mixture targeting a specific pathogen would have a synergistic effect on the success of biological control because of additive or synergistic effects. To test this hypothesis, the antagonistic potential of 20 native T. asperellum strains was tested against Forc in vitro. The potential of two selected strains was further evaluated, either separately or as a mixture, in controlling FRSR and improving cucumber growth in vivo. This study may reveal potential strains for developing effective biocontrol agents against FRSR in semi-arid regions.
Materials and methods
Fungal strains and inoculum preparation
We used a strain of Fusarium oxysporum f. sp. radicis-cucumerinum (GenBank accession no. MW471132), previously characterized as a highly virulent pathogen of cucumber plants (El_komy et al., 2021). Twenty strains of Trichoderma asperellum (TAS10 to TAS30), isolated from the rhizosphere of healthy cucumber plants in production fields in the Riyadh region, Saudi Arabia, were chosen as antagonistic fungi. These strains were identified using morphological characteristics (Samuels et al., 1999). To confirm this morphological identification, the rDNA internal transcribed spacer (ITS), translation elongation factor 1-α (tef1-α) and RNA Polymerase II subunit (rpb2) genes were PCR-amplified. A single fragment of ∼ 550-bp, ∼ 700-bp, and ∼1100-bp was amplified from each strain for ITS, tef1-α, and rpb2 regions, respectively. Amplicons were sent for sequencing by Macrogen, (Seoul, Korea), and the DNA sequences were searched against the GenBank database using BLAST and hence were identified as T. asperellum (data not published). All fungal strains were routinely cultivated on potato dextrose agar (PDA) plates at 28 °C. For long-term storage, the fungi were preserved in 30% glycerol conidial suspension at -80 °C.
Fungal inocula were prepared by culturing fungal strains individually on plates containing PDA (Difco Laboratories, Detroit, MI, USA) for 2 weeks at 28 °C. Conidia were harvested by adding 5 mL water to the plates and scraping the culture with a sterile glass rod. These suspensions were filtered through a double layer of cheesecloth to separate fungal hyphae and residue from the conidia. The concentration of the conidial suspension was adjusted to 1 × 107 conidia mL−1 using a hemocytometer. The quantity of the inoculum to be used was calculated to achieve a final concentration of 103 conidia g−1 soil for Forc and 106 conidia g−1 of soil for the T. asperellum strains (El_komy et al., 2016).
Antagonistic potential of Trichoderma asperellum strains
Dual culture technique
T. asperellum strains were tested for their antagonism against Forc using the dual-culture technique, as described by Morán-Diez et al. (2019), with some modifications. The experiment was performed in a completely randomized design with five replicates for each strain. The PDA plates were inoculated with two 5 mm-diameter mycelial discs of 7-day-old cultures, one of the pathogenic strain and one, of the biocontrol strain, placed 7 cm apart from each other. The Forc discs were placed in the plates 2 days before the Trichoderma discs. The cultures of Forc growing alone were used as the control treatment. All the plates were incubated at 25 °C in the dark. After 12 days of dual culture, pathogen colony diameters were measured, and the biocontrol strains were classified with regard to antagonism according to the scale by Bell et al. (1982). The percentage of fungal inhibition (FI) was calculated in relation to the growth of the controls as follows: FI = (C-T/C) × 100, where C = colony area of Forc (cm2) alone in the control, and T = colony area of Forc (cm2) in the presence of Trichoderma strains. The interaction zone between the two fungal cultures, was also observed by scanning electron microscopy (SEM, JSM-6380-LA, JEOL, Japan), looking for signs of mycoparasitism. The experiment was repeated twice.
Antibiosis assays
To determine the effect of hydrolytic enzymes and metabolites secreted on PDA, the antagonistic capacity of Trichoderma strains was analyzed using cellophane membranes assays Morán-Diez et al. (2019). Briefly, mycelial discs (5 mm) of 7-day-old cultures of each Trichoderma strain were first cultured individually on PDA plates overlaid with cellophane and incubated for 3 days at 25 °C. After incubation, the cellophane membrane with Trichoderma mycelia was removed. On the same medium, a Forc culture disc (diameter, 0.5 cm) was placed at the center of the plate. Forc cultures grown on PDA were used as controls. Five replicates were maintained for each treatment. Pathogen colony diameters were measured 5 days after incubation, and the FI % was determined as described above. The experiment was repeated twice.
Biocontrol assays under greenhouse conditions
Biocontrol trials were conducted to investigate the potential of T. asperellum TAS23 and TAS27 separately or as a mixture to control FRSR and enhance growth in cucumber plants under greenhouse conditions. The compatibility between the two strains was identified in vitro using the dual culture method, with the absence of inhibition halos or overgrowth (Supplementary Fig. S1). The trials were conducted in a completely randomized design with eight treatments: (T1) non-infested soil (healthy control), (T2) non-infested soil treated with TAS23, (T3) non-infested soil treated with TAS27, (T4) non-infested soil treated with TASMix (TAS23 + TAS27), (T5) soil infested with Forc, (T6) soil infested with Forc and treated with TAS23, (T7) soil infested with Forc and treated with TAS27, and (T8) soil infested with Forc and treated with TASMix. Each treatment consisted of 15 replicates, with two plants per replicate (pot). Cucumber Beta Alpha (Cucumis sativus L. ‘Beit Alpha’), which is susceptible to Forc, was used in this study (El_komy et al., 2021). To check for potential pathogens, seeds were surface-sterilized by immersion in 1% sodium hypochlorite for 30 s, washed thrice with sterile distilled water, and then placed aseptically on PDA plates. Ten replicates were prepared, each with 10 seeds. The plates were incubated at 23 °C and observed daily for 7 days. Seedborne pathogens were not detected throughout the cucumber seeds. Plastic pots (15 cm in diameter) were filled with a sterilized mixture (2:1 sandy loam-peat moss, v/v). For soil infestation, Forc conidial suspension was added to the potting soil to obtain 103 conidia g−1 soil and left undisturbed for one week for pathogen establishment. For the soil Trichoderma application, the inoculum of the biocontrol strains was applied (106 conidia g−1 of soil) and left for one week for establishment. After treatment, the surface-sterilized cucumber seeds were sown in the soil (four seeds per pot) and watered as needed. Eight days after planting (DAP), the cucumber seedlings were reduced to two plants per pot. Plants in all treatments were fertilized with basal doses of NPK (40, 90, and 60 mg kg−1 of soil, respectively) in the form of ammonium sulfate (20.5% N), superphosphate (15% P2O5), and potassium sulfate (48% K2O). To prevent outbreaks of downy mildew, plants were sprayed with copper oxychloride (2 g L−1) when the climate was highly humidity. The pots were kept under greenhouse conditions at 25 °C with a 12 h photoperiod and 60% relative humidity for 45 DAP. The greenhouse assay was repeated once.
Disease assessment
At the end of the experiments (45 DAP), the cucumber plants were checked for infection by examining their roots and crowns for any rot or discoloration of the vascular system and possible isolation of the pathogen on Komada’s medium. The plants were classified as healthy when disease symptoms were absent, and the pathogen was not isolated (Pavlou & Vakalounakis, 2005). Disease incidence (DI) was expressed as a percentage of the number of diseased plants divided by the total number of plants (Pavlou & Vakalounakis, 2005). Disease severity was rated on a scale of 0–4, where 0 = symptomless tissue, 1 = discoloration and rotting (< 25%), 2 = discoloration and rotting (20–50%), 3 = discoloration and rotting (51–75%), and 4 = discoloration and rotting (76–100%). The disease severity index (DSI) was calculated using the formula: DSI = [Σ (rating × number of plants rated)/(total number of plants × the highest rating)] × 100 (Chen et al., 2019).
The synergistic effect of TASMix (TAS23 + TAS27) in controlling FRSR was estimated using Abbott’s formula (Levy et al., 1986). First, control efficiency (E) was determined for each treatment using the estimates of DI and DSI in treated and control plants (Yti and Yc, respectively) using the following formula: E = [(Yti – Yc)/Yc] × 100. Second, the expected control efficiency (Ee) for the TASMix treatment was calculated using the formula: Ee = (ETAS23 + ETAS27) – (ETAS23 × ETAS27)/100, where ETAS23 and ETAS27 are the E data observed for the single application of T. asperellum strains TAS23 and TAS27, respectively. The observed control efficiency (Eobs) was estimated from the E data for the TASMix treatment. A synergism factor (SF) was estimated using the equation: SF = Eobs / Ee. As a decision rule, when SF > 1, the interaction is defined as synergistic, when SF = 1, the interaction is defined as additive, and when SF < 1, the interaction is defined as antagonistic (Levy et al., 1986).
Plant growth measurements
For each treatment, the daily germination percentage of seeds was recorded for 8 DAP. The germination rate index (GRI) was evaluated using the formula GRI = G1/1 + G2/2 + Gx/x, where G1 = germination percentage at 1 DAP; G2 = germination percentage at 2 DAP; and Gx = germination percentage at x DAP (Esechie, 1994). At the end of the experiments (45 DAP), lengths (cm) of both shoot and root systems as well as their respective dry weights (g; after oven drying at 65 °C for 72 h), were recorded for 10 plants in each treatment. The synergistic effect of TASMix in promoting plant growth was estimated, as described above.
Extraction of total genomic DNA
Plant DNA
Stem portions of 2 cm length immediately above ground were cut from each plant for each treatment at 45 DAP, washed with tap water to remove soil particles, surface sterilized with sodium hypochlorite (1%) for 30 s, washed with sterile water, blotted with paper, individually packed in aluminum foil, and finally frozen at -80 °C until DNA extraction. Briefly, DNA was extracted from 200 mg of cucumber stems (three separate stem portions for each treatment) using the DNeasy Plant Mini Kit (QIAGEN, Hilden, Germany).
Soil DNA
Total soil DNA was extracted from 200 mg rhizosphere soil samples (three replicates for each treatment) using the PowerSoil® DNA Isolation Kit (MoBio Laboratories Inc., Carlsbad, CA, USA) according to the manufacturer’s instructions.
Fungal DNA
Fungal genomic DNA was extracted from a pure culture of the fungus using the DNeasy Plant Mini Kit and diluted to 50 ng μl−1 with Milli-Q water. Qualitative and quantitative analyses of the DNA were performed using a NanodropND-2000 spectrophotometer (Thermo Fisher, USA). All isolated DNA was dissolved in 100 μl of nuclease-free water and stored at -20 °C for further analysis.
Quantitative real-time PCR assay
Quantification of Forc DNA in cucumber stem and rhizosphere soil samples was performed by quantitative PCR (qPCR) using specific ForcF1 (5′-GGTGACGCAGCAGTCTAGA-3′) and ForcR2 (5′-GTGACGCAGGGTAGGCAT-3′) primers (Lievens et al., 2007). The qPCR assay was performed with the GeneAmp 7900HT Sequence Detection System (Applied Biosystems) in 20 μL reaction mixture containing 10 μL of 2 × SYBR Green I Master Mix (Roche, Basel, Switzerland), 100 nM of each primer, and 12.5 ng total DNA. Amplification was done at 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s, and 60 °C for 1 min. The qPCR reactions were performed for the three biological replicates of each treatment, with three technical replicates each. Each assay also included a negative control with sterile water in place of the DNA template. For template DNA quantification, a standard curve based on threshold cycles (Ct) for serial dilution of the fungal DNA was conducted (Filion et al., 2003).
In situ reactive oxygen species (ROS; superoxide) accumulation
In situ accumulation of superoxide (O2•−) was visualized by staining of cucumber roots with nitro blue tetrazolium chloride (NBT) following the method of Chen et al. (2019). Briefly, freshly harvested roots were vacuum infiltrated directly with 0.1 mg mL−1 NBT in 25 mM phosphate buffer (pH 6.0) for 5 h. The roots were then rinsed in 80% (v/v) ethanol for 10 min at 70 °C, mounted in lactic acid/phenol/water (1:1:1, v/v), and photographed with a stereomicroscope (Leica MZ125).
Electrolyte leakage (EL) and malondialdehyde (MDA) assays
At 45 dpi, 1 cm long root fragments from four plants from each treatment (representing four biological replicates) were used for the EL assay as described by Yan et al. (2020). Washed root fragments were incubated in 20 ml of deionized water at 30 °C for 4 h, following which, the electrical conductivity (EC) of the solution was measured at 25 °C (reading one). The samples were then boiled for 10 min and cooled to room temperature (25 °C), and the conductivity was measured (reading two). The EL, in percentage, was calculated as follows: EL (%) = (value of reading one/value of reading two) × 100. To measure lipid peroxidation (LP) in the root tissues, malondialdehyde (MDA) content was quantified based on the total 2-thiobarbituric acid (TBA) reaction according to Heath and Packer (1968). First, root tissues were ground in liquid nitrogen, and then 500 mg of tissue powder was extracted in 0.1% trichloroacetic acid (TCA) in a ratio of 1:5 (w/v). The mixture was centrifuged at 12,000 g for 30 min at 4 °C, and the supernatant was collected. Briefly, the reaction was initiated after the addition of 500 μL of root extract to 2 mL of 20% TCA containing 0.5% TBA for 30 min in a boiling water bath. The reaction was then stopped in an ice bath. Subsequently, the samples were centrifuged for 15 min at 9000 g, and the specific and non-specific absorption values were recorded at 532 and 600 nm, respectively. LP concentration was expressed as mmol g−1 FW of the MDA-TBA complex formed using an extinction coefficient of 155 mM−1 cm−1.
Determination of enzyme activities
At 45 dpi, four separate root samples for each treatment were ground into a fine powder in a mortar with a pestle with liquid nitrogen. Then, a total of 200 mg of tissue powder was homogenized in 2 mL of 100 mM TRIS–HCl buffer (pH 8) containing 100 mM ethylenediaminetetraacetic acid (EDTA), 100 mM sodium borate, 1 mM phenylmethylsulfonyl fluoride (PMSF), 10% glycerol, and 2% (wt vol−1) polyvinylpolypyrrolidone (PVPP). After centrifugation at 12,000 × g for 15 min at 4 °C, and the supernatant was assayed for enzymatic activities (Velikova et al. 2000). Total protein concentrations of the extracts were measured according to Bradford (1976). For the blank treatments, the reaction mixtures containing heat-inactivated enzyme extract (boiled for 5 min) were used. The catalase (CAT) activity was assayed by measuring the rate of H2O2 disappearance at the absorbance of 240 nm over 1 min at 25 °C Kar and Mishra (1976). Briefly, the reaction was started by adding 50 μL of the crude enzyme extract to 750 μL of a reaction mixture consisting of 25 mM phosphate buffer (pH 7.0) and 10 mM H2O2. Based on an extinction coefficient of 36 M−1 cm−1, enzyme activity was expressed as μmol of H2O2 oxidized mg−1 of protein min−1 (Anderson et al. 1995). Peroxidase (POX) activity was assayed following the method of Kar and Mishra (1976), which measures the colorimetric determination of pyrogallol oxidation at 420 nm for 1 min at 25 °C. Briefly, the reaction was started by additing 50 μL of the crude enzyme extract to 750 μL of a reaction mixture containing of 25 mM potassium phosphate buffer (pH 6.8), 150 μL of 20 mM pyrogallol, and 50 μL of 20 mM H2O2. Based on an extinction coefficient of 2.47 mM−1 cm−1, the POX activity was expressed in μmol of purpurogalin mg−1 of protein min−1 (Chance and Maehley 1955). Following the same method for POX activity, polyphenoloxidase (PPO) enzymatic activity was measured in cucumber roots but H2O2 was not used in the substrate mixture (Fortunato et al., 2012). The phenylalanineammonia lyase (PAL) was assayed following the method of Zucker (1965). The enzymatic reaction was started by adding 50 μL of the plant extract to 400 μL of a reaction mixture containing of 50 mM sodium borate buffer (pH 8.8) and 400 μL of 20 mM L-phenylalanine and, then the reaction mixture was incubated in a water bath for 1 h at 25 °C. Next, the reaction was ended by adding 10 μL of 6 N HCl before measuring the absorbance of the transcinnamic acid derivatives at 560 nm. Based on an extinction coefficient of 104 nm−1 cm−1, PAL activity was expressed as nmol min−1 mg−1 protein.
Statistical analysis
Two independent experiments were conducted. The in vitro experiments were conducted in a completely randomized design while the greenhouse trials were conducted in a completely randomized block design. All statistical analyses were performed using SAS Version 9.1 software (SAS Institute Inc., 2003). Prior to analysis of variance (ANOVA), data were validated for assumptions of normality (Kolmogorov–Smirnov test, P > 0.05) and homogeneity of variance across treatments (Levene’s test, P > 0.05). A preliminary analysis of the data showed non-significant (P > 0.05) treatment × experiment interactions, and data from repeated experiments were pooled (Gomez & Gomez, 1984). Data from all variables were subjected to ANOVA using SAS Version 9.1 software (SAS Institute Inc., 2003). Data expressed as percentages were analyzed using the arcsine transformation method (Gomez & Gomez, 1984). All the data were presented as mean values, and Fisher’s least significant difference was used for the analysis of mean separation at a 5% level of probability.
Results
Antagonistic activity against pathogens
The results revealed that all the 20 Trichoderma strains were antagonistic to Forc as they significantly (P < 0.05) inhibited colony radial growth of the pathogen; however, the extent of growth reduction varied significantly among the different strains (Table 1). Classification of the Trichoderma strains with respect to antagonism and the overlapping of the colony was performed according to the scale by Bell et al. (1982) (Table 1). In dual culture assays, the strains TAS23 and TAS27 showed the highest inhibition values (P < 0.05; 85% and 89.2%, respectively) with antagonism class 1, while other Trichoderma strains had antagonism classes of 2–4 (Table 1). These two Trichoderma strains were able to overgrow and sporulate on the colony of Forc within a week (Fig. 1). After hyphae of TAS23 or TAS27 and that of Forc came in contact, the coilings by Trichoderma strains around Forc hyphae, or degradation of pathogenic hyphae as well as parallel growth of these antagonists strains closely associated with the pathogen was detected (Fig. 1).
Visualization of overgrowth and heavy sporulation by Trichoderma asperellum strains TAS23 or TAS27 on the colony of Fusarium oxysporum f. sp. radicis-cucumerinum (Forc) through the dual culture assays on potato dextrose agar medium 12 days after inoculation at 28 ± 1 °C (a). Higher growth can be showed in the control plates on the right, where Forc grows unchallenged by the biocontrol strains. Mycoparasitic action of T. asperellum strains on the mycelium of Forc observed under scanning electron microscopy (b and c); showing the coiling of TAS23 around Forc hyphae (arrows in yellow), degradation (arrows in red) and penetration (arrows in white) of pathogen hyphae as well as parallel growth of TAS27 closely associated with the pathogen (arrows in green)
Testing of the T. asperellum strains for their capability to produce diffusible hydrolytic enzymes and metabolites against Forc revealed that TAS23 and TAS27 showed a significantly higher FI (P < 0.05; 79.6% and 85.2%, respectively) than that of other Trichoderma strains. Hence, the biocontrol strains of T. asperellum TAS23 and TAS27, were selected for further in vivo antagonistic experiments.
Biocontrol of FRSR
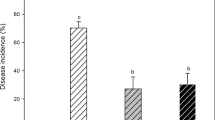
Cucumber plants grown in the presence of Forc alone presented the highest mean values (P < 0.05) for DI (96.8%) and DSI (79.2%) (Table 2). These plants showed wilting, root rot, and reduced length of the main root, along with a dense, pinkish orange mycelial growth and rotting at the base of the stem, all of which can result in severe damage or death of the plant (Fig. 2). Plants treated with the Trichoderma strains, separately or as a mixture, showed less prominent symptoms and slower disease development than the non-treated plants grown on infested soil (Fig. 2). In this regard, DI and DSI reduced significantly when TAS27 (34.0% and 40.0%, respectively), and TAS23 (20.2% and 26.3%, respectively), were incorporated into the pathogen-infested soil, indicating that application of these biocontrol agents could reduce the occurrence of disease (Table 2). However, the best control efficacy of DI (51%) and DSI (59.6%) was obtained in TASMix-treated plants (Table 2). No symptoms were observed in healthy control plants (Fig. 2). According to Abbott’s formula, TASMix had a synergistic effect (SF > 1) in controlling FRSR (Supplementary Table. S1).
Disease symptoms on cucumber plants grown in soil infested with F. oxysporum f. sp. radicis-cucumerinum (Forc) under greenhouse conditions (a). The infected control plants showed wilting, root rot, and reduction in the length of the main root, along with a dense, pinkish-orange mycelial growth and rotting at the base of the stem. Biological control of Fusarium root and stem rot disease in cucumber by T. asperellum strains TAS27 and TAS23 applied separately or as a mixture (TASMix) under greenhouse conditions (b). Healthy control plants (c)
Plant growth measurements
In the greenhouse trials, plant growth parameters were negatively affected by Forc infection, generating a significant (P < 0.05) decrease in all the treatments compared to non-infected controls (Table 3 and Fig. 2). Conspicuously, treatment with only Forc seriously decreased the in the germination index and plant dry weight, and the length of the shoot and root systems, which were significantly (P < 0.05) suppressed compared with the control treatment (43.3, 40.0, 13.4, and 49.0%, respectively; Table 2). Plants treated with both Trichoderma strains, separately or in a mixture, showed a significantly higher (P < 0.05) impact on plant growth parameters, irrespective of the presence or absence of Forc (Table 2). Under infection stress, treatment of TASMix + Forc was the best of all the treatments individually as it generated the highest (P < 0.05) increase in germination index (65.7%), plant dry weight (112%), and the length of the root and shoot systems (35.6 and 134%, respectively). There were synergistic effects of TASMix (SF > 1) on most plant growth parameters under Forc stress (Supplementary Table. S1).
Quantification of Forc in cucumber stem and rhizosphere soil
To confirm the bio-control efficacy on pathogen colonization, the amount of Forc in cucumber stem and rhizosphere soil samples was estimated using specific primers by real-time PCR at 45 DPI. The presence of Forc DNA was detected only in treatments with the pathogen (Fig. 3), indicating that the primer pair ForcF1/ForcF2 for Forc did not cross-react with non-target DNA. Moreover, qPCR detected high amounts of fungal-specific DNA in the treatment of Forc-only on all days post-inoculation, indicating a strong pathogenic infection (Fig. 2). Nevertheless, Forc amounts were significantly (P < 0.05) reduced in all Trichoderma-inoculated treatments (Fig. 2). Treatment with TASMix showed the strongest inhibitory effect on Forc population in cucumber stem and rhizosphere soil (2.6-and fourfold, respectively), followed by TSA27 (2-and 2.8-fold, respectively) and TSA23 (1.4-and 2.7-fold, respectively), as compared to the infected control (Fig. 2). This result further supports the presents synergistic effects of the application of TAS23 with TAS27 as a mixture, on suppressing Forc populations.
The effects of Trichoderma asperellum strains, TAS23 and TAS27, applied separately or as a mixture on Forc genomic DNA detected by real-time PCR in stem and rhizosphere soil samples of cucumber plants grown in not-infested and Fusarium oxysporum f. sp. radicis-cucumerinum-infested soil under greenhouse conditions. Each bar represents the average of two experiments, each with three replicates per treatment. Columns marked by an inverted triangle (▼) indicate the absence of Forc genomic DNA. Different letters above the bars indicate significant differences at P < 0.05 according to the Fisher’s LSD test. Error bars denote the standard deviations of the mean
ROS accumulation and EL and MDA levels
NBT staining showed that Forc-infection stress caused an intensive accumulation of O2•− in cucumber roots, as shown by the deep blue color deposits (Fig. 4). However, Trichoderma-inoculated treatments remarkably attenuated Forc-induced O2•− accumulation in roots and TASMix showed the best effect (Fig. 4). To examine the membrane damage caused by excessive ROS, EL and MDA levels were measured in cucumber roots. The results showed that Forc infection resulted in a 2.4- and 3.7-fold increase in root EL and MDA, respectively as compared to that of the non-infected control (Fig. 5). Meanwhile, MDA concentrations reduced significantly with the incorporation of TAS23 and TAS27 individually into the pathogen-infested soil, with values of 9.2 and 7.1 mmol g−1 FW, respectively (Fig. 5). However, these treatments showed insignificant decreases in root EL compared to the infected control (Fig. 5). Interestingly, TASMix treatment was more effective (P < 0.05) in reducing root EL and MDA levels by 1.6-and 2.2-fold, respectively than those of the Forc-only treatment (Fig. 3). No significant differences (P > 0.05) in EL and MDA levels were detected among the treatments in the non-infected plants (Fig. 5).
In situ accumulation of reactive oxygen species in cucumber roots. The roots were stained with nitro blue tetrazolium chloride to indicate superoxide (O2•−) content. Infection with Fusarium oxysporum f. sp. radicis-cucumerinum (Forc) caused an intensive accumulation of O2•− as reflected by the deep blue color deposits, compared with that of healthy control treatment. Treatments with Trichoderma asperellum strains TAS27 and TAS23 applied separately or as a mixture (TASMix) remarkably attenuated Forc-induced ROS accumulation in roots. Bar = 4 mm
The effects of Trichoderma asperellum strains, TAS23 and TAS27, applied separately or as a mixture on relative electrolyte leakage and malondialdehyde (MDA) concentrations in roots of cucumber plants grown in not-infested and Fusarium oxysporum f. sp. radicis-cucumerinum-infested soil under greenhouse conditions. Each bar represents the average of two experiments, each with four replicates per treatment. Different letters above the bars indicate significant differences at P < 0.05 according to the Fisher’s LSD test. Error bars denote the standard deviations of the mean
Enzymatic activity
We found that the enzymatic activities were significantly higher (P < 0.05) in Forc-infected roots than in non-infected roots (Fig. 6). Meanwhile, Trichoderma-treated plants showed greater enzymatic activities at various extents, irrespective of Forc-infection (Fig. 6). All Trichoderma treatments led to increased CAT activity, however, only the treatment of TASMix + Forc statistically outperformed the infected control (Fig. 6). Plants treated with TASMix + Forc showed the highest POX activity (80.7 μmol min−1 mg−1 protein), which was 2.3 times higher than in those with Forc-only treatment while the treatments with TAS23 + Forc and TAS27 + Forc, were 1.8 and 1.6 times higher, respectively. The TAS27 + Forc and TAS23 + Forc treatments also led to a significant increase (P < 0.05) in PPO activity by 1.7-and 1.5 times higher, respectively than the infected control (Fig. 6). However, the dual inoculation of TASMix + Forc showed the highest (P < 0.05) PPO activity (55.3 μmol min−1 mg−1 protein), which was two-fold greater than that in plants with Forc-only treatment (Fig. 6). For PAL activity, Plants treated with TASMix + Forc and TAS27 + Forc showed the highest PAL activity (7.8 and 6.9 nmol min−1 mg−1 protein, respectively), which were 1.8 and 1.6 times higher, respectively than that of plants with Forc-only treatment (Fig. 6).
The effects of Trichoderma asperellum strains, TAS23 and TAS27, applied separately or as a mixture on the enzymatic activities of catalase (CAT), peroxidase (POX), polyphenol oxidase (PPO), and polyphenol oxidase (PPO) in roots of cucumber plants grown in not-infested and Fusarium oxysporum f. sp. radicis-cucumerinum–infested soil under greenhouse conditions. Each bar represents the average of two experiments, each with four replicates per treatment. Different letters above the bars indicate significant differences at P < 0.05 according to the Fisher’s LSD test. Error bars denote the standard deviations of the mean
Discussion
To develop a biocontrol management strategy in semi-arid regions, suitable native antagonistic Trichoderma strains should be selected, as they are naturally adapted to harsh arid ecosystems and are expected to exhibit better performance than exotic strains in the rhizosphere soil (Khan et al., 2020; Yu et al., 2021). However, the success of biological control of plant diseases and improvement of plant growth by Trichoderma can be enhanced by an ideal antagonistic strain mixture (Aleandri et al., 2015; Cong et al., 2019; Guetsky et al., 2002). This study describes a management strategy for Fusarium root and stem rot disease caused by Forc in cucumber plants using a mixture of selected native T. asperellum strains, which were recovered from rhizosphere soil samples of healthy cucumber plants in production fields. In dual culture assays, we screened 20 T. asperellum strains against Forc. Of these, TAS23 and TAS27 showed remarkable inhibition of the pathogen by more than 85%. These strains were able to grow faster than the other Trichoderma strains and colonize a large area of PDA, inhibiting pathogen growth. They also overgrew on Forc culture and exhibited profuse sporulation, indicating that they can be aggressive mycoparasites. The ability to grow rapidly is an important feature that offers great potential for Trichoderma strains to compete with Forc for space and nutrients (El_komy et al., 2015). Our microscopic observations showed coilings of Trichoderma strains around Forc hyphae as well as parallel growth with close hyphal association, which are essential for the mycoparasitism to be successful (Howell, 2003; Benítez et al., 2004; Pimentel et al. 2020). Moreover, the Forc mycelia had abnormal morphology and were lysed, which could be owing to antagonistic compounds produced by TAS23 and TAS27 in the interaction region. Subsequently, in vitro tests showed that these two strains secreted metabolites that inhibited the mycelial growth of Forc on PDA plates by 79.6- 85.2%. Accordingly, in plate assays, the strains exhibited various mechanisms of action, including producing hydrolytic enzymes such as chitinase and b-1,3-glucanase, and siderophores, which might have contributed to their direct antifungal activity (Benítez et al., 2004; Hermosa et al., 2014). These results suggest that in the Trichoderma– pathogen interaction, TAS23 and TAS27 use multiple mechanisms that could be affecting the ability of the Forc to cause disease in cucumber plants. The antagonistic effect exhibited by Trichoderma strains against Forc seems to be associated not only with direct competition and mycoparasitism but also antibiosis.
In this study, we found that the application of TAS23 or TAS27 alone can effectively control FRSR in cucumber plants under greenhouse conditions. However, the TASMix treatment was more (P < 0.05) effective in reducing FRSR incidence and the severity index (51 and 59.6% as compared to Forc-only control plants, respectively) than applying either T. asperellum strains alone. Similarly, Forc colonization within cucumber stems and the rhizosphere reduced significantly in the presence of TASMix. According to Abbott’s formula, the effect of the Trichoderma strain mixture was synergistic (SF > 1). This synergistic disease-suppressing effect represents a promising choice for improving biological control tools and controlling FRSR in semi-arid regions. Trichoderma spp. are effective against soilborne diseases through the use of multiple antagonistic mechanisms, including mycoparasitism, competition, production of antifungal compounds, and induced systemic resistance (Benítez et al., 2004; Hermosa et al., 2014; Howell, 2003). TASMix-cucumber interactions may involve any of these mechanisms acting individually or synergistically, to suppress FRSR. Our hypothesis is supported by many previous studies suggesting that mixed inoculations of antagonistic strains, including Trichoderma spp., were more effective than the use of single strains in the management of plant diseases (Aleandri et al., 2015; Chemeltorit et al., 2017; Chien & Huang, 2020; Cong et al., 2019; Jangir et al., 2019; Karuppiah et al., 2019; Singh & Singh, 2012; Spadaro & Gullino, 2005).
In addition to disease suppression, all Trichoderma treatments greatly enhanced cucumber plant growth, irrespective of the presence or absence of Forc. However, TASMix-treated plants showed the highest increases (P < 0.05) in seed germination index, root and shoot length, and plant dry weight. These beneficial effects on plant growth may also be owing to the synergistic effect of the two Trichoderma strains, which exerted considerable growth over the control plants. The promotion of plant growth by Trichoderma has also been documented (Aleandri et al., 2015; El_komy et al., 2016; Rojo et al., 2007; Singh & Singh, 2012). The interactions of Trichoderma spp. with plants accelerate their growth, enhance the uptake and solubilization of nutrients, change the microflora composition around roots, and activate the host defense system so that the plant is poised to resist potential pathogens (Benítez et al., 2004; Hermosa et al., 2014). Moreover, Trichoderma spp. can produce auxins and gibberellins, which can directly or indirectly modulate plant growth and development (Martínez-Medina et al., 2014). Similar results were reported by Mei et al. (2019) using T. asperellum 525, which significantly promoted the quality and yield of cucumbers. However, understanding the mechanism by which strains TAS23 and TAS27 promote cucumber growth warrants further investigation.
During pathogenesis, the necrotrophic fungus Forc induces excessive production of ROS, including O2•− in the root tissues, which promotes cellular oxidative damage and facilitates the invasion and colonization of Forc in dead tissues (Fortunato et al., 2012). The MDA produced during lipid peroxidation and EL, are reliable indicators of cellular damage that results in infection stress (Fortunato et al., 2012). In this study, minimum levels of MDA and EL were found in TASMix + Forc-treated plants, suggesting that roots of cucumber plants treated with Trichoderma strains reduced Forc colonization. This has been confirmed by the intensive in situ accumulation of O2•− in Forc-only treated roots, which led to extensive pathogen colonization and severe disease symptoms. This is in agreement with Chen et al. (2019), who found that Trichoderma-colonized cucumber roots remarkably decreased MDA content and EL, enhancing resistance to Fusarium wilt disease. In addition, Yu and Luo (2020) showed lower levels of LP in Pinus massoniana seedlings treated with T. koningiopsis, which contributed to better resistance to damping-off and root rot caused by F. oxysporum.
Our results showed that the application of Trichoderma mixture strains TASMix stimulated the biochemical defense responses of cucumber plants, leading to enhanced tolerance to Forc. CAT activity was significantly higher (P < 0.05) in cucumber roots infected with Forc while that in the TASMix + Forc plants was greater than that of the infected control. Such an increase is important as this antioxidant enzyme neutralizes the oxidative stress induced by Forc (De Gara et al., 2003). Thus, higher CAT activity in TASMix + Forc plants could be a strategy to restrict pathogen colonization for attenuating intensive Forc-induced ROS accumulation (Chen et al., 2019). In plants, PAL is directly related to tolerance to pathogenic infection (El_komy et al., 2020; Fortunato et al., 2012). This enzyme modulates L-phenylalanine with the formation of trans-cinnamic acid, which in turn forms phenylpropanoids, leading to the synthesis of several defense-related secondary compounds, such as phenols and lignin (Dallagnol et al., 2011). This suggests that the better protection against Forc development in TASMix plants may be attributed to higher PAL activity in the root tissues. In line with this, Trichoderma-enhanced resistance to Ralstonia solanacearum has been attributed to higher PAL activity in tomato plants (Konappa et al., 2018). Similarly, POX activity was significantly higher (P < 0.05) in the roots of plants treated with TASMix + Forc than in those treated with Forc alone. POX is an antioxidant enzyme commonly involved in host defense against oxidative stress induced by the pathogen, and it also increases cell wall reinforcement and lignification (Campbell & Sederoff, 1996; Chen et al., 2019; Fortunato et al., 2012). Accordingly, the reduced development of FRSR symptoms in TASMix-treated cucumber plants may be linked to high POX activity. Our results also confirmed that the synergistic effect of FRSR by Trichoderma strain mixture may be associated with the high production of PPO, because this enzyme catalyzes the oxidation of phenolics to fungitoxic compounds called o-quinones, and hence, protects cucumber roots from pathogen invasion (El_komy et al., 2020; Vanitha et al., 2009). Likewise, Trichoderma-colonized cucumber plants showed decreased MDA content and increased POX and PPO activity, leading to enhanced tolerance to Fusarium wilt (Mei et al., 2019).
In conclusion, our hypothesis was verified in the sense that exploiting the potential synergistic effects of organisms is an important aspect of the effective and efficient use of biocontrol agents under organic farming conditions in semi-arid regions. The Trichoderma strain mixture, TASMix was superior to individual strains in eliciting protection against FRSR and promoting cucumber growth. However, more efforts are needed to implement this approach in field trials to confirm the above findings.
References
Abo-Elyousr, K. A., Hashem, M., & Ali, E. H. (2009). Integrated control of cotton root rot disease by mixing fungal biocontrol agents and resistance inducers. Crop Protection, 28, 295–301.
Aleandri, M. P., Chilosi, G., Bruni, N., Tomassini, A., Vettraino, A. M., & Vannini, A. (2015). Use of nursery potting mixes amended with local Trichoderma strains with multiple complementary mechanisms to control soil-borne diseases. Crop Protection, 67, 269–278.
Bell, D. K., Wells, H. D., & Markham, C. R. (1982). In vitro antagonism of Trichoderma species against six fungal plant pathogens. Phytopathology, 72, 379–382.
Benítez, T., Rincón, A. M., Limón, M. C., & Codon, A. C. (2004). Biocontrol mechanisms of Trichoderma strains. International Microbiology, 7, 249–260.
Bradford, M. M. (1976). A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of proteindye binding. Analytical Biochemistry, 72, 1151–1154.
Campbell, M. M., & Sederoff, R. R. (1996). Variation in lignin content and composition (mechanisms of control and implications for the genetic improvement of plants). Plant Physiology, 110, 3.
Chemeltorit, P. P., Mutaqin, K. H., & Widodo, W. (2017). Combining Trichoderma hamatum THSW13 and Pseudomonas aeruginosa BJ10–86: A synergistic chili pepper seed treatment for Phytophthora capsici infested soil. European Journal of Plant Pathology, 147, 157–166.
Chen, S. C., Ren, J. J., Zhao, H. J., Wang, X. L., Wang, T. H., Jin, S. D., & Ahammed, G. J. (2019). Trichoderma harzianum improves defense against Fusarium oxysporum by regulating ROS and RNS metabolism, redox balance, and energy flow in cucumber roots. Phytopathology, 109, 972–982.
Chien, Y. C., & Huang, C. H. (2020). Biocontrol of bacterial spot on tomato by foliar spray and growth medium application of Bacillus amyloliquefaciens and Trichoderma asperellum. European Journal of Plant Pathology, 156, 995–1003.
Cong, Y., Fan, H., Ma, Q., Lu, Y., Xu, L., Zhang, P., & Chen, K. (2019). Mixed culture fermentation between Rhizopus nigricans and Trichoderma pseudokoningii to control cucumber Fusarium wilt. Crop Protection, 124, 104857.
Dallagnol, L. J., Rodrigues, F. A., DaMatta, F. M., Mielli, M. V., & Pereira, S. C. (2011). Deficiency in silicon uptake affects cytological, physiological, and biochemical events in the rice–Bipolaris oryzae interaction. Phytopathology, 101, 92–104.
De Gara, L., PintoMC, De., & Tommasi, F. (2003). The antioxidant system Visà- Vis reactive oxygen species during plant-pathogen interaction. Plant Physiology and Biochemistry, 41, 863–870.
El_Komy, M. H., Hassouna, M. G., Abou-Taleb, E. M., Al-Sarar, A. S., & Abobakr, Y. (2020). A mixture of Azotobacter, Azospirillum, and Klebsiella strains improves root-rot disease complex management and promotes growth in sunflowers in calcareous soil. European Journal of Plant Pathology, 156, 713–726.
El_Komy, M. H., Saleh, A. A., Eranthodi, A., & Molan, Y. Y. (2015). Characterization of novel Trichoderma asperellum isolates to select effective biocontrol agents against tomato Fusarium wilt. The Plant Pathology Journal, 31, 50–60.
El_komy, M. H., Al-Qahtani, R. M., Widyawan, A., Molan, Y., & Almasrahi, A. (2021). First Report of Fusarium Root and Stem Rot Caused by Fusarium oxysporum f. sp. radicis-cucumerinum on Greenhouse Cucumbers in Saudi Arabia. Plant Disease, https://doi.org/10.1094/PDIS-01-21-0122-PDN.
El_Komy, M. H., Saleh, A. A., Ibrahim, Y. E., Hamad, Y. K., & Molan, Y. Y. (2016). Trichoderma asperellum strains confer tomato protection and induce its defense-related genes against the Fusarium wilt pathogen. Tropical Plant Pathology, 41, 277–287.
Esechie, H. (1994). Interaction of salinity and temperature on the germination of sorghum. Journal of Agronomy and Crop Science, 172, 194–199.
Filion, M., St-Arnaud, M., & Jabaji-Hare, S. H. (2003). Quantification of Fusarium solani f. sp. phaseoli in mycorrhizal bean plants and surrounding mycorrhizosphere soil using real-time polymerase chain reaction and direct isolations on selective media. Phytopathology, 93, 229–235.
Fortunato, A. A., Rodrigues, F. Á., & do Nascimento, K. J. T. (2012). Physiological and biochemical aspects of the resistance of banana plants to Fusarium wilt potentiated by silicon. Phytopathology, 102, 957–966.
Gomez, K. A., & Gomez, A. A. (1984). Statistical procedures for agricultural research. Wiley, New York. https://doi.org/10.1017/S0014479700014496
Guetsky, R., Shtienberg, D., Elad, Y., Fischer, E., & Dinoor, A. (2002). Improving biological control by combining biocontrol agents each with several mechanism of diseased suppression. Phytopathology, 92, 976–985.
Heath, R. L., & Packer, L. (1968). Photoperoxidation in isolated chloroplasts: I. Kinetics and stoichiometry of fatty acid peroxidation. Archives of Biochemistry and Biophysics, 125, 189–198.
Hermosa, R., Cardoza, R. E., Rubio, M. B., Gutiérrez, S., & Monte, E. (2014). Secondary metabolism and antimicrobial metabolites of Trichoderma. In Biotechnology and biology of Trichoderma (pp. 125–137). Elsevier.
Howell, C. R. (2003). Mechanisms employed by Trichoderma species in the biological control of plant diseases: The history and evolution of current concepts. Plant Disease, 87, 4–10.
Jambhulkar, P. P., Sharma, P., Manokaran, R., Lakshman, D. K., Rokadia, P., & Jambhulkar, N. (2018). Assessing synergism of combined applications of Trichoderma harzianum and Pseudomonas fluorescens to control blast and bacterial leaf blight of rice. European Journal of Plant Pathology, 152(3), 747–757.
Jangir, M., Sharma, S., & Sharma, S. (2019). Target and non-target effects of dual inoculation of biocontrol agents against Fusarium wilt in Solanum lycopersicum. Biological Control, 138, 104069.
Janousek, C. N., Lorber, J. D., & Gubler, W. D. (2009). Combination and rotation of bacterial antagonists to control powdery mildew on pumpkin. Journal of Plant Diseases and Protection, 116, 260–262.
Kar, M., & Mishra, D. (1976). Catalase, peroxidase, and polyphenoloxidase activities during rice leaf senescence. Plant Physiology, 57, 315–319.
Karuppiah, V., Sun, J., Li, T., Vallikkannu, M., & Chen, J. (2019). Co-cultivation of Trichoderma asperellum GDFS1009 and Bacillus amyloliquefaciens 1841 causes differential gene expression and improvement in the wheat growth and biocontrol activity. Frontiers in Microbiology, 10, 1068.
Khan, A. R., El-Komy, M. H., Ibrahim, Y. E., Hamad, Y. K., Molan, Y. Y., & Saleh, A. A. (2020). Organic Management of Tomato Fusarium wilt using a Native Bacillus subtilis Strain and Compost Combination in Saudi Arabia. International Journal of Agriculture and Biology, 23, 1003–1012.
Konappa, N., Krishnamurthy, S., Siddaiah, C. N., Ramachandrappa, N. S., & Chowdappa, S. (2018). Evaluation of biological efficacy of Trichoderma asperellum against tomato bacterial wilt caused by Ralstonia solanacearum. Egyptian Journal of Biological Pest Control, 28, 1–11.
Levy, Y., Benderly, M., Cohen, Y., Gisi, U., & Bassand, D. (1986). The joint action of fungicides in mixtures: Comparison of two methods for synergy calculation. EPPO Bulletin, 16, 651–657.
Lievens, B., Claes, L., Vakalounakis, D. J., Vanachter, A. C., & Thomma, B. P. (2007). A robust identification and detection assay to discriminate the cucumber pathogens Fusarium oxysporum f. sp. cucumerinum and f. sp. radicis-cucumerinum. Environmental Microbiology, 9, 2145–2161.
Liu, K., McInroy, J. A., Hu, C. H., & Kloepper, J. W. (2018). Mixtures of plant-growth-promoting rhizobacteria enhance biological control of multiple plant diseases and plant-growth promotion in the presence of pathogens. Plant Disease, 102, 67–72.
Martínez-Medina, A., Alguacil, M. D. M., Pascual, J. A., & Van Wees, S. C. (2014). Phytohormone profiles induced by Trichoderma isolates correspond with their biocontrol and plant growth-promoting activity on melon plants. Journal of Chemical Ecology, 40, 804–815.
Martinez, A., Obertello, M., Pardo, A., Ocampo, J. A., & Godeas, A. (2004). Interactions between Trichoderma pseudokoningii strains and the arbuscular mycorrhizal fungi Glomus mosseae and Gigaspora rosea. Mycorrhiza, 14, 79–84.
Mei, L. I., Hua, L. I. A. N., Su, X. L., Ying, T. I. A. N., Huang, W. K., Jie, M. E. I., & Jiang, X. L. (2019). The effects of Trichoderma on preventing cucumber Fusarium wilt and regulating cucumber physiology. Journal of Integrative Agriculture, 18, 607–617.
Mendoza García, R. A., Ten Hoopen, G. M., Kass, D. C. J., Sánchez Garita, V. A., & Krauss, U. (2003). Evaluation of mycoparasites as biocontrol agents of Rosellinia root rot in cocoa. Biological Control, 27, 210–227.
Morán-Diez, M. E., Carrero-Carrón, I., Rubio, M. B., Jiménez-Díaz, R. M., Monte, E., & Hermosa, R. (2019). Transcriptomic analysis of Trichoderma atroviride overgrowing plant-wilting Verticillium dahliae reveals the role of a new M14 metallocarboxypeptidase CPA1 in biocontrol. Frontiers in Microbiology, 10, 1120.
Pavlou, G. C., & Vakalounakis, D. J. (2005). Biological control of root and stem rot of greenhouse cucumber, caused by Fusarium oxysporum f.sp. radicis-cucumerinum, by lettuce soil amendment. Crop Protection, 24, 135–140.
Rojo, F. G., Reynoso, M. M., Ferez, M., Chulze, S. N., & Torres, A. M. (2007). Biological control by Trichoderma species of Fusarium solani causing peanut brown root rot under field conditions. Crop Protection, 26, 549–555.
Rose, S., Parker, M., & Punja, Z. K. (2003). Efficacy of biological and chemical treatments for control of Fusarium root and stem rot on greenhouse cucumber. Plant Disease, 87, 1462–1470.
Samuels, G. J., Lieckfeldt, E. L. K. E., & Nirenberg, H. I. (1999). Trichoderma asperellum, a new species with warted conidia, and redescription of T. viride. SYDOWIA-HORN-, 51, 71–88.
Singh, S. P., & Singh, H. B. (2012). Effect of consortium of Trichoderma harzianum isolates on growth attributes and Sclerotinia sclerotiorum rot of brinjal. Vegetable Science, 39, 144–148.
Spadaro, D., & Gullino, M.L. (2005). Improving the efficacy of biocontrol agents against soilborne pathogens. Crop Protection, 24, 601e613.
Tandon, A., Fatima, T., Gautam, A., Yadav, U., Srivastava, S., & Singh, P. C. (2018). Effect of Trichoderma koningiopsis on chickpea rhizosphere activities under different fertilization regimes. Open Journal of Soil Science, 8, 261–275.
Vanitha, S. C., Niranjana, S. R., & Umesha, S. (2009). Role of phenylalanine ammonia lyase and polyphenol oxidase in host resistance to bacterial wilt of tomato. Journal of Phytopathology, 157, 552–557.
Yan, G., Fan, X., Peng, M., Yin, C., Xiao, Z., & Liang, Y. (2020). Silicon improves rice salinity resistance by alleviating ionic toxicity and osmotic constraint in an organ-specific pattern. Frontiers in Plant Science, 11, 260.
Yu, C., & Luo, X. (2020). Trichoderma koningiopsis controls Fusarium oxysporum causing damping-off in Pinus massoniana seedlings by regulating active oxygen metabolism, osmotic potential, and the rhizosphere microbiome. Biological Control, 150, 104352.
Yu, Z., Wang, Z., Zhang, Y., Wang, Y., & Liu, Z. (2021). Biocontrol and growth-promoting effect of Trichoderma asperellum TaspHu1 isolate from Juglans mandshurica rhizosphere soil. Microbiological Research, 242, 126596.
Zucker, M. (1965). Induction of phenylalanine deaminase by light and its relation to chlorogenic acid synthesis in potato tuber tissue. Plant Physiology, 40, 779–784.
Acknowledgements
The authors extend their appreciation to the Deanship of Scientific Research at King Saud University for funding this work through research group No. RG-1440-029.
Author information
Authors and Affiliations
Contributions
MHK designed and performed the experiments, collected the data and wrote the manuscript. RMQ helped in greenhouse experiments. YEI, AAA,and MAS investigation, review & editing the manuscript. All authors reviewed the manuscript critically.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical statement
Authors declared that this manuscript has not published elsewhere. All authors read and approved the final version of this manuscript. The authors declare that the present work was developed without any potential conflict of interest, with no human or animal participants.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
El-Komy, M.H., Al-Qahtani, R.M., Ibrahim, Y.E. et al. Soil application of Trichoderma asperellum strains significantly improves Fusarium root and stem rot disease management and promotes growth in cucumbers in semi-arid regions. Eur J Plant Pathol 162, 637–653 (2022). https://doi.org/10.1007/s10658-021-02427-0
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10658-021-02427-0