Abstract
Rhizoctonia solani, a devastating soil borne fungus inciting banded leaf and sheath blight (BLSB) disease is a constraint in maize production and improvement program. Rhizoctonia isolates collected from seven diverse maize cropping zones of India were examined for morphological and molecular variability. All the tested isolates caused symptoms of BLSB on maize and were also cross infective on rice and sugarcane hosts, but showed significant variability in hyphal diameter, mean hyphal cell size, weight, size and distribution of scleorotia, culture pigmentation, incubation period, pathogenicity and expression of symptoms. Neighbour joining cluster analysis placed the 62 isolates of R. solani into four major groups, A, B, C and D. Group A was more diverse and included isolates of diverse agro-ecological zones. The cluster analysis corresponded well with principle component analysis. Pathogenicity testing of R. solani isolates on maize genotype (CM 501) revealed highly variable virulence pattern of the pathogen population suggesting its high evolutionary potential, and hence adaptability to diverse geographical regions. The study reveals a strong evidence of inherent potential of the R. solani isolates to survive in diverse ecological zones and its probable spread to other maize cultivars across India. Sequence comparisons of the internal transcribed sequence-ribosomal DNA region of 62 isolates did not reveal much diversity among the isolates. Majority of the isolates (n = 61) clustered together with anastomosis group (AG) AG1-IA used as reference strain in the phylogram, distinct from AG1-IB, AG2–2IIIB and Waitea circinata used as reference strains. BLSB isolates representing distinct geographical locations shared identical sequences indicating long-distance dispersal of the pathogen. The study confirms that the genetic flexibility of the pathogen allows for its adaptation to variable ecological niches and long-distance introduction of new genotypes into the region. The study emphasizes that epidemiological studies may complement the molecular studies.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Maize (Zea maysL.) is an important cereal crop worldwide, with a wide adaptability to diverse agro-climatic conditions. In India, the annual production of maize is approximately 24.2 million tonnes produced on an area of about 9 million hectares (Mahajan et al. 2015). Soil borne fungus Rhizoctonia solani Kühn [teleomorph Thanatephorus cucumeris (Frank) Donk] is a major threat to maize cultivation in India, causing Banded leaf and sheath blight (BLSB) disease on Maize (Singh et al. 2014). The disease was first reported by Payak and Renfro (1966) from the Tarai region (foot hill plain areas) of Uttar Pradesh (now Uttarakhand), India. Owing to its increasing severity, it is gaining epiphytotic proportions since last two decades. BLSB has the potential for extensive yield losses up to 40–70% in currently grown cultivars (Sharma et al. 2002). The main bottleneck in development of disease resistant cultivars has been the non-existence of host resistance to BLSB. The devastating epidemics of BSLB have also been reported from Bhutan, China, Indonesia, Nepal, the Philippines, and Vietnam (Sharma et al. 1993, 2002). Additionally, the disease has been reported in several African and Latin American countries (Pascual and Hyakumachi 2000). Rhizoctonia solani is divided into 13 anastomosis groups (AGs) on the basis of hyphal anastomosis between different isolates. The AGs are genetically isolated, non-interbreeding populations with distinct host ranges (Carling et al. 2002). The phylogenetic reconstruction among AGs and their subgroups is done by determining complete DNA sequence, its alignment and processing. By comparison of rDNA-ITS region phylogenetic relationationship between the subgroups and geographically separated isolates can be studied (Fenille et al. 2003).
The diversity of Rhizoctonia species has been extensively examined in India but, most of the studies were restricted to a region (Sharma et al. 2005; Akhtar et al. 2009; Rajput and Harlapur 2014; Lore et al. 2015). An understanding of the genetic diversity and pathogenic variability of Rhizoctonia spp. (sensu lato) is critical for developing disease management practices and better understanding of epidemiology. Therefore, studies on the Rhizoctonia species complex should mainly focus on the accurate characterisation and epidemiology of plant diseases attributed to Rhizoctonia spp. (Lakshman and Amaradasa 2014).
Management strategies for Rhizoctonia are based on the use of systemic fungicides, soil fumigants and antagonists (Montealegre et al. 2003; Naeimi et al. 2010; Howell and Stipanovic 1995; Murdoch and Leach 1993). However, these strategies often yield inconsistent results in the management of Rhizoctonia diseases. Various reports confirm that R. solaniAGs and subgroups differ in sensitivity to different fungicides and also to bio-control agents (Lakshman and Amaradasa 2014; Kataria et al. 1991).
The objective of the present study was to characterize and identify species of Rhizoctonia causing BLSB from diverse agro-climatic zones of India. Our experimental approach for characterization and identification involved morphotyping and sequencing of ITS-rDNA region, followed by pathogenicity evaluation for biological typing of the pathogen. The study presents broader information about the pathogenic and molecular variability among the Rhizoctonia isolates infecting maize in India. The findings of the study will help breeders to screen the available germplasm and pathologists to further examine the fungicides and bio controls for devising pest management modules.
Materials and methods
Survey, collection and maintenance of Rhizoctonia isolates
Extensive surveys were conducted for collection of Rhizoctonia isolates during the 2013–2015 cropping season, in all the maize producing regions of India (Table 1). Soil types ranged from sandy/clayey alluvium, clayey loam/desert loam and acidic/lateritic soils. Majority of collections (n = 41 isolates) were made from trans-Gangetic plains which forms the major maize cultivation zone. The infected leaf sheaths and sclerotia were collected in butter paper bags, properly labelled and stored in cool buckets, to avoid secondary microbial invasion. Sixty two isolates of Rhizoctonia were characterized from 114 diseased samples based on cultural, morphological and pathogenic variability tested under controlled environment (Table 1). Cross infectivity of the Rhizoctonia isolates was also tested on rice and sugarcane which are cultivated as collateral crops.
The infected leaf sheaths (5 mm2 pieces) and sclerotia of the fungus from each infected sample were washed in sterile distilled water, surface sterilized in 0.5% NaOCl for 3 min., rinsed 3 times in sterile distilled water, dried on filter paper, and plated on to 1.2% water agar. The plates were incubated at 25 ± 2 °C in BOD incubator for 24–48 h in the dark room. Single hyphal tips growing from plated tissue were transferred to fresh Potato Dextrose Agar (PDA, Hi Media Pvt. Ltd., India) plates using a sterile dissecting needle after examining the mother culture under stereomicroscope for diagnostic features of Rhizoctonia spp. (González et al. 2006). Fresh PDA slants of each isolate were maintained after 48 h of growth on PDA. To maintain the vigour and virulence of the pathogen throughout the study, pure cultures were inoculated on sterile barley seeds prepared by autoclaving for 90 min. at 121 °C; 0.117 MPa.,twice. Mycelia plugs 5 mm in diameter of Rhizoctonia isolates grown for 7 days on PDA were added to 100 g of sterile barley seeds in conical flasks and incubated for 7 days in the dark, with shaking for uniform colonisation and avoiding formation of clumps. The colonised barley seeds were dried for 48 h in a laminar air flow hood and stored at −5 °C for long term storage. The colonised barley seeds were used to prepare PDA plates for further studies. The pure cultures were also maintained on PDA slants at 10 °C ± 2 °C and stored in a B.O.D. Incubator. The virulence of the pathogen was maintained every year on pot culture of the host (Zea mays. L. cultivar CM 501) and the fungi isolated from symptomatic tissues.
Cultural and morphometric characteristics
The isolates of Rhizoctonia were examined for colony texture, colour, growth pattern and radial growth of mycelium. Growth pattern of mycelium was examined by visual observation as compact (cmp), sparse (spr), fluffy (ffy), or cottony growth (ctn). The radial growth was obtained by measuring the mycelium diameter after every 24 h of growth for 5 days. The isolates were categorised as fast (+++), medium (++) and slow (+) based on colony growth. The colour of mycelium was determined with the help of Munsell’s Soil Colour Chart (Munsell 1954). The culture and key colour card were placed side by side and colour of the mycelium was observed from bottom side of the culture plate. Culture plates were maintained in triplicates and average values for each parameter was recorded (Table S1).
Two days old pure cultures of Rhizoctonia isolates were used for morphometric studies. Mycelium of 48 h old cultures was stained with aniline blue (0.5%) in lacto-phenol and covered by a glass cover slip. Hyphal characters were examined under Olympus BH–2 compound microscope attached with photomicrography unit 40X eyepiece andoil immersion (100X) objectives. The observations were made at 100X magnification and images taken. Visual observations were also made for colour of hyphae assigned as hyaline and coloured, presence or absence of constriction at the base of branching and angle of branching, by method described by Burpee et al. (1980). Data based on the distance between the two septa, diameter of the vegetative hyphae and branching of the fungal hyphae were recorded and characterized. Hyphal diameter and cell size were expressed as average of 10 hyphae from the mycelial mat in the microscopic field. The data was statistically analysed.
Sclerotial characteristics
The sclerotial characteristics were studied under a stereo-binocular microscope (Olympus SZ61). Three discs (9 mm2 diameter), containing sclerotia of Rhizoctonia isolates (Table 1) were punched from each Petri plate. The discs were transferred to a beaker containing 10 ml of sterile distilled water and stirred for 30 min. to remove sclerotia from the medium. The contents were filtered through cheese cloth, washed in five changes of distilled water, and collected in a glass vial containing 2.5 ml of 2.5% ammonium sulphate and kept undisturbed for 10 min. The sclerotia, which floated on the liquid surface, were filtered through Whatman No. 42 filter paper and washed with the distilled water. The number of sclerotia was counted under a stereo binocular microscope. Pattern of sclerotial distribution was examined after 2 weeks of culture growth on PDA. Sclerotial weight was recorded 3 month after inoculation. The time required for the first sclerotia formation was recorded at 12 h intervals. For observations on sclerotial size, 20 sclerotia were collected randomly from each isolate and their size measured using an ocular micrometer in a calibrated microscope. The shape and number of sclerotia produced was also recorded (Table S2). Ten sclerotia from each isolate were harvested and weighed in an electronic balance and the average weight of a single sclerotium was calculated.
Pathogenic and cross infectivity tests
Mass inoculum of R. solani was prepared as described by Tsror (2010). Barley grains (250 g) were soaked in 500 ml of water containing 100 μg/ml of chloramphenicol in Erlenmeyer flasks and heated to boiling for 10 min. The grains were washed thrice with tap water and excess water drained off. The flasks were then autoclaved twice for 1 h at 24 h intervals (75 ml water was added before second autoclaving and excess water drained off). The sterile grains were inoculated with ten PDA plugs of 5 mm diameter, from 3 day-old R. solani. During incubation the grains were mixed by shaking daily to promote uniform colonization and prevent formation of grain clumps. The colonized grains were air dried in sterile trays in a laminar flow cabinet for 24–48 h for use in inoculating test plants in subsequent experiments.
Maize cultivar CM 501 was also maintained to test the pathogenic behaviour of the isolates. Sheath inoculation was done by placing three Rhizoctonia colonized barley seeds in the lowest sheath of 30 day-old-plants. Twelve plants were inoculated for each isolate viz., six pots with two plants each. Another 12 plants were treated with sterile water and this served as controls. The observations recorded were for incubation period, size of lesion, length of infected area and disease progress and expression of symptoms from the time of appearance of symptoms up to 35 days (Table S3; S4; Fig. 3). Observation on disease severity was recorded on 1–5 rating scale (Payak and Sharma 1983). The experiment was repeated twice.
All 62 Rhizoctonia isolates maintained on maize host were also inoculated on rice and sugarcane to examine the cross infectivity as previously described for maize. The reaction was recorded as poor (+), average (++) and strong (+++). The experiment was repeated twice.
The data from cultural, morphometric and pathogenicity tests was analyzed by clustering of isolates using GenStat Release 17.1 software and statistical analysis carried out by SAS Ver 10.0. Descriptive statistics score summary was arrived at using PROC MEANS procedure with desired statistic estimates such as Means, Standard Error (SE), Standard Deviation (SD), and Coefficient of Variation (CV) (Tables S6).
Pearson’s correlation coefficients were determined using PROC CORR procedure of SAS (Tables 2 and 3). Isolates-by-characteristics interaction was analyzed with linear mixed model procedure of SAS and their interactions were considered as random effects, where the isolates represented the different characteristics. Principal component analysis (PCA) and Clustering was done by DARwin Ver 6.0 for discussion of results.
Fungal DNA extraction, amplification, sequencing and phylogenetic analysis
Mycelia of Rhizoctonia isolates were grown in Potato Dextrose Broth for four days at 25 °C ± 2 °C with shaking at 100 rpm. Mycelium was collected by filtration through sterilized filter paper lining a Buchner funnel and extensively washed with distilled water, blotted dry between the layers of tissue, immediately frozen in liquid nitrogen in aluminium foil packets and stored at -70 °C for molecular studies.
Total genomic DNA was extracted using the CTAB method (Murray and Thompson 1980). Quantification of DNA was done by loading 1 μl of DNA in Pico-drop Spectrophotometer (Pico-drop Ltd. Cambridge, UK) and a dilution 200 ng/μl was used as template DNA for amplification. The stock DNA was stored at −80 °C until further use.
The ITS region of rDNA of 62 Rhizoctonia isolates was amplified with ITS-1 (5’-TCCGTAGGTGAACCTGCGG-3′) and ITS-4 (5’-TCCTCCGCTTATTGATATGC-3′) primer pair (White et al. 1990), with 200 ng/μl of DNA. The 50 μl reaction mixture consisted of 50 mΜ each dATP, dCTP, dGTP, and dTTP, 40 pmol of oligonucleotide primer pair ITS-1 and ITS-4, 10 mM Tris-HCl, pH 8.3, 50 mM KCl, 1.5 mM MgCl2 and 1.25 units Taq DNA polymerase (GoTaq Flexi DNA Polymerase, Promega Corpn., USA). The DNA Thermal Cycler (MyCycler™ Bio-Rad) was programmed for, 1 cycle of 3 min. at 94 °C; followed by 35 cycles of 1 min. at 94 °C, 1 min. at 55 °C and 1 min. at 72 °C; and 1 cycle of 10 min. at 72 °C for final extension. The amplicons were separated on 1% agarose gel by electrophoresis (GX 300C, Genetix Biotech Asia Pvt. Ltd., India) with Tris-acetic acid EDTA (TAE) buffer. The agarose gel was stained with ethidium bromide and visualized by gel documentation system (Syngene Bio imaging Pvt. Ltd., India). The amplicons were excised from the gel and purified by Wizard® SV Gel and PCR Clean-Up System (Promega Corpn.,USA). Quality check of purified amplicons was done by gel electrophoresis to examine the purity and integrity before sequencing. Quantification of 50 μl purified samples was done by pico-drop (Pico-drop Ltd. Cambridge, UK), samples were visualized on 1% agarose gel stained in ethidium bromide and subsequently sequenced by ABI 3730 XL Sequencer (XclerisLabs Pvt. Ltd.). The sequences were assembled using codon code aligner software (Trapnell et al. 2011).
Sequence data for the partial ITS-5.8S rDNA of Rhizoctonia isolates was manually checked and edited using Chromas software (Technelysium) and aligned using Clustal-W (Thompson et al. 1994). The sequences were deposited in GenBank (Accessions KF959669 -KF959730). The sequence similarity among isolates for phylogenetic studies was examined.
The MUSCLE algorithm of MEGA 5.2 was used for multiple alignments for constructing Neighbour joining (NJ) and Maximum likelihood (ML) tree. Alignment statistics and genetic distances among sequences were estimated by a pair-wise comparison using p-difference model. Confidence intervals in tree topologies were estimated by bootstrap analysis with 1000 replicates. Only nodes with bootstraps values over 60% were considered to be significant.
Results
Isolation, characterization and pathogenicity of isolates
The maize plants sampled from the surveyed areas (Fig. 1) showed symptoms of banded leaf and sheath blight disease characterised by alternate bleached areas or zones appearing as purplish brown to olive brown bands on the leaf sheaths. One hundred and fourteen Rhizoctonia (sensu lato) were isolated from diseased samples of which, 62 isolates were identified based on colony surface (upper/reverse), colony texture, colour of hyphae, and colony margins which are characteristic of the genus. The isolates represented distinct geographical locations in 7 agro-ecological zones under maize cropping (Table 1). All the 62 isolates maintained for study were cream-colored on the upper surface and light yellow on reverse surface, fluffy/compact growth, dense/sparse texture with regular/irregular growing margins and hyaline/light brown hyphae. The vegetative mycelium was hyaline to brownish having hyphae partitioned into multinucleate cells separated by a septum containing a dough-nut shaped pore, branching at a 90oangles (Fig. 2A). Cultural variability was irrespective of the agro-climatic zone; however, hyphal diameter, mean hyphal cell size and radial growth at optimum temperature showed a significant variation.
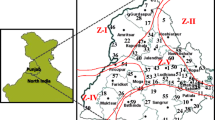
(a) Map showing 15 agroecological zones of India under maize cropping; (b) Locations in India (n = 62), shown in numbers for collection of Rhizoctonia isolates from maize fields in different agroecological zones under maize cropping (details given in Table 1)
The sclerotial characteristics showed variations for 4 of the 9 variables selected, viz., initiation time on PDA, distribution pattern, weight and size. Sclerotial morphology ranged from smooth to rough (Fig. 2B), MG-Bpn isolate formed beaked sclerotia. For rest of the characters viz., pigmentation, number per plate, aggregation, texture and appearance in culture (surface/submerged/aerial) significant variations could not be observed (Table S2).
One isolate out of the 62 (MG-Bpn from Barapani) formed light/dark orange (upper/reverse) colony, pinkish hyphae, cottony growth with highly irregular growing margins on PDA. The MG-Bpn showed mean hypha cell size (length × width) 147.34 × 4.593 μm and slow radial growth and formed orange coloured, smooth textured pear shaped sclerotia on PDA of size 0.6 mm. The MG-Bpn was the slowest in producing sclerotia (23 days), incubation period for disease (96 h).
The pathogenic behaviour of all the Rhizoctonia isolates showed significant variation for all the 5 variables viz.; incubation period, type of symptoms, length of area infected on sheath, disease progress and severity. All the Rhizoctonia isolates investigated exclusively produced banded leaf and sheath blight on maize.
The pathogenic behaviour of isolates on maize susceptible cultivar Zea mays cv. CM 501 revealed significant variability. Isolates HR-Dtp, HR-Ldw, RJ-Lhs, HP-Kul, HR-Kth, RJ-Jhr, RJ-Srm, HP-Ahr, PJ-Lud and JH-Dmk were the most aggressive (disease severity >80%) and isolates MG-Bpn; HR-Dbw; RJ-Ftn; HR-Ymn; UP-Cnd; MH-Klp; HR-Tek; HR-Pti; HR-Jhk; and HR-Kau(M) were the least aggressive (disease severity <30%) (Table S3). The variation in symptom expression was categorized into seven groups (Fig. 3a–g; Table S4). All the Rhizoctonia isolates investigated caused banded leaf and sheath blight on maize and were cross infective to rice and sugarcane (Fig. 3A–C; Table S5) .
Symptoms expressed by Rhizoctonia isolates inoculated to pot culture of Zea mays c. v. CM 501 under controlled conditions (a-g; type I-VII, details given in Table S4). Symptoms expressed on cross infection of Rhizoctonia isolates on rice and sugarcane. A (i-iii) maize to maize, maize to rice, maize to sugarcane; B (i-iii) rice to rice, rice to maize, rice to sugarcane; C (i-iii) sugarcane to sugarcane, sugarcane to maize sugarcane to rice
Statistical analysis of morphological characters
Summary of the descriptive statistics reveals a higher mean values for hyphal length and diameter compared to rest of the nine colony characters (angle of branching, constriction at base, colony growth, colony texture, colour of hyphae, lower surface of colony, margin of colony, radial growth and upper surface of colony). Of these, significant variations were observed for hyphal diameter, hyphal cell size and radial growth which can be useful for characterizing the morphological variability in R. solani. The weight of sclerotia reveals highest mean values followed by time for initiation of sclerotia in culture, indicating a significant variation in these two variables out of nine variables selected. Also, disease severity and incubation period have a very significant role in characterizing pathogenic behaviour of Rhizoctonia isolates as revealed by higher mean values for these two variables out of 8 variables studied (Table S6).
Principal component analysis of morphological characters
Principal component analysis (PCA) was conducted to extract the independent variables that matter the most and give direct measurement of total variance explained by a few of the important components. Principal components and their respective proportion of the variation explained by eigenvectors are presented (Tables S7). The high amount of variation explained by PCA reveals the impact of morphology and pathogenicity of the isolates. Biplots of PCA analysis with first two principal components were derived for extent of phenotypic variations among the isolates for cultural characteristics, sclerotial variability and pathogenic behaviour respectively (Fig. 4A–C) and the measurement of variance among the variables selected for morphological variability in terms of cultural and sclerotial characteristics; and the pathogenic behaviour respectively. The graphs clearly demarcate the characteristics that are dispersed along two principal components axis and emphasize the extent of phenotypic variation explained by these clusters. The characters that positively correlated and performed best in respective variables selected are on the upper right side of the quadrant. Among 62 isolates, HR-Srs, SKM-Skm and MG-Bpn clustered with positive loading of eigen values for first two principal components. The experimental results show that PCA analysis was able to identify hyphal diameter, mean hyphal cell size and radial growth as suitable for discussing cultural variability. Size, weight and time for initiation of sclerotia were the best variables for sclerotial variability, whereas; disease progress and incubation period, best emphasized the pathogenic behaviour of isolates (Fig. 4A–C). The dimensionality of dataset was reduced using PCA analysis to assess the multiple characters and screen isolates that exhibit similar attributes.
The correlation matrix was utilized in determining the eigenvector values. The number of components was extracted using screen plot of eigenvalues. Biplot was developed from first two principal components (PRN1 and PRIN2) (Table S7). Eigen values were scored to determine the characters showing significant variation among the isolates with respect to characters. The number of variables that can best fit to describe the characters for observed variation was identified from eigenvector values of PCA analysis. The first two major axes of differentiation (PC1 and PC2) explained 65.4% of the total variation for cultural characteristics, 31.54% and 43.31% of the total variation for sclerotial characteristics and pathogenic behaviour respectively.
Cluster analysis of morphological characters
Cluster analysis was carried out for 62 isolates based on 11 cultural, 9 sclerotia and 8 pathogenic characters. Neighbour joining cluster analysis put the 62 isolates of R. solani into four major groups, A, B, C and D for morphological characteristics (Fig. 5). Group A, includes a total 11 isolates, Group B includes a total 12 isolates; Group C includes 20 isolates and Group D includes 19 isolates. Group A was more diverse than the group B. It includes isolates of diverse agro-ecological zones. In general, isolates clustered together irrespective of their agro-climatic zones. The isolates, though clustered close to group, formed a separate sub-group within the group. Among the isolates, MG-Bpn (62) was distinct from rest of the isolates. Isolates DL-Kbg (1) and HR-Ppl (37) were distinct in their respective groups. Majority of the isolates of trans-gangetic plains clustered in Group C and D, except HR-Znd, HP-Ahr, PJ-Lud UP-Cnd, MH-Klp, and HR-Ymn. Isolate MG-Bpn separated as distant branch. This indicated distinctive nature of these isolates over remaining isolates.
Phylogenetic tree using neighbour joining (NJ) method for 62 isolates of Rhizoctonia based on morphological, sclerotial and pathogenic variability. Isolate numbers are given on the branch termini. Branch robustness was tested using 1000 bootstrap (details of isolates presented in Table 1)
PCA did not reveal much diversity among the Rhizoctonia isolates investigated though isolates MG-Bpn, HR-Srs, and SKM-Skm clustered far apart from all other isolates indicating their dissimilarity with others. The cluster analysis also corresponded well with principle component analysis.
Phylogenetic analysis using ITS region
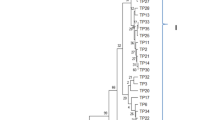
Amplification of rDNA-ITS of the Rhizoctonia isolates using primers ITS-1 and ITS-4 (White et al. 1990) produced DNA fragments ranging from 650 to 750 bp. Sequence alignment and phylogenetic analysis of the ITS 5.8S–rDNA region grouped 62 R. solani AG1-1A isolates into two clades. Data subjected to analysis by distance and character state methods confirmed that majority of BLSB isolates characterized as AG1-IA, were genetically similar. Among 61 isolates of AG1-IA, the ITS region showed minor differences due to frequent indels. Phylogenetic analysis by neighbour joining (NJ) and maximum likelihood methods supported very similar phylogenetic trees with bootstrap values of 100%. The maximum likelihood tree (Fig. 6) and the NJ tree (Fig. S2) confirmed separation of the Rhizoctonia isolates in clades A and B and excluded both clades from known R. solani AG1 subgroups AG1-IB. The phylogenetic tree, based on the rDNA-ITS region sequences, revealed that AG1-IA formed a clade that differed from the cluster of each of the other groups.
A maximum likelihood (ML) tree showing phylogenetic relationships for Rhizoctonia solani isolates using rDNA ITS sequences. CLUSTALW (v. 1.6) was used for nucleotide sequence alignment. Phylogenetic distances were calculated with the Kimura 2-parameter nucleotide substitution model. Bootstrap values of >60% assessed by 1000 replications are shown next to the branches. Bar indicates one base change per 100 nucleotide positions
The final alignment of sequences showed 726 positions with 237/726 conserved sites; 470/726 variable sites; 322/726 parsimony informative sites and 145/726 singleton sites. The 470 variable characters were uninformative and thus excluded from the analysis. The average frequencies of four nucleotides for all the samples were A = 26.50%; T = 32.40%; C = 21.70%, G = 19.40%. The average G + C content was estimated as 41.10% with the highest G + C content was estimated as 43.00% in Maharastra isolate (MH-Klp) and lowest G + C content was found as 40.10% in Meghalaya (MG-Bpn). As expected, average transitional pairs (si =16) were more frequent than transversional pairs (sv = 23) with an average ratio of 0.72. Pair-wise genetic distance values and evolutionary divergence of sequences based on the Kimura 2-parameter are provided in Table 4. Of the 62 isolates only 22 isolates showed variations as indicated by bootstrap values in the phylogeny tree (Fig. 6).
Discussion
Banded leaf and sheath blight (BLSB) caused by R. solani is a major threat to maize cultivation in India. The environmental conditions, lack of genetic resistance, repeated cultivation and lack of awareness to disease management practices among the farmers are among the primary reasons for increased incidence of the disease.
In the current study, 62 isolates collected from seven major maize cropping zones of India were characterized for morphological and molecular variability. Although previous studies have addressed pathogenic profiles from specific regions, a knowledge gap remains for the distribution and occurrence of most widespread Rhizoctonia species and anastomosis groups in the maize growing zones of India.
Our study revealed that, all tested isolates, caused symptoms of BLSB on maize and were also cross infective on rice and sugarcane hosts, but showed significant variability in hyphal diameter, mean hyphal cell size, the size/weight/distribution pattern of their sclerotia, culture pigmentation incubation period, pathogenicity and expression of symptoms. Rhizoctonia isolates formed sclerotia ranging in size from 0.6–4.8 mm; single or aggregated, fluffy to compact colony, dense to sparse culture growth with regular to irregular growing margins on PDA. However, HR-Ymn was non-sclerotia producing and showed poor symptom expression in pathogenicity tests. As far as the maize leaf and sheath blight pathogen in India is concerned, most of the taxonomic characters such as effused white to buffbrown colonies on PDA, hyphae 3–11 mm (average 7.4 mm) in diameter, hyaline at first, later brown,septate, branched at right angle, constriction and septum near origin of branch; multinucleate hyphae, with 3–16 nuclei (mostly 4–8) per cell; sclerotia of primary rind type, differentiated into loose medullary region with acompact marginal rind of 3–4 layers of dark pigmentation have been documented by Ahuja and Payak (1988). The sclerotia were variable in size and shape but mostly discoid and irregularly curved, optimum growth at 25–30 °C, no growth below 15 °C and above 35 °C, and growth rate of about 45 mm/24 h.
Morphologically, Rhizoctonia AG 1-1A isolates clustered into four major groups (Fig. 5) representing diverse agro-ecological zones pointing towards high genetic diversity in Rhizoctonia isolates. These results are different from earlier studies by Singh et al. (2002) and Guleria et al. (2007) where clustering of Rhizoctonia isolates was observed on the basis of geographical location. Pillai and Singh (1994) demonstrated differential reactions of rice cultivars to R. solani collected from different locations in India. Virulence pattern of R. solani isolates carried out on a single maize genotypes (CM 501) revealed highly variable virulence pattern of the pathogen population suggesting its high evolutionary potential, and hence ability to adapt to diverse geographical regions. The results of our research provided strong evidence that the isolates have an inherent potential to survive in diverse ecological zones which may be the reason for wide spread disease incidence on almost all maize cultivars across India. The symptoms of Waitea isolate collected from Barapani (Meghalaya) were distinct, appeared as light orange spots on the leaf sheath. It formed a cottony light orange to pinkish colony on PDA and pin-head sized orange sclerotia.
The cultural characteristic of Rhizoctonia isolates on PDA and the sequence of rDNA-ITS revealed unique characteristics, to place Indian isolates in AG1-IA group. Thus, AG1-IA was the predominant Rhizoctonia pathogen associated with maize surveyed from all the agro-ecological zones in this study. However, further field studies are needed on the ecology and pathogenicity of Rhizoctonia spp. with larger sample sizes.
Analysis of the rDNA-ITS region has been a useful method for identification or grouping of many fungal species. Phylogenetically distinct clusters of 22 isolates were observed based on rDNA-ITS sequence analysis. This indicated distinctive nature of these isolates over remaining isolates. The isolates more closely associated within a sub-group share close similarity, possibly due to common gene pool. Characterization of this group will require more extensive sequencing of a larger collection of isolates, and determination of their pathogenicity to various crops. Some more set of primers could be used to amplify sequence conservation within the group and sequence diversity so that nucleotide polymorphisms could be analysed. In R. solani, many reports demonstrated that analysis of ITS sequences is useful for classification of intra groups within AGs (Kuninaga et al. 1997; Salazar et al. 2000). The maize crop is reported to be infected by several Rhizoctonia AGs and species such as AG1-IA, AG1-IB, AG2–2IIIB, R. zeae, R. oryzae (Lakshman et al. 2016). In the phylogenetic tree, W. circinata isolated in this study did not group with W. circinata var. zeae and W. circinata var. oryzae though it is W. circinata. However, it needs to be determined if it may be an altogether new species. Of the 62 isolates only 22 isolates showed variations as indicated by bootstrap values in the phylogeny tree (Fig. 6). These isolates could be of interest to examine further for sub-grouping. For a more rigorous investigation into genetic diversity, the sequencing of additional genes needs to be considered. However, establishing the AG for the isolates, the primary objective of sequencing and phylogeny, was achieved. The AG1-IA isolates formed a distinct clade in the phylogram that was distinctly separated from the AG1-IB and AG2–2IIIB reference strain based on previous studies. Ahvenniemi et al. (2009) correlated such variations, with genetic incompatibility due to the compensatory base changes found in the secondary structure of ITS-2.The morphological variation within isolates from a single AG can be due to the multinucleate nature of the cells which contain 3–10 nuclei (Sneh et al. 1991).
From the study it appears that sheath blight disease of maize is caused by Rhizoctonia AG1-IA, with the exception of Waitea circinata isolate from Meghalaya. This may be due to similar temperature and nutrition available during the growth and development periods of each crop (Li et al. 1998). The rice and maize crop rotation in the same fields is likely to increase the incidence of BLSB due to R. solaniAG1-IA. Hence, a combination of rotation or inter planting with other crops viz., potato, onion, toria, millets, mustard, legumes and pulses is recommended to minimise cross-infection.
Most of the BLSB isolates within subgroups AG1 IA show identical sequences at the ITS-5.8S rDNA region although collected from geographically different gene pools. This supports the widespread distribution of similar genotypes in the region. Hence, there seems to be no relationship between geographical origin of the isolates and the genetic profiles because the Rhizoctonia isolates within a clade represented most of the sampling sites. Long-distance dispersal of mycelia and sclerotia as documented in earlier studies for AG1, AG3 and AG8 can be one reason for occurrence of isolates of R. solani from diverse geographical regions (Wang et al. 2015).
Significant differences were detected between isolates from diverse agro-ecologies cultured in common environment (culture conditions/glass house). The virulence of isolates from domesticated origin and wild origin can be further examined for an accurate picture of inherent survival potential of the isolates and the role of isolate origin in pathogenicity. This would also provide information on genetic basis of the ecological adaptation of the pathogen, its virulence, and the possible correlations among the different parameters of pathogenic efficiency in a given pathosystem.
Hyakumachi et al. (2005) discussed the prevalence of local adaptations among the different pathogen populations, in addition to other genetic and environmental factors affecting pathogen aggressiveness. Their study revealed that isolates from wild habitats were less aggressive than isolates from domesticated habitats, but presented a wider variance in aggressiveness, probability due to selection of variations in pathogen phenotypes in man-made agro-ecosystems. In the present study all the isolates sampled were from the domesticated habitats and from summer cropping season which has higher seasonal temperatures and optimum humidity due to good amount of rainfall which are conducive for pathogen. Also, long term cropping may cause selection to some extent which may trigger population divergence. This may cause a change in ecological requirements and also development of partial host specialization in parallel, under the different environmental conditions prevailing in the wild and domesticated habitats.
The results of this investigation identified four phylogenetic clusters of maize pathogenic AG1-IA isolates of Rhizoctonia solani based on morphological characters. This demonstrates that pathogenic variations of the isolates exist within the same geographic regions. The information gained may provide insights into the natural variability of the pathogen and calls for necessity of more comprehensive studies of pathogenic Rhizoctonia isolates covering all maize growing agro-climatic regions of India.
References
Ahuja, S. C., & Payak, M. M. (1988). A field inoculation technique for evaluating maize germplasm to banded leaf and sheath blight. Indian Phytopathology, 31, 517–520.
Ahvenniemi, P., Wolf, M., Lehtonen, M. J., Wilson, P., & Valkonen, K. M. (2009). Evolutionary diversification indicated by Compensatory Base changes in ITS2 secondary structures in a complex fungal species, Rhizoctonia solani. Journal of Molecular Evolution, 69, 150–163.
Akhtar, J., Jha, V. K., Kumar, A., & Lal, H. C. (2009). Occurrence of banded leaf and sheath blight of maize in Jharkhand with reference to diversity in Rhizoctonia solani. African Journal of Agricultural Sciences, 1, 32–35.
Burpee, L.L., Sanders, P.L., Cole, H. Jr., &Sherwood, R.T., (1980). Anastomosis groups among isolates of Ceratobasidium cornigerum and related fungi. Mycologia, 72, 689–701.
Carling, D. E., Kuninaga, S., & Brainard, K. A. (2002). Hyphal anastomosis reactions, rDNA-internal transcribed spacer sequences, and virulence levels among subsets of Rhizoctonia solani anastomosis group-2 (AG-2) and AB-I. Phytopathology, 92, 43–50.
Fenille, R. C., Ciampi, M. B., Kuramae, E. E., & Souza, N. L. (2003). Identification of Rhizoctonia solani associated with soybean in Brazil by rDNA-ITS sequences. Fitopatologia Brasileira, 28, 413–419.
González, G. V., Portal Onco, M. A., & Rubio, V. S. (2006). Review: Biology and systematics of the form genus Rhizoctonia. Spanish J. of Agric. Res., 4(1), 55–79.
Guleria, S., Aggarwal, R., Thind, T. S., & Sharma, T. R. (2007). Morphological and pathological variability in rice isolates of Rhizoctonia solani and molecular analysis of their genetic variability. Journal of Phytopathology, 155, 654–661.
Howell, C. R., & Stipanovic, R. D. (1995). Mechanisms in the bio control of Rhizoctonia solani-induced cotton seedling disease by Gliocladium virens: Antibiosis. Phytopathology, 85, 469–472.
Hyakumachi, M., Priyatmojo, A., Kubota, M., & Fukui, H. (2005). New anastomosis groups, AG-Tand AG-U, of binucleate Rhizoctonia spp. causing root and stem rot of cut-flower and miniature roses. Phytopathology, 95, 784–792.
Kataria, H. R., Verma, P. R., & Gisi, U. (1991). Variability in the sensitivity of Rhizoctonia solani anastomosis groups to fungicides. Journal of Phytopathology, 133, 121–133.
Kuninaga, S., Natsuaki, T., Takeuchi, T., & Yokosawa, R. (1997). Sequence variation of the rDNA ITS regions within and between anastomosis groups in Rhizoctonia solani. Current Genetics, 32, 237–243.
Lakshman, D. K., & Amaradasa, B. S. (2014). The pathogen biology, identification and management of Rhizoctonia species with emphasis on isolates infecting turf grasses. Indian Phytopathology, 67, 327–345.
Lakshman, D. K., Jambhulkar, P. P., Singh, V., Sharma, P., & Mitra, A. (2016). Molecular identification, genetic diversity, population genetics, and genomics of Rhizoctonia solani, In Principles of plant pathology in genomic era (pp 55–89). New Delhi: Today and Tomorrow Printers and Publishers.
Li, H. R., Wu, B. C., & Yan, S. Q. (1998). Aetiology of Rhizoctonia in sheath blight of maize in Sichuan. Plant Pathology, 47, 16–21.
Lore, J. S., Jain, J., Hunjan, M. S., Gargus, G., Mangat, G. S., & Sandhu, J. S. (2015). Virulence spectrum and genetic structure of Rhizoctonia isolates associated with rice sheath blight in the northern region of India. European Journal of Plant Pathology, 143, 847–860.
Mahajan, V., Yadav, O.P., Kumar, P., Kumar, B., Chikkappa, G. K., Kaul, J., Singh, A.K., Parihar, C.M., Jat, S.L., Sekhar, J.C., Shekhar, M., Hooda, K.S., Paul, D., & K.P. Singh. (2015). Annual Progress Report Kharif Maize 2015. All India Coordinated Research Project on Maize. Eds. Indian Institute of Maize Research, Pusa Campus, New Delhi 110 012, India. pp. 999.
Montealegre, J. R., Reyes, R., Pérez, L. M., Herrera, R., Silva, P., & Besoain, X. (2003). Selection of bio antagonistic bacteria to be used in biological control of Rhizoctonia solani in tomato. Electronic Journal of Biotechnology, 6, 115–127.
Munsell. (1954). Munsell soil color charts. Baltimore: Munsell Color Company Inc..
Murdoch, C. W., & Leach, S. S. (1993). Evaluation of Laetisaria arvalis as a biological control agent of Rhizoctonia solani on white potato. American Journal of Potato Research, 70, 625–634.
Murray, M. G., & Thompson, W. F. (1980). Rapid isolation of high molecular weight plant DNA. Nucleic Acids Research, 8, 4321–4325.
Naeimi, S., Okhovvat, S. M., Javan-Nikkhah, M., Vágvölgyi, C., Khosravi, V., & Kredics, L. (2010). Biological control of Rhizoctonia solani AG1-1A, the causal agent of rice sheath blight with Trichoderma strains. Phytopathologia Mediterranea, 49, 287–300.
Pascual, C. B., & Hyakumachi, M. (2000). Distribution of vegetatively compatible populations of Rhizoctonia solani AG-1 IA in a field planted with different host species. Journal of General Plant Pathology, 66, 206–209.
Payak, M. M., & Renfro, B. L. (1966). Diseases of maize new to India. Indian Phytopathology, 19, 122 (abstr.)
Payak, M. M., & Sharma, R. C. (1983). Techniques of scoring for resistance to important diseases of maize. I.A.R.I., New Delhi: AICMIP.
Pillai, P. K., & Singh, V. P. (1994). Differential reaction of rice cultivars to isolates of Rhizoctonia solani. Indian Phytopathology, 47, 96–98.
Rajput, L. S., & Harlapur, S. I. (2014). Status of banded leaf and sheath blight of maize in North Karnataka. Karnataka Journal of Agricultural Sciences, 27(82–84).
Salazar, O., Julian, M. C., Hyakumachi, M., & Rubio, V. (2000). Phylogenetic grouping of cultural types of Rhizoctonia solani AG 2-2 based on ribosomal ITS sequences. Mycologia, 92, 505–509.
Sharma, R. C., Leon, C. D., & Payak, M. M. (1993). Diseases of maize in south and South-East Asia: Problems and progress. Crop Protection, 12, 414–422.
Sharma, R. C., Srinivas, P., & Batsa, B. K. (2002). Banded leaf and sheath blight of maize its epidemiology and management. In N. P. Rajbhandari, J. K. Ransom, K. Adhikari, & A. F. E. Palmer (Eds.), Proc. of a maize symposium, 2001 (pp. 108–112). NARC and CIMMYT: Kathmandu.
Sharma, M., Gupta, S. K., & Sharma, T. R. (2005). Characterization of variability in Rhizoctonia solani by using morphological and molecular markers. Journal of Phytopathology, 153, 449–456.
Singh, V., Singh, U. S., Singh, K. P., Singh, M., & Kumar, A. (2002). Genetic diversity of Rhizoctonia solani isolates from rice differentiation by morphological characteristics, pathogenicity, anastomosis behaviour and RAPD fingerprinting. Journal of Mycology and Plant Pathology, 32, 332–334.
Singh, V., Kumar, S., Lal, M., & Hooda, K. S. (2014). Cultural and morphological variability among Rhizoctonia solani isolates from trans-gangetic plains of India. Research on Crops, 15, 644–650.
Sneh, B., Burpee, L., & Ogoshi, A. (1991). Identification ofRhizoctoniaspecies. APS press, St (p. 133). Paul.
Thompson, J. D., Higgins, D. G., & Gibson, T. J. (1994). CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Research, 22, 4673–4680.
Trapnell, C., Williams, B. A., Pertea, G., Mortazavi, A., Kwan, G., van Baren, M. J., Salzberg, S. L., Wold, B. J., & Pachter, L. (2011). Transcript assembly and abundance estimation from RNA-Seq reveals thousands of new transcripts and switching among isoforms. Nature Biotechnology 2010, 28(5), 511–515. https://doi.org/10.1038/nbt.1621.
Tsror, L. (2010). Biology, epidemiology and management of Rhizoctonia solani on potato. Journal of Phytopathology, 158, 649–658.
Wang, L., Liu, L. M., Hou, L. L., & Huang, S. W. (2015). Pathotypic and genotypic diversity of Rhizoctonia solani AG-1-IA causing rice sheath blight in china. Plant Pathology, 64, 718–728.
White, T. J., Bruns, T., Lee, S., & Taylor, J. W. (1990). Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In M. A. Innis, D. H. Gelfand, J. J. Sninsky, & T. J. White (Eds.), PCR protocols: A guide to methods and applications (pp. 315–322). New York: Academic.
Acknowledgements
The authors thankfully acknowledge the generously provided materials of R. solani from the researchers from centres of maize as well as technical support and advice from experts in the field. Financial support for this research was provided by the Department of Science and Technology, Ministry of Science & Technology, India under OYS Scheme (Grant: SR/FT/LS-073/2009). There is no conflict of interest in the present investigation.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
In compliance with the Ethical standards of the Journal, I declare that the manuscript is being submitted with the informed consent and due contribution from all the authors. No animal/human was used as research specimen in the study. On behalf of all the co authors, the corresponding author declares that there is no conflict of interests. I confirm that the information provided is accurate
Rights and permissions
About this article
Cite this article
Singh, V., Amaradasa, B.S., Karjagi, C.G. et al. Morphological and molecular variability among Indian isolates of Rhizoctonia solani causing banded leaf and sheath blight in maize. Eur J Plant Pathol 152, 45–60 (2018). https://doi.org/10.1007/s10658-018-1447-2
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10658-018-1447-2