Abstract
Race 2SA55 of the wheat stem rust pathogen, Puccinia graminis f. sp. tritici, was described for the first time during 2000 in South Africa. This race is of particular interest as it was the first local report of increased virulence towards barley cultivars by wheat stem rust. Using three original accessions of 2SA55 from the rust collection of the University of the Free State, nine single pustule isolates were established. Phenotyping of isolates to characterise 2SA55 according to the North American nomenclature system revealed variation in virulence for Sr9g on the tester lines Kubanka and Acme, which was not detected in the initial description of 2SA55. Seven isolates coded as BNGSC, which except for virulence on Sr9b resembled the initial avirulence/virulence description for 2SA55. The remaining two isolates with Sr9g virulence coded as BPGSC. The seedling infection types for the two 2SA55 derived races BNGSC and BPGSC on the Stakman differential set as well as on an extended set of Sr lines and 144 South African wheat cultivars and advanced breeding lines revealed no further distinction in reaction between them. Microsatellite analysis indicated that while race BN/PGSC shares phenotypic similarity with several of the South African non-Ug99 races, it was genetically distinct.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Nine races of Puccinia graminis f. sp. tritici (Pgt), three of which were new, were detected in South Africa (SA) from 2009 to 2013 (Terefe et al. 2016). Although no major epidemics have been experienced recently, Pgt continues to evolve and threaten wheat production in SA. The persistent change in variability can be ascribed to foreign incursions and mutation of existing races. Using microsatellite markers, Visser et al. (2009, 2011) showed that the South African Pgt population consists of two major groups, one containing four South African members of the Ug99 race group and another containing several non-Ug99 races, presumed to have developed from races detected in SA at the turn of the nineteenth century. The first variant in the Ug99 group was detected in SA in 2000 (Boshoff et al. 2002), followed by three others in 2007, 2009 and 2010, respectively (Visser et al. 2011; Pretorius et al. 2012b).
One of the older non-Ug99 Pgt races, reported according to the Agricultural Research Council-Small Grain (ARC-SG) notation as race 2SA55, was first collected in SA in 2000 (Boshoff et al. 2002). This race has appeared infrequently in recent years and except for its original avirulence/virulence description has not been considered in a genetic context with other Pgt isolates from the region. In addition, the original description lacks details from an avirulence/virulence perspective for additional stem rust (Sr) resistance genes. The objective of this study was to characterise the race more comprehensively in terms of pathogenicity and microsatellite profile.
Methods
The ARC-SG carries out annual rust surveys in the major wheat producing areas of SA. Phenotyping of field isolates on a standard SA stem rust differential set is done following procedures customary for rust race analysis. Any isolate differing in its phenotype from previously identified SA stem rust races is further characterised using a more complete set of international differential lines.
Phenotyping
Three single pustule isolates of each of three accessions of 2SA55, stored at the University of the Free State as UVPgt54.A, UVPgt54.B and UVPgt54.C, were phenotyped with the appropriate stem rust differential set. In addition, seedling infection types (ITs) for isolates UVPgt54.C1 and UVPgt54.B1 were determined on the Stakman differentials, an extended set of 56 Sr gene tester lines, 144 SA wheat cultivars and advanced breeding lines, and one barley cultivar. Seedling inoculation, incubation in the dew chamber and glasshouse, and infection type evaluations were performed as previously described (Pretorius et al. 2012a).
Microsatellite analysis
The genetic relationship among nine isolates established from the original race 2SA55 and 11 other South African Pgt races was determined using 22 polymorphic microsatellite markers (Karaoglu et al. 2013). The primer pairs used were PgSUN3, PgSUN7, PgSUN8, PgSUN11, PgSUN14, PgSUN16, PgSUN17, PgSUN20, PgSUN22, PgSUN24, PgSUN25, PgSUN28, PgSUN29, PgSUN33, PgSUN35, PgSUN38, PgSUN39, PgSUN40, PgSUN42, PgSUN44, PgSUN47 and PgSUN53.
The 11 SA Pgt races were 2SA4 (SA race annotation), TTKSF (North American race annotation; 2SA88), TTKSF+Sr9 h (2SA88 + Sr9 h), 2SA100, BPGSC+Sr27 (2SA102), BNGSC+Sr27 (2SA103), BPGSC+Sr27,Kw (2SA104), BPGSC+Sr27,Kw,Satu (2SA105), TTKSP (2SA106), PTKST (2SA107) and BFBSC (2SA108) (Terefe et al. 2016). Race TTKSK (Ug99) that was collected in Uganda in 1998 (Pretorius et al. 2000), was also included. For these isolates, genomic DNA prepared from germinated urediniospores and germ tubes was used. For race 2SA55, genomic DNA was extracted from three well separated pustules on infected leaf tissue for each of three isolates using the cetyltrimethylammonium bromide method (CTAB; Saghai-Maroof et al. 1984; Visser et al. 2009).
Each 10 μl PCR reaction contained 2 ng genomic DNA, 1 μM of each primer, and a 1× concentration of KAPA Taq ReadyMix (KAPABIOSYSTEMS, Sigma-Aldrich, Johannesburg, SA). The amplification regime was one cycle of 4 min at 94 °C, followed by 35 cycles of 94 °C for 30 s, recommended annealing temperature for 30 s and 72 °C for 30 s. A 5 min final elongation step at 72 °C was added. After confirming successful amplification on an 1.2% (w/v) agarose gel, the amplified DNA was separated on a 5% (w/v) denaturing polyacrylamide gel using 1× TBE (89 mM Tris-borate pH 8, 2.0 mM EDTA) as running buffer (Sambrook et al. 1989). DNA fragments were visualized using silver staining (Silver Sequence™ DNA Sequencing System; Promega).
Statistical analysis of microsatellite data
A multi-locus allelic data matrix based on the microsatellite data was generated for all genotypes. The genetic relationships amongst the South African Pgt races were determined using an unrooted dendrogram generated by the unweighted neighbor-joining (NJ) cluster analysis in DARwin 5.0.158 (Perrier et al. 2003). A total of 30,000 bootstraps were included for the data set.
STRUCTURE 2.23 software was used to determine the genetic structure of the South African Pgt population based on the nine single pustule 2SA55 isolates and the 11 described races according to Bayesian model-based clustering. The admixture model was used to calculate the exact number of sub-populations according to Evanno et al. (2005) using the ad hoc ΔK statistic. The K-values ranged from 1 to 10, with 10 replicate runs included per K value. The burn-in period length and Monte Carlo Markov Chain (MCMC) iterations were initially both set at 10000. Once the definitive K value was determined, the analysis was repeated with a burn-in length of 100,000 and 100,000 MCMC iterations.
Analysis of molecular variance (AMOVA; Excoffier et al. 1992) was done using the Arlequin 3.11 software package (Excoffier et al. 2005) to assess the genetic variation between sub-populations, between groups within sub-populations and within groups based on the STRUCTURE results. The significance level for all AMOVA tests was set at 0.05 and the number of permutations used was 16,000. The fixation index FST that was calculated provided a measure of the genetic differentiation of sub-populations and groups with a value greater than 0.25 indicating significant genetic differentiation (Hart and Clark 1997).
Results
Occurrence and distribution
From 2000 to 2013 a total of 1083 stem rust isolates were phenotyped in SA with only 38 isolates being classified as race 2SA55 (Table 1). This represents less than 4% of the total number of stem rust isolates phenotyped. This race has not been found in subsequent surveys conducted during 2014 to 2016. The occurrence of 2SA55 was restricted to the Western Cape.
Phenotyping
Single pustule isolates originating from the original accessions of race 2SA55 revealed variation for virulence on Sr9g, which was not detected with the initial description of this race. Seven isolates coded to race BNGSC whereas two were virulent for Sr9g and thus classified as BPGSC (Table 2). All isolates were avirulent for the triticale cultivars Coorong, Kiewiet and Satu and therefore differed from three other South African BPGSC variants that attack one or more of the Sr genes in these triticales. Race 2SA55 isolates belonging to BNGSC were virulent only for Little Club and Spelmar whereas isolates classified as BPGSC were also virulent on Kubanka and Acme (Table 3). Seedling ITs for race BNGSC on the entries (from left to right) Little Club, Marquis, Reliance, Kota, Arnautka, Mindum, Spelmar, Kubanka, Acme, Einkorn, Vernal and Khapli are shown in Online Resource 1a and 1b. Comparative seedling infection types for races BNGSC and BPGSC on seedlings carrying Sr9g is shown in Online Resource 2.
ITs recorded for races BNGSC and BPGSC on 56 additional tester lines are shown in Table 4. Representative isolates of races BNGSC and BPGSC did not vary in their response on a set of 144 South African wheat cultivars and breeding lines. The majority of the wheat entries were resistant to races BNGSC and BPGSC with 88 exhibiting a 0; to a;1 reaction. Only three entries were susceptible as seedlings. Thirty entries showed intermediate responses with ITs of 2 to 2+. The remainder of the lines were mixed in their response to races BNGSC and BPGSC. BNGSC and BPGSC produced X+ seedling ITs on the barley cv. SSG564.
Genetic analysis of race 2SA55
The genetic comparison of races BNGSC and BPGSC with the other SA Pgt races and TTKSK revealed the presence of four main groups within the dendrogram (Fig. 1). All four South African members of the Ug99 race group, together with TTKSK, formed the first group supported by a 100% bootstrap value. The second and third groups contained races BNGSC+Sr27, BPGSC+Sr27,Kw, BFBSC and BPGSC+Sr27 (second group) and 2SA4, 2SA100 and BPGSC+Sr27,Kw,Satu (third group), respectively. Several of the tree branches were supported by high bootstrap values.
An unrooted dendrogram indicating the genetic relationships between Pgt race 2SA55 (BNGSC and BPGSC), 11 other South African Pgt races and race TTKSK (Ug99). The dendrogram was constructed using neighbor-jointing cluster analysis based on allelic data of 22 SSR markers (Karaoglu et al. 2013). Bootstrap values above 70% are indicated
The fourth group contained all nine single pustule isolates of races BNGSC and BPGSC of 2SA55. Included amongst the seven genetically identical single pustule isolates were five isolates that were phenotyped as BNGSC and two that were phenotyped as BPGSC. Isolates 2SA55.B3 and 2SA55.C3 that were both phenotyped as BNGSC shared 95.5 and 93.2% genetic similarity respectively with the other seven isolates. Collectively, isolates of races BNGSC and BPGSC shared the closest genetic similarity of 84.1% with 2SA100, 2SA4 and BPGSC+Sr27,Kw,Satu and 81.2% with BPGSC+Sr27.
When all 21 Pgt isolates were used to determine the genetic structure of the South African Pgt population, two distinct sub-populations were found (results not shown). The analysis grouped all Ug99 race group members into the first sub-population and all non-Ug99 isolates into the second. AMOVA analysis confirmed this grouping when 73.5% of the genetic variation was attributed to variation between the two sub-populations, 12.7% to variation between groups within the sub-populations and 13.8% to variation within groups. An FST value of 0.86 confirmed the significant variation between the two sub-populations.
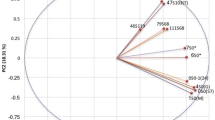
Since the AMOVA results suggested the presence of separate groups within the sub-populations, STRUCTURE analysis was repeated using only the non-Ug99 isolates (Fig. 2). Two groups were found. The first group consisted of the nine single pustules of race BN/PGSC while the second contained all the other non-Ug99 races. Isolate 2SA55.C3 was admixed containing equal amounts of genetic contribution from both groups. This confirmed the dendrogram results with 2SA55 isolates being genetically distinct from the other non-Ug99 races.
STRUCTURE analysis of eight South African non-Ug99 Pgt races. The analysis was based on allelic data of 22 SSR markers (Karaoglu et al. 2013)
Discussion
Race 2SA55 (BNGSC) was first reported in SA during 2000 and constituted the majority of Pgt isolates pathotyped during that year. However, this race was not found in 193 isolates pathotyped during 2001–2003. From 124 isolates collected in 2004, only four (3%) were identified as 2SA55. Similarly in 2007 and 2008, race 2SA55 was rare and detected at frequencies of 3 and 5%, respectively. Furthermore, the race was not found among 517 isolates collected during surveys of six major wheat growing regions from 2009 to 2013 (Terefe et al. 2016). In addition, the occurrence of race 2SA55 was restricted to the Western Cape Province where wheat, barley and various forage crops are grown under rain fed conditions. ITs obtained for BNGSC in the present work support the original description (Boshoff et al. 2002), except for Sr9b which was previously considered effective, most likely because of the use of differential line W2402 (Sr9b + Sr7b).
Except for the Sr9b response, 2SA55 (BNGSC) resembles Pgt race 2SA32 (standard race 326) described from SA between 1981 and 1985 (Le Roux and Rijkenberg 1987; Le Roux 1989). Lombard (1986) reported race 326 from the 1960’s but his ITs on the Stakman set do not match those obtained in the present study.
The infrequent occurrence, restricted distribution and relatively low pathogenicity to current South African wheat germplasm of 2SA55, suggest that this race is currently of little importance within SA. However, the current characterisation of 2SA55 revealed interesting observations. Firstly, we have shown that the original field isolates collected and described as 2SA55 coded predominantly to BNGSC but also contained variants with undescribed virulence for Sr9g, which coded to BPGSC. Races with a BPGSC notation have often been detected in SA, but with virulence for one or more of the Sr genes in Coorong, Kiewiet and Satu triticale. All 2SA55 isolates, irrespective of their BNGSC or BPGSC code, were avirulent on these triticale tester lines. From common Pgt races detected in SA, 2SA102 (BPGSC+Sr27), virulent on Coorong triticale (Sr27) shows a close phenotypic resemblance to 2SA55 (BNGSC or BPGSC). Race 2SA55 (BNGSC) differs from 2SA102 being avirulent for Sr9g and Sr27 but 2SA55 (BPGSC) and 2SA102 differ only at the Sr27 locus. Other common Pgt races related to 2SA55 in virulence profiles include 2SA104 (BPGSC+Sr27,Kw) virulent on Coorong (Sr27) and Kiewiet (SrKw), 2SA105 (BPGSC+Sr27,Kw,Satu), virulent on Sr27, SrKw and SrSatu in Satu, 2SA103 (BNGSC+Sr27) and 2SA108 (BFBSC+Sr27).
The avirulence/virulence profiles of BNGSC and BPGSC, both without triticale virulence, appear to be related to ancestral SA stem rust races which acquired virulence for Sr9b, Sr9g, Sr11, Sr27, SrKw and SrSatu. However, despite being phenotypically similar to other SA stem rust races, microsatellite analysis showed low genetic similarity amongst them. This suggests the existence of different genetic lineages within the broad BN/PGSC phenotype and warrants a more comprehensive study.
References
Boshoff, W.H.P., Pretorius, Z.A., Van Niekerk, B.D., & Komen, J.S. (2002). First report of virulence in Puccinia graminis f. sp. tritici to wheat stem rust resistance genes Sr8b and Sr38 in South Africa. Plant Disease 86, 922.
Evanno, G., Regnault, S., & Goudet, J. (2005). Detecting the number of clusters of individuals using the software structure. A simulation study. Molecular Ecology, 14, 2611–2620.
Excoffier, L., Smouse, P. E., & Quattro, J. M. (1992). Analysis of molecular variance inferred from metric distances among DNA haplotypes: Application to human mitochondrial DNA restriction data. Genetics, 131, 479–491.
Excoffier, L., Laval, G., & Schneider, S. (2005). Arlequin ver. 3.0: An integrated software package for population genetics data analysis. Evolutionary Bioinformatics Online, 1, 47–50.
Hart, D. L., & Clark, A. G. (1997). Principles of population genetics (3rd ed.). Sunderland: Sinauer Associates.
Karaoglu, H., Lee, C., & Park, R. (2013). Simple sequence repeats in Puccinia graminis: Abundance, cross-formae speciales and intra-species utility and development of novel markers. Australasian Plant Pathology, 42, 271–281.
Le Roux, J. (1989). Physiologic specialization of Puccinia graminis f. sp. tritici in South Africa during the period 1981-1985. Phytophylactica, 19, 456–472.
Le Roux, J., & Rijkenberg, F. H. (1987). Occurrence and pathogenicity of Puccinia graminis f. sp. tritici with increased virulence for Sr24. Plant Disease, 71, 1115–1119.
Lombard, B. (1986). Host–pathogen interactions involving wheat and Puccinia graminis f. sp. tritici in South Africa. PhD thesis, University of Stellenbosch, South Africa.
Perrier, X., Flori, A., & Bonnot, F. (2003). Data analysis methods. In P. Hamon, M. Seguin, X. Perrier, & J. C. Glazmann (Eds.), Genetic diversity of cultured tropical plants (pp. 43–76). Enfield, UK: Science Publishers.
Pretorius, Z. A., Singh, R. P., Wagoire, W. W., & Payne, T. S. (2000). Detection of virulence to wheat stem rust resistance gene Sr31 in Puccinia graminis f. sp. tritici in Uganda. Plant Disease, 84, 203.
Pretorius, Z. A., Jin, Y., Bender, C. M., Herselman, L., & Prins, R. (2012a). Seedling resistance to stem rust race Ug99 and marker analysis for Sr2, Sr24 and Sr31 in south African wheat cultivars and lines. Euphytica, 186, 15–23.
Pretorius, Z. A., Szabo, L. J., Boshoff, W. H. P., Herselman, L., & Visser, B. (2012b). First report of a new TTKSF race of wheat stem rust (Puccinia graminis f. sp. tritici) in South Africa and Zimbabwe. Plant Disease, 96, 590.
Saghai-Maroof, M. A., Soliman, K. M., Jorgensen, R. A., & Allard, R. W. (1984). Ribosomal DNA spacer-length polymorphisms in barley: Mendelian inheritance, chromosomal location and population dynamics. Proceedings National Academy Science USA, 81, 8014–8018.
Sambrook, J., Fritsch, E. F., & Maniates, T. (1989). Molecular cloning, a laboratory manual (2nd ed.). New York: Cold Spring Harbour Laboratory Press.
Terefe, T. G., Visser, B., & Pretorius, Z. A. (2016). Variation in Puccinia graminis f. Sp. tritici detected on wheat and triticale in South Africa from 2009 to 2013. Crop Protection, 86, 9–16.
Visser, B., Herselman, L., & Pretorius, Z. A. (2009). Genetic comparison of Ug99 with selected south African races of Puccinia graminis f. sp. tritici. Molecular Plant Pathology, 10, 213–222.
Visser, B., Herselman, L., Park, R. F., Karaoglu, H., Bender, C. M., & Pretorius, Z. A. (2011). Characterization of two new Puccinia graminis f. sp. tritici races within the Ug99 lineage in South Africa. Euphytica, 179, 119–127.
Funding
The microsatellite analysis was funded by the National Research Foundation, SA (grant number 87692).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
We hereby declare that our submission is in compliance with the ethical responsibilities and standards of the EJPP as well as those of the University of the Free State.
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic Supplementary Materials
ESM 1
(DOC 5230 kb)
Rights and permissions
About this article
Cite this article
Boshoff, W.H.P., Pretorius, Z.A., Terefe, T.G. et al. Phenotypic and genotypic description of Puccinia graminis f. sp. tritici race 2SA55 in South Africa. Eur J Plant Pathol 152, 783–789 (2018). https://doi.org/10.1007/s10658-018-1527-3
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10658-018-1527-3