Abstract
Fusarium culmorum can affect plants including cereals or develop saprophytically in the soil. It is unknown whether its saprophytic ability is different in strains with a different aggressiveness. This knowledge could be used for effective breeding of resistant cultivars. Here, we aimed to study the development of two F. culmorum strains with a different aggressiveness in the soil under barley, to compare their saprophytic ability and to reveal the influence, if any, of the host plant on the development of strains in the soil and on the roots. Saprophytic development of the strains was studied on membranes inoculated with macroconidia and placed into non-sterile soil under barley of two genotypes. The fungus was identified on the membranes and on barley roots by indirect immunofluorescent method. Both strains could develop saprophytically. The more aggressive strain (Fc538) showed a lesser saprophytic fitness than the less aggressive strain (Fc885): its mycelial density was lower and the number of chlamydospores was greater. Barley genotypes influenced the development of the fungal strains. Interestingly, conditions for saprophytic development of both strains were more favourable in the soil under barley of the resistant genotype. The more aggressive strain colonized barley of the resistant genotype more actively. Rot symptoms appeared earlier in the barley of the resistant genotype, but the number of diseased plants was greater in the barley of the susceptible genotype. The presence of a saprophytic stage in life cycle should slow down the accumulation of aggressive races in field populations of F. culmorum. Possible interactions between F. culmorum strains and barley plants are discussed.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Fusarium culmorum (W. G. Smith) Saccardo is an anamorphic facultative plant pathogen without a distinct substrate specialization, which can affect a wide range of hosts including most cereals (Cook and Baker 1983; Jenkinson and Parry 1994). In cereals it causes Fusarium head blight (FHB), Fusarium root rot and Fusarium crown rot (Paulitz et al. 2002; Wang et al. 2006). These diseases result in reductions in yield and grain quality as well as in grain contamination with mycotoxins (Parry et al. 1995; McMullen et al. 1997; Tóth et al. 2004). Colonising the roots of cereals, F. culmorum may grow to the crown (Paulitz 2006). When F. culmorum infected the wheat seedlings, the development of the crown rot was followed with the translocation of deoxynivalenol (DON) towards the head, which resulted in the accumulation of the mycotoxin in the grain (Covarelli et al. 2012).
The damage caused by F. culmorum can be mitigated by breeding cultivars with increased resistance. However, the pathogen may reduce resistance level of the host by evolving new, more aggressive races. One prerequisite for this is that the pathogen has a large genetic variation. In Gibberella zeae (anamorph: Fusarium graminearum), which is closely related to F. culmorum, sexually-derived progeny could be even more aggressive than the parental isolates (Voss et al. 2010). F. culmorum is also known to have a large molecular and phenotypic diversity (Miedaner and Schilling 1996; Miedaner et al. 2001; Muthomi et al. 2000; Mishra et al. 2003).
At the same time F. culmorum is known to be a competitive saprotrophic coloniser of plant remains in the soil (Burgess and Griffin 1967). An aggressive strain F. culmorum 30 can develop saprophytically in sterile soil without plants, forming mycelium, macroconidia and chlamydospores, while in non-sterile soil its development is suppressed by competitors (Shakhnazarova et al. 2000). In the presence of plants the intensity of saprophytic development of Fc30 was higher than in the soil without plants (Shakhnazarova et al. 1999). However, it is unknown whether F. culmorum strains with a different aggressiveness also differ in their saprophytic ability. As a hypothesis, less aggressive strains may survive better in the soil then more aggressive ones because the ability to cause diseases in plants should not give any adaptive advantages in the soil. The knowledge about this could be used to assess the rate of accumulation of aggressive races in the soil population of the fungus and to predict the stability of selected plant cultivars.
In this work we studied the development of two strains of F. culmorum in the soil and on the roots of barley differing in resistance to root rot. One of the strains was more aggressive than the other. The objectives of our research were: (1) to compare the saprophytic ability of a more and a less aggressive strain, and (2) to reveal the influence, if any, of the host plant on the behaviour of the strains in the soil and their development on the roots.
Mаterials and methods
Experimental design
In the experiment we aimed to determine: 1) whether strains with a different aggressiveness differed in their ability to saprophytic development in the soil under barley, and 2) whether barley plants could influence the pathogen when the latter was still in the soil, before the colonisation of roots (taking into account that F. culmorum is a facultative phytopathogen without a distinct substrate specialisation). Barley plants with a different susceptibility to root rot were used in order to find out whether the intensity of development of a fungal strain in the soil and its colonisation ability depended on the influence of a certain genotype.
We tried to make experimental conditions as close as possible to natural ones. For this reason, the experiment was conducted in non-sterile soil in the presence of plants. The soil was inoculated with macroconidia of the fungal strains. In this way, we could assess the colonisation ability of each strain (registering the amount of fungus on the roots) and its influence on plant susceptibility to disease. To assess the saprophytic ability of the strains in the soil under plants, we used membranes inoculated with macroconidia and placed in the soil inoculated with the corresponding fungal strain. Barley seeds were placed into the soil. The state of fungal strains on the membranes, the amount of fungus on the roots and root rot symptoms were assessed on the 3rd, 5th, 7th, 12th and 20th day of the experiment. At each sampling date three pots were emptied completely because partial sampling would have damaged plant roots and membranes. To level conditions for plants of different age, we used pots with a different volume of soil. Fungal strains were identified on the membranes and the root surface with the help of an indirect immunofluorescence method.
It was expected that the data obtained in the experiment would fill the gap in our knowledge about the earliest stages of interactions between host plants and strains with a different aggressiveness, starting from germination of macroconidia and the development of strains in the soil under barley, till their colonisation the roots and effect on plants.
Study objects and fungal inoculum production
Barley seeds of two genotypes were obtained from the collection of the N.I. Vavilov All-Russia Research Institute for Plant Industry. Barley k-3454 Hordeum vulgare L., convariation vulgare, was characterised as resistant to root rot. Barley k-27652 H. vulgare L., convariation distichon, was characterised as susceptible to root rot.
Fusarium culmorum strains (Fc538 and Fc885) were chosen from 25 strains obtained from the collection of the All-Russia Research Institute for Plant Protection based on their ability to cause root rot of various intensity on the barley described above. In sterile vermiculite Fc538 caused root rot symptoms in 100% of barley plants of the susceptible genotype (k-27652) and in 85% plants of the resistant genotype (k-3454). Inoculation of the vermiculite with Fc885 caused root rot symptoms in 50% plants of the susceptible genotype and in 20% plants of the resistant genotype. PCR with the use of species-specific primers OPT18 F/R confirmed that the isolates belonged to the species F. culmorum. Species-specificity of OPT18 F/R primers for F. culmorum was shown by Schilling et al. (1996). DNA of Fc885 biomass did not amplify with the help of N 1–2 F/R primers, which restrict the intergenic fragment of the DNA region responsible for the synthesis of the DON. Based on this, we assumed that this strain probably did not produce deoxynivalenol. It was shown that DNA of the high-producing DON F. culmorum strains was amplified with N1–2 and N1–2R primers, while no amplification was observed for the low-producing strains (Bakan et al. 2002). The DNA of Fc538 biomass was amplified with the use of the primers mentioned. Fc538 was isolated from the seeds of winter wheat (Nizhny Novgorod Region) in 1995. Fc885 was isolated from the pumpkin stem (Leningrad Region) in 1995.
The strains were grown for 10 days on Czapek-Dox agarized medium (Singleton et al. 1992) at 25оC in the dark. Noteworthy, Fc538 grown in culture formed abundant macroconidia (by an order of magnitude more than Fc885). Macroconidia of both strains were washed off the medium with sterile water, filtered through nylon cloth to separate the mycelium and concentrated by centrifuging (4000×g, 10 min). A suspension of concentrated macroconidia in sterile water was made. The inoculum was examined microscopically to assess its quality (lack of mycelium and chlamydospores) and to count the number of macroconidia.
Study of saprophytic behaviour of F. culmorum strains in the soil under barley
Saprophytic development of F. culmorum strains was studied on membranes placed into the soil. The method of preparing membranes was described earlier (Strunnikova et al. 2015). Briefly, sterile membrane filters made of nitrocellulose (Millipore, pore diameter 0.23 μ) were cut into pieces with an area of ca. 3 cm2. The prepared membranes were inoculated under sterile conditions with the suspension of macroconidia of each strain (30 μl of suspension per membrane), the concentration being 5∙105 conidia/ml. Altogether, 33 membranes with macroconidia of each F. culmorum strain were prepared. The membranes were dried at room temperature immediately after inoculation and inserted into sterile nylon bags (pore diameter 200 μm). Three membranes inoculated with Fc538 and three membranes inoculated with Fc885 were left as inoculation control for the subsequent assessment of the state of each of the strains on the starting day of the experiment (‘Day 0’ time point).
Conditions of experiment
Experiment was conducted in non-sterile soil with two barley genotypes (k-3454 and k-27652) and two treatments. The experiment was conducted in three biological replications. The soddy-podzolic sandy loam soil was analysed at the Department of Agrochemistry of the St Petersburg State University (pH 5.38. Content (mg/kg soil) NO3 – traces; NH4–90.0; P – 70.0; K – 101.2. Humus – 3.26%; total N – 0.14%; total C – 1.89%). Inoculation on the Czapek-Dox medium did not reveal any native F. culmorum in the soil. In one case (the first treatment) we added to the soil the suspension of Fc538 macroconidia (10 ml of water with 106 macroconidia per 100 g of soil). In another case (the second treatment), the same amount of suspension of Fc885 macroconidia were added to the soil. The soil was thoroughly mixed and put, in the amount of 700 g, 1200 g and 1500 g, into pots with a volume of 500 ml, 900 ml and 1100 ml, respectively. In this way, 30 pots inoculated with Fc538 and 30 pots inoculated with Fc885 were prepared (in total, 60 pots). Membranes with macroconidia of Fc538 and Fc885 prepared at the time of the experiment establishment were placed into each pot at a depth of 3 cm from the surface. The membrane with macroconidia was put into the pot where the corresponding fungal strain had already been introduced. After that, presterilised and swollen barley seeds of each genotype were placed into the pots, 6 seeds per pot. In addition, barley of both genotypes was grown in the soil not inoculated with the F. culmorum strains (negative controls). Barley was grown under natural light for 3 and 5 days in 500 ml pots, for 7 days in 900 ml pots, 12 and 20 days in 1100 ml pots. Soil moisture was sustained at a level of 60% of complete water capacity by daily watering. Membranes with strains of F. culmorum and barley plants were sampled out 3, 5, 7, 12 and 20 days post inoculation (dpi). Three pots were completely emptied in each case on the sampling day. Membranes with F. culmorum strains were dried at room temperature immediately after taking them out of the soil and stored dry until the immunofluorescent staining of the fungus on their surface.
Assessing the incidence of barley disease
Barley roots sampled on the 3, 5, 7, 12 and 20 dpi were thoroughly and carefully washed to remove soil by submerging them into water 3–4 times. Eighteen barley plants were sampled on each day in each case. Roots of all plants were examined for the presence of root rot symptoms. The disease was expressed as the percentage of plants with root rot symptoms. After diagnosing the symptoms of disease, roots were air-dried at room temperature and stored dry until the identification of F. culmorum strains on their surface.
Drying the membranes and the roots fixed the samples, making it possible to stain them not on the day of sampling but later.
Obtaining antibodies against F. culmorum and testing their specificity
To obtain polyclonal antibodies, rabbits were injected with a suspension of homogenised mycelia and macroconidia of F. culmorum subcutaneously and a solution of water-soluble proteins intravenously. Water-soluble proteins were extracted and antibodies were obtained, purified and concentrated following the procedure described in Pantou et al. (2005). The specificity of the anti-rabbit antibodies obtained was determined by indirect immunofluorescence with the use of goat anti-rabbit IgG conjugated to fluorescein isothiocyanate (FITC). Mycelia and macroconidia of F. culmorum fluoresced brightly if 1:64-diluted immunoglobulins were used. No cross reactions were observed with fungal structures of different Fusarium species: (F. oxysporum, F. solani, F. gibbosum, F. heterosporum, F. sambucinum, F. semitectum, F. moniliforme, F. aquaeductuum) and isolates of other soilborne fungi (Verticillium, Pythium, Paecilomyces, Mucor, Gliocladium, Rhizoctonia, Penicillium, Sclerotinia, Mycogone, Acremonium, Cladosporium).
Identification of F. culmorum strains on roots and membranes
The procedure of identifying the fungus on the membranes and on the roots was described earlier (Strunnikova et al. 2015). In order to assess the colonisation density of barley roots of both genotypes with F. culmorum strains, nine barley plants (three randomly selected plants from each of the three pots) were used in each case. The roots of three-day-old plants were stained and their entire surface was examined. Two root pieces with a length of at least 3 cm were cut from each of the nine plants sampled later. At least 6 cm of each of the nine plants were stained and then microscopically examined in each case.
Dry roots were dipped in water for 30–40 min, depending on the root diameter, and placed on filter paper to remove water excess. Then the roots were stained with a 1% aqueous solution of neutral red for 20–30 min (to quench autofluorescence), rinsed thrice with water and gently dried again. The roots were then dipped for 20 min into solutions of 1:64-diluted rabbit antibodies against F. culmorum, rinsed with buffered physiological solution (900 ml 0.2 M Na2HPO4, 100 ml 0.2 M KH2PO4, 0.85% NaCl, pH 7.6) and slightly dried. Then they were treated with goat anti-rabbit immunoglobulins conjugated to FITC for 20 min, rinsed again and dried. The density of root colonisation by F. culmorum strains was judged upon by the frequency of occurrence of fungal microcolonies in the optical field of the microscope. The number of microcolonies in each viewed area of the root was recalculated for the linear centimetre of the root.
The membranes were stained with 1% solution of aniline blue on 70% ethanol for 6 min. Excessive stain was removed by washing in water. The partially dried membranes were treated with bovine serum albumin conjugated to rhodamine for 15 min, washed and partly dried. These treatments quenched autofluorescence of the membranes and some soil microorganisms. Species-specific antibodies and anti-rabbit antibodies conjugated to FITC were then successively applied for 20 min each. After each treatment the membranes were rinsed with buffered physiological solution. Staining was performed at room temperature. Stained membranes and roots were examined in an Imager A1 fluorescence microscope (Carl Zeiss, Germany) at a magnification of 400 × .
The entire area of the membranes was examined microscopically and the amount of F. culmorum on each membrane was registered in 30 fields of view, randomly selected in different membrane areas. The intensity of mycelium growth was estimated using a 1 to 5 rating scale developed previously, which corresponded to the following amount of mycelium on membranes (in metres): 1 = < 1 m/cm2; 2 = 1 to 2.3 m/cm2; 3 = 2.3–3.6 m/cm2; 4 = 3.6–5.5 m/cm2; and 5 = > 5.5 m/cm2. Formed macroconidia and chlamydospores were counted.
Statistical analysis
The average values of growth parameters of F. culmorum strains on membranes and roots during the whole observation period are represented in the graphs. The density of mycelium and the number of fungal structures were assessed in 90 optical fields in each case (n = 90). To assess the amount of Fc538 and Fc885 on the roots of barley of both genotypes, roots of nine plants were used in each case (n = 9). Standard errors of the average values for each time point are represented in the graphs with a vertical line. To determine statistical significance of the differences, each mean was contrasted pairwise with every other mean at each time point, followed by post hoc multiple comparison Fisher LSD test (ANOVA). In this way, at each time point we compared the development of two strains under barley and on the roots of barley of each genotype as well as the development of a strain under barley of various genotypes and on the roots. If P ≤ 0.05, these points were denoted by different letters. To assess the ratio of mycelium and chlamydospores in the populations of Fc538 and Fc885 formed on the membranes in the soil, the number of chlamydospores was recalculated per unit of mycelial density based on the data obtained in each case (Fig. 5).
The influence of barley genotypes on the development Fc538 and Fc885 on membranes and on the roots over all considered time points was analysed using repeated measurements ANOVA (Table 1). For factorial effect, Fisher statistics (F) was calculated for each strain separately with the corresponding significance level (p). For mathematical processing we used the data of repeated measurements of the development indices of fungal structures on the membranes and on barley roots for the entire observation period. The result is presented as a dispersion table giving the F-test, the number of degrees of freedom (df time, df error) and p-value. All statistical analyses were performed using the Statistica v. 10.0, StatSoft Inc. 1984–2011.
Results
Saprophytic development of the fungal strains on membranes in the soil under barley of two genotypes
Macroconidia of both strains introduced into the soil on membranes germinated. Mycelium, new macroconidia and chlamydospores could be seen on the membranes by 3 dpi.
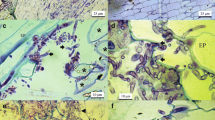
Microscopic examination of the membranes showed that the development of Fc538 and Fc885 in the soil under two barley genotypes was characterised by several common features. In both strains, some of the germinated macroconidia increased in size and became chlamydospores (Fig. 1c, d, e). This transformation of the germinated macroconidia into chlamydospores explains the increase in the number of resting structures early in the development of both strains (Fig. 4). In single cases the fungal hyphae were distended and chlamydospores were formed inside them (Fig. 1f). As early as by 5 dpi, barley roots could be seen on some of the membranes but no intensification of the fungal growth near the roots was observed (Fig. 1b). Bacterial colonies were observed on the membranes by 5 dpi and later. However, there were few areas where the fungal hyphae were distinctly seen to be colonised by the bacteria. Starting from the 5th day, we could observe an active lysis of the mycelium, accompanied by an increase in the diameter of the hyphae and their subsequent complete destruction (Fig. 1e). Usually, the lysis of the mycelium was expressed in the fragmentation of the hypha, with only some of its areas being destroyed (Fig. 1a).
Fluorescence light microscopy images of Fusarium culmorum strains developing on membranes in the soil under barley of susceptible genotype (a, d, f) and of resistant genotype (b, c, e). a Fragmentation of F. culmorum 885 mycelium (FM), 7 days post inoculation (dpi). b Development of F. culmorum 885 mycelium (M) near a root hair (RH); intensification of fungal growth near the root was not observed, 7 dpi. c Transformation of germinated macroconidia of F. culmorum 885 into chlamydospore (FCh), 3 dpi; germinated macroconidia (GMc), mycelium (M). d Transformation of germinated macroconidia of F. culmorum 538 into chlamydospore (FCh), 3 dpi. e Lysis of F. culmorum 538 mycelium (LM), 5 dpi; formation of chlamydospores in the germinated macroconidia (FCh). f Formation of chlamydospore in the hypha of F. culmorum 538 (ChFH); germinated macroconidia (GMc), forming chlamydospore (FCh), 5 dpi. Scale bar is 10 μm in all panels
Lysis (possibly, autolysis) was observed all the time. It was mostly mycelium and the germinated macroconidia that were being destroyed. However, the development of both strains was also observed all the time. It was expressed in the germination of the macroconidia formed already on the membranes in the soil as well as in mycelium growth. After 7–12 days, the destruction prevailed over growth, resulting in a decreased population density of each strain (Figs. 2, 3 and 4). However, brightly fluorescing fungal mycelium as well as occasional germinating macroconidia could be seen on the membranes as late as on 20 dpi, indicating that the fungus grew actively.
Change with time in mycelium growth of F. culmorum 538 (straight lines) and F. culmorum 885 (dashed lines) on membranes in the soil under susceptible barley genotype (SG) and resistant barley genotype (RG). Bars indicate standard errors (n = 90). Different letters indicate significant differences (P ≤ 0.05)
Change with time in macroconidia formation by F. culmorum 538 (straight lines) and F. culmorum 885 (dashed lines) on membranes in the soil under susceptible barley genotype (SG) and resistant barley genotype (RG). Bars indicate standard errors (n = 90). Different letters indicate significant differences (P ≤ 0.05)
Change with time in chlamydospores formation by F. culmorum 538 (straight lines) and F. culmorum 885 (dashed lines) on membranes in the soil under susceptible barley genotype (SG) and resistant barley genotype (RG). Bars indicate standard errors (n = 90). Different letters indicate significant differences (P ≤ 0.05)
The strains of different aggressiveness varied in number of the structures they formed during the development in the soil. Mycelial density of the less aggressive Fc885 on certain days of the experiment was higher than that of the more aggressive Fc538 regardless of the barley genotype (Fig. 2). The difference in the mycelial density between the strains was more pronounced under plants of the susceptible genotype. By 3 and 5 dpi the less aggressive Fc885 formed more macroconidia than the more aggressive Fc538 (Fig. 3). In subsequent days the number of macroconidia formed by the strains differed depending on the barley genotype and the time of sampling. The number of chlamydospores formed per unit of mycelial density was much higher in the more aggressive Fc538 than in the less aggressive Fc885 (Fig. 5).
Barley genotypes influenced the development of the fungal strains. Thus, the mycelial density of both strains on the whole was higher under plants of the resistant genotype than under plants of the susceptible genotype (Fig. 2). Abundant formation of macroconidia by both strains was observed earlier on the membranes in the soil under barley of the resistant genotype (Fig. 3). Areas of lysed mycelium were more often observed on membranes under plants of the resistant genotype. It appeared during the microscopic examination of the membranes that the process of development (growth/lysis) of F. culmorum, regardless of the strain, was more intense under barley plants of the resistant genotype. A considerable influence of barley genotypes on the saprophytic development of Fc538 and Fc885 in the soil was confirmed by dispersion analysis of the data (Table 1).
Colonisation of the roots of barley of two genotypes by the fungal strains
Control plants of barley of both genotypes grown in the soil not inoculated with strains of F. culmorum were colonised by this fungus (Fig. 6). Microscopic examination showed that on the 3rd day and later the fungus was present mostly at the root base. It did not spread towards the root tip and did not observe on root hairs. The amount of the fungus changed little throughout the experiment and was the same on the roots of barley of both genotypes.
Change with time in barley root colonisation of susceptible genotype (SG) and of resistant genotype (RG) by F. culmorum 538 (straight lines) and F. culmorum 885 (dashed lines). Colonisation of control plants of barley of both genotypes by F. culmorum is shown by dotted lines. Bars indicate standard errors (n = 9). Different letters indicate significant differences (P ≤ 0.05)
In the soil inoculated with the pathogen, the fungal strains were observed on the roots of barley of both genotypes also by 3 dpi (Fig. 6). A similar dynamics of colonisation of barley roots of both genotypes by the two fungal strains was observed, which can be seen from the curves on the graphs. The density of F. culmorum on the roots decreased by 5 dpi, increased again by 7 dpi and remained stable until the end of the experiment in case of colonisation of the roots of barley with Fc885. It was in case of colonisation of the roots of barley of both genotypes with a more aggressive Fc538 that the density of the fungal mycelium on the roots statistically significantly decreased by 20 dpi (Fig. 6).
The process of colonisation of barley roots had several common features, which did not depend on the genotype of the host or the strain of the fungus. The fungus colonised the roots mostly in the zone of the root hairs. By 3–5 dpi the fungus was most often observed on the root hairs (Fig. 7a). It should be noted that the appearance of F. culmorum on the root hairs was observed on barley roots even on 20 dpi. The colonisation of the roots by the fungus coming from the soil may be a prolonged process. Not often but sometimes fungal microcolonies could be observed on 3-days-old roots and in the elongation zone. Both fungal strains were mostly represented on the barley roots by the mycelium and were usually localised as microcolonies of various size, usually small ones (Fig. 7b). Sometimes the fungus was present as a hypha situated along the root in a groove, the root tissues in the vicinity being somewhat necrotized (Fig. 7c). In some cases F. culmorum was present at the area of formation of lateral roots (Fig. 7d). It was in these areas that the fungus was the most harmful to the plant, sometimes resulting in the death of the lateral root.
Fluorescence light microscopy images of Fusarium culmorum strains developing on barley roots of susceptible genotype (b, c) and of resistant genotype (a, d). a hyphae of F. culmorum 885 colonising barley root hairs, 3 days post inoculation (dpi). b, a small microcolonia of F. culmorum 538 formed on barley root surface, 5 dpi. c hyphae (H) of F. culmorum 538 growing along the barley root; a necrosis zone was observed (arrows), 7 dpi. d The microcolony of F. culmorum 538 was formed near the lateral root; the death of the lateral root (DLR), 12 dpi. Scale bar is 10 μm in all panels
A more aggressive Fc538 colonised the roots of barley of both genotypes more actively than a less aggressive Fc885 (Fig. 6). This was evidently associated with a greater virulence of Fc538. However, barley genotypes influenced the colonisation of the roots by Fc538: its density on the roots of barley of the resistant genotype was much higher on 3, 7 and 12 dpi than on the roots of barley of the susceptible genotype (Fig. 6). Statistically significant influence of barley of both genotypes was shown only in the case of Fc538 on the roots; the influence on colonisation by Fc885 was not statistically significant (Fig. 6, Table 1).
Influence of strains of Fusarium culmorum on susceptibility of barley to root rot
Control barley plants grown in the soil not inoculated with strains of F. culmorum did not have any symptoms of root rot (data not shown).
On the whole, regardless of the host genotype, the number of diseased plants was higher when the soil was inoculated with a more aggressive Fc538 as compared to a less aggressive Fc885 (Fig. 8). The greatest number of diseased plants was noted in case of barley of the susceptible genotype colonised with Fc538. Colonisation of roots by the less aggressive Fc885 caused root rot symptoms in approximately the same number of plants of both genotypes.
Development of root rot in barley plants of susceptible genotype (SG) and of resistant genotypes (RG) after inoculation of soil with F. culmorum 538 (straight lines) and F. culmorum 885 (dashed lines). Bars indicate standard errors (n = 18). Different letters indicate significant differences (P ≤ 0.05)
The time when the first plants with root rot symptoms were noted differed depending on the barley genotype. By 3 dpi, root rot symptoms were observed only in plants of barley of the resistant genotype colonised with Fc538 (Fig. 8). In subsequent days, in case of inoculation with this strain the number of diseased plants of the resistant genotype increased gradually, reaching the peak by 20 dpi. On the roots of plants of the susceptible genotype colonised with Fc538, the first rot symptoms appeared by 5 dpi, but the disease developed more intensely from then on. The less aggressive Fc885 caused root rot symptoms by 5 dpi also in plants of the resistant genotype. At that time, the barley of the susceptible genotype did not have any root rot symptoms yet. Colonisation of plants with Fc885 resulted in a fast development of root rot in plants of barley of the resistant genotype and a slower development in plants of barley of the susceptible genotype.
Discussion
Our work is the first study of the behaviour of two strains of F. culmorum with a different aggressiveness in the soil and their interactions with barley plants differing in susceptibility to root rot under conditions close to natural ones.
We found that both studied strains of F. culmorum could develop saprophytically in the soil under barley. They formed mycelium, macroconidia and chlamydospores (Figs. 1, 2, 3, 4 and 5). A similar saprophytic development of the aggressive strain 30 of F. culmorum under various soil conditions has been observed in our earlier study (Strunnikova 2011).
The structures formed by F. culmorum in the soil have different functions. Mycelium is the structure with the help of which F. culmorum feeds and spreads in the soil. The fact that the fungal strains in our study formed mycelium in the soil is an unequivocal indication of their saprophytic development. Mycelium is more easily lysed than macroconidia and especially chlamydospores, as shown for Fc538 and Fc885 (present study) and Fc30 (Shakhnazarova et al. 2004). However, it is also constantly renewed owing to the growth of the fungal hyphae and the germination of macroconidia newly formed on the mycelium.
The function of F. culmorum macroconidia in the soil is not entirely clear. They are formed on the mycelium, and it would seem that their abundance should directly reflect the mycelial density, but this is not always the case. In our previous experiments, the number of macroconidia formed by Fc30 depended not only on the mycelial density but also on the soil type (Shakhnazarova et al. 2004). In the present study, differences in the number of macroconidia formed in the soil in Fc538 and Fc885 (Fig. 3) could be associated with the individual characteristics of the strains. Even when the strains were grown on the nutrient medium, Fc538 formed much more conidia than Fc885. However, we cannot rule out that barley of the two genotypes also had some influence on the differences in the number of macroconidia formed by the fungus in the soil.
Chlamydospores of F. culmorum are resting structures, ensuring the survival of the fungus under adverse conditions. They are less easily lysed than mycelium and macroconidia. As shown in our experiments with Fc30, the number of chlamydospores in the soil population of the fungus usually increased at the times when the mycelium and the macroconidia were actively lysed (Shakhnazarova et al. 2004). In another experiment, the most abundant formation of chlamydospores in the introduced macroconidia of Fc30 was noted when the same membrane was сo-inoculated with the fungus and the antagonistic bacterium Pseudomonas fluorescens, which suppressed the fungal growth considerably (Strunnikova et al. 2007).
There were fewer structures of Fc538 and Fc885 by 20 dpi but, importantly, their number never fell down to the zero level (Figs. 2, 3 and 4). Apparently, as many fungal structures remained on the membranes as it was possible under the existing conditions in the soil. We know that Fc30 introduced in the soil on the membranes can develop there throughout the vegetation period of barley (100 days), forming all the structures characteristic of this fungus (Strunnikova et al. 2008).
We found that the two F. culmorum strains with a different aggressiveness in our experiment showed a different saprophytic fitness. It was higher in the less aggressive Fc885 than in the more aggressive Fc538. This was indicated by the fact that the population of Fc885 was mostly represented by the mycelium, which is a vegetative structure, while that of Fc538 was mostly represented by chlamydospores — resting structures (Figs. 2, 4, and 5). The mycelium of Fc885 may be more resistant to lysis than that of Fc538.
However, the question of most interest to both breeders and farmers is likely to be how fast highly aggressive races, capable of reducing the resistance level of new cultivars, may accumulate in the field populations of F. culmorum. A lower saprophytic ability of the more aggressive Fc538 as compared with the less aggressive Fc885 does not mean that the saprophytic stage would not exercise a selection pressure on highly aggressive strains. Our results indicated that the more aggressive Fc538 existed as a saprophyte even in the soil under affected barley. On the membranes in the soil Fc538 formed mycelium and macroconidia, that is, structures that are the most susceptible to the attacks of soil microflora. As a result, the mycelial density of the aggressive Fc538 decreased considerably by 12 dpi (Fig. 2), the number of macroconidia decreased by 20 dpi (Fig. 3) and chlamydospores were also subject to lysis though less actively than the mycelium. Earlier we have shown that antagonistic soil microflora could considerably decrease the population density of another aggressive fungal strain, Fc30 (Strunnikova et al. 2007, 2008, 2015).
All these considerations indicate that soil antagonistic microflora may be a powerful restraining factor of the accumulation of aggressive strains in the field populations of F. culmorum.
The absence of root rot symptoms in control barley plants grown in the soil not inoculated with the pathogen as well as the negative result of the inoculation experiment may indicate that the presence of the fungus on the roots of control plants was associated with the seed infection rather than the presence of native F. culmorum in the soil.
Assessing the effect of two barley genotypes on the development of F. culmorum strains in the soil and their colonisation of the roots, we obtained several new and rather unexpected results. The genotypes of barley influenced the development of Fc538 and Fc885 in the soil (Figs. 2, 3 and 4, Table 1). The most pronounced differences in the number of structures formed by the strains were observed in the soil under barley of the susceptible genotype (Figs. 2, 3 and 4). Interestingly, however, the soil conditions under barley of the resistant genotype were apparently more favourable for the saprophytic development of F. culmorum. The density of mycelium of both strains and the number of macroconidia (in case of Fc885) were greater in the soil under barley of the resistant genotype as compared with the soil under the barley of the susceptible genotype (Figs. 2 and 3). A more intensive fungal growth in the soil under resistant barley might be associated with an additional source of nutrition — possibly, root exudates. Root exudates of the barley of the resistant genotype may contain some components attractive for the fungus. The result is an increased density of the mycelium of both strains on the membranes under barley of the resistant genotype.
Another surprising finding was the ability of the more aggressive strain to colonise actively the roots of barley of the resistant genotype (Fig. 6). Though the more aggressive strain Fc538 colonised barley of both genotypes in greater amounts that the less aggressive Fc885, Fc538 colonised more actively the roots of plants of the resistant rather than the susceptible genotype. Active colonisation by Fc538 of the roots of barley of the resistant genotype may be due to a higher mycelial density of this strain in the soil under this barley (Fig. 2). However, the mycelial density of the less aggressive Fc885 in the soil under barley of the resistant genotype also increased but this was not accompanied by an enhanced colonisation of the roots of the resistant barley by this strain (Fig. 6). It might be that the less aggressive Fc885, being better adapted to the development in the soil, did not “strive” to occupy another niche (plant roots) in contrast to the more aggressive Fc538 less adapted to the existence in the soil.
Finally, though root rot symptoms appeared earlier in barley of the resistant genotype colonised by both strains, these plants were later less affected by rot than the susceptible barley (Fig. 8). The fungus may have colonised the roots of the barley of the resistant genotype not on 3 dpi, when we started observations, but earlier. We observed a more active development of the mycelium on the membranes in the soil under barley of the resistant genotype (Fig. 2). F. culmorum, introduced directly into the soil, may also have developed actively under barley of the resistant genotype. An active development of mycelium in the soil might have resulted in an earlier colonisation of the roots of barley of the resistant genotype. As soon as the fungus colonised the roots, it could induce defence responses in barley. The emergence of the first root rot symptoms as early as on 3 dpi (Fig. 8) may be an indication that the fungus colonised the roots of barley of the resistant genotype earlier that the roots of the susceptible barley. However, it is also possible that host defence responses may develop at different rates, hence the differences in the disease level in barley with a different susceptibility to root rot.
Differences in the pathogenesis of Fc538 on the roots of barley of the resistant and the susceptible genotypes accompanied by a different colonisation level might be associated not only with the abundance of the fungus on the roots but also with the amount of the toxins it produces. The role of DON produced by F. culmorum in the pathogenesis in FHB disease of wheat has been shown (Mesterhazy et al. 1999). A direct correlation was found between the disease index and DON concentration in barley seedlings infected with F. culmorum (Hestbjerg et al. 2002). At the same time, in case of wheat infected with Gibberella zeae no significant correlation was found between the biomass of the fungus (measured as Fusarium exoantigen absorbance) and DON production (Voss et al. 2010).
Differences in the intensity of root rot caused by Fc538 in the barley of the resistant and the susceptible genotypes may be due to the complex of defence reactions in the resistant barley. So, it was found that different intensity of FHB in the susceptible and the resistant wheat cultivars was associated with the restriction of the spreading of the hyphae of F. culmorum, a lower concentration of toxin production by the pathogen and with an impaired diffusion of toxins in the tissues of the heads of the resistant cultivar (Kang and Buchenauer 2000).
In conclusion, a decreased density of populations of F. culmorum introduced into the soil on the membranes, which was shown in this study, gives some reasons to believe that aggressive isolates of the pathogen would not accumulate in the soil under affected cultivars. A lesser density of mycelium in Fc538 might indicate that the more aggressive strain, when it developed in non-sterile soil, was more vulnerable than the less aggressive Fc885.
The comparison of data on the development of strains of varying aggressiveness on the membranes in the soil under barley of different genotypes with the level of root colonisation and susceptibility to disease provides a new insight into early stages of interactions between F. culmorum and barley and into the influence of conditions on these interactions.
References
Bakan, B., Giraud-Delville, C., Pinson, L., Richard-Molard, D., Fournier, E., & Brygoo, Y. (2002). Identification by PCR of Fusarium culmorum strains producing large and small amounts of deoxynivalenol. Applied and Environmental Microbiology, 68, 5472–5479.
Burgess, L. W., & Griffin, D. M. (1967). Competitive saprophytic colonization of wheat straw. Annals of Applied Biology, 60, 137–142.
Cook, J. R., & Baker, F. K. (1983). The nature and practice of biological control of plant pathogens. St. Paul: American Phytopathology Society Press.
Covarelli, L., Beccari, G., Steed, A., & Nicholson, P. (2012). Colonization of soft wheat following infection of the stem base by Fusarium culmorum and translocation of deoxynivalenol to the head. Plant Pathology, 61, 1121–1129. https://doi.org/10.1111/j.1365-3059.2012.02600.x.
Hestbjerg, H., Felding, G., & Elmholt, S. (2002). Fusarium culmorum infection of barley seedlings: correlation between aggressiveness and deoxynivalenol content. Journal of Phytopathology, 150, 308–312.
Jenkinson, P., & Parry, D. W. (1994). Isolation of Fusarium species from common broad-leaved weeds and their pathogenicity to winter wheat. Mycological Research, 98, 776–780.
Kang, Z., & Buchenauer, H. (2000). Ultrastructural and immunocytochemical investigation of pathogen development and host responses in resistant and susceptible wheat spikes infected by Fusarium culmorum. Physiological and Molecular Plant Pathology, 57, 255–268.
McMullen, M., Jones, R., & Gallenberg, D. (1997). Scab of wheat and barley: a re-emerging disease of devastating impact. Plant Disease, 81, 1340–1348.
Mesterhazy, A., Bartok, T., Mirocha, C. G., & Komoroczy, R. (1999). Nature of wheat resistance to Fusarium head blight and the role of deoxynivalenol for breeding. Plant Breeding, 118, 97–110.
Miedaner, T., & Schilling, A. G. (1996). Genetic variation of aggressiveness in individual field populations of Fusarium graminearum and Fusarium culmorum tested on young plants of winter rye. European Journal of Plant Pathology, 102, 823–830.
Miedaner, T., Schilling, A. G., & Geiger, H. H. (2001). Molecular genetic diversity and variation for aggressiveness in populations of Fusarium graminearum and Fusarium culmorum sampled from wheat fields in different countries. Journal of Phytopathology, 149, 641–648.
Mishra, P. K., Fox, R. T. V., & Culham, A. (2003). Inter-simple sequence repeat and aggressiveness analyses revealed high genetic diversity, recombination and long-range dispersal in Fusarium culmorum. Annals of Applied Biology, 143, 291–301.
Muthomi, J. W., Schutze, A., Dehne, H. W., Mutitu, E. W., & Oerke, E. C. (2000). Characterization of Fusarium culmorum isolates by mycotoxin production and aggressiveness to winter wheat. Journal of Plant Diseases and Protection, 107, 113–123.
Pantou, M. P., Strunnikova, O. K., Shakhnazarova, V. Y., Vishnevskaya, N. A., Papalouka, V. G., & Typas, M. A. (2005). Molecular and immunochemical phylogeny of Verticillium species. Mycological Research, 109, 889–902.
Parry, D. W., Jenkinson, P., & McLeod, L. (1995). Fusarium ear blight (scab) in small grain cereals – a review. Plant Pathology, 44, 207–238.
Paulitz, T. C. (2006). Low input no-till cereal production in the Pacific Northwest of the U.S.: the challenges of root disease. European Journal of Plant Pathology, 115, 271–281.
Paulitz, T. C., Smiley, R. W., & Cook, R. J. (2002). Insights into the prevalence and management of soilborne cereal pathogens under direct seeding in the Pacific Northwest, USA. Canadian Journal of Plant Pathology, 24, 416–428.
Schilling, A. G., Moller, E. M., & Geiger, H. H. (1996). Polymerase chain reaction-based assays for speciesspecific detection of Fusarium culmorum, F. graminearum, and F. avenaceum. Phytopathology, 86, 515–522.
Shakhnazarova, V. Y., Strunnikova, О. K., & Vishnevskaya, N. A. (1999). Effect of soil water content on development of Fusarium culmorum. Mycologia and Phytopathologia, 33, 53–59 (rus).
Shakhnazarova, V. Y., Strunnikova, О. K., Vishnevskaya, N. A., Stefanova, N. A., & Muromtsev, G. S. (2000). Structure and dynamics of Fusarium culmorum population in soil of different textures. Eurasian Soil Science, 33, 86–91.
Shakhnazarova, V. Y., Strunnikova, О. K., & Vishnevskaya, N. A. (2004). Development of introduced population of Fusarium culmorum in soil: formation and lysis of fungal structures. Mycologia and Phytopathologia, 38, 79–88 (rus).
Singleton, L. L., Mihail, J. D., & Rush, C. M. (Eds.). (1992). Methods for research on soilborne phytopathogenic fungi. St. Paul: American Phytopathological Society Press.
Strunnikova, O. K. (2011). Study of Fusarium culmorum development in soil for search of protective actions against barley root rot. Agricultural Biology, 3, 98–101 (rus).
Strunnikova, O. K., Shakhnazarova, V. Y., Vishnevskaya, N. A., Chebotar, V. K., & Tikhonovich, I. A. (2007). Development and relations of Fusarium culmorum and Pseudomonas fluorescens in soil. Microbiology, 76, 596–602. https://doi.org/10.1134/S002626170705013X.
Strunnikova, O. K., Shakhnazarova, V. Y., Vishnevskaya, N. A., Chebotar, V. K., & Tikhonovich, I. A. (2008). Interactions between Fusarium culmorum and Pseudomonas fluorescens in the rhizosphere and rhizoplane of barley. Mycologia and Phytopathologia, 42, 70–77 (rus).
Strunnikova, О. K., Vishnevskaya, N. A., Ruchiy, A. S., Shakhnazarova, V. Y., Vorobyov, N. I., & Chebotar, V. K. (2015). The influence of soils with different textures on development, colonization capacity and interactions between Fusarium culmorum and Pseudomonas fluorescens in soil and on barley roots. Plant and Soil, 389, 131–144. https://doi.org/10.1007/s11104-014-2351-y.
Tóth, B., Mesterházy, A., Nicholson, P., Teren, J., & Varga, J. (2004). Mycotoxin production and molecular variability of European and American isolates of Fusarium culmorum. European Journal of Plant Pathology, 110, 587–599.
Voss, H.-H., Bowden, R. L., Leslie, J. F., & Miedaner, T. (2010). Variation and transgression of aggressiveness among two Gibberella zeae crosses developed from highly aggressive parental isolates. Phytopathology, 100, 904–912.
Wang, H., Hwang, S. F., Eudes, F., Chang, K. F., Howard, R. J., & Turnbull, G. D. (2006). Trichothecenes and aggressiveness of Fusarium graminearum causing seedling blight and root rot in cereals. Plant Pathology, 55, 224–230.
Acknowledgements
This work was supported by the Russian Foundation for Basic Research (grant 15-04-09023-a). The Russian Science Foundation supported identification of F. culmorum strains (grant 16-16-00080) and pot experiments with soil (grant 14-16-00137). We thank the anonymous reviewers and the subject editor for their highly valuable comments and suggestions.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Research involving human participants and/or animals
Our research did not involve either Human Participants or Animals.
Informed consent
Non-applicable.
Rights and permissions
About this article
Cite this article
Strunnikova, O., Vishnevskaya, N., Shakhnazarova, V. et al. Development of two strains of Fusarium culmorum with a different aggressiveness in the soil and on the roots of barley of two genotypes. Eur J Plant Pathol 151, 579–592 (2018). https://doi.org/10.1007/s10658-017-1396-1
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10658-017-1396-1