Abstract
Fungal species comprising the Fusarium graminearum species complex (FGSC) may cause disease in maize and wheat. Host preference within the FGSC has been suggested, in particular F. boothii towards maize ears. Therefore, the disease development and mycotoxin production of five FGSC species in maize and wheat grain was determined. Eighteen isolates representing F. acaciae-mearnsii, F. boothii, F. cortaderiae, F. graminearum and F. meridionale were used. Each isolate was inoculated on maize ears and wheat heads to determine host preferences. Disease severity and disease incidence was measured for maize and wheat, respectively. Fungal colonisation and mycotoxins, deoxynivalenol (DON), nivalenol and zearalenone, was also quantified. Isolates differed significantly (P < 0.05) in their ability to produce symptoms on maize ears, however, no significant differences between FGSC species were determined. Similarly, significant differences (P < 0.05) between isolates but not between FGSC species in disease incidence on wheat were determined. The isolates also differed significantly (P < 0.05) in their ability to colonise maize and wheat grain. No significant differences in fungal colonisation, among the five FGSC species, were determined in field grown maize. However, under greenhouse conditions, F. boothii was the most successful coloniser of maize grain (P < 0.05). In wheat, F. graminearum colonised the grain more successfully and produced significantly more (P < 0.05) DON than the other species. Fusarium boothii isolates were the best colonisers and mycotoxin producers in maize, and F. graminearum isolates in wheat. The selective advantage of F. boothii to cause disease on maize was supported in this study.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
At least 33 mycotoxin-producing Fusarium species have been associated with grain crops in South Africa (Beukes et al. 2017), of which Fusarium graminearum sensu lato (s.l.) Schwabe is one of the most ubiquitous fungi associated with grain diseases, not only in South Africa, but also worldwide. High fungal infection rates lead to reduced seed germination, seedling blight and low kernel weight in small-grain cereals drastically reducing profits. The economic impact is further aggravated by price deductions due to reduced seed quality (Windels 2000). The most common diseases caused by F. graminearum include Gibberella ear rot (GER) of maize (Munkvold 2003; Boutigny et al. 2011, 2012) and Fusarium head blight (FHB) of small-grain cereals such as wheat and barley (Boutigny et al. 2011).
Infection of grain by F. graminearum also raises food safety concerns as the fungus is able to produce toxic secondary metabolites known as mycotoxins. Two groups of mycotoxins are commonly associated with F. graminearum s.l., namely zearalenone (ZEA) and the type B-trichothecenes (TCT-B), most commonly deoxynivalenol (DON) and, at a lower frequency, nivalenol (NIV) (Lee et al. 2009; Boutigny et al. 2012; Desjardins and Proctor 2011; Malbrán et al. 2014). The TCT-B group of mycotoxins are phytotoxic and act as virulence factors on sensitive plant hosts (Proctor et al. 1995, 2002; Jansen et al. 2005). Fusarium graminearum s.l. produces one of three strain-specific TCT-B profiles (chemotypes), namely: (i) NIV and its acetylated derivatives (NIV chemotype), (ii) deoxynivalenol and the acetylated derivative 3-acetyldeoxynivalenol (3-ADON chemotype), or (iii) deoxynivalenol and the acetylated derivative 15-acetyldeoxynivalenol (15-ADON chemotype) (Ward et al. 2002; Alexander et al. 2011).
Since 2000, at least 16 distinct phylogenetic lineages have been described within the F. graminearum morphospecies (O’Donnell et al. 2000, 2004, 2008; Starkey et al. 2007; Yli-Mattila et al. 2009; Sarver et al. 2011; Aoki et al. 2012). These lineages have been designated phylogenetic species status based on high-throughput multi-locus genotyping with DNA sequences from 13 nuclear genes, combined with genealogical concordance of phylogenetic species recognition (Taylor et al. 2000; Aoki et al. 2012). These lineages are collectively referred to as the F. graminearum species complex (FGSC).
An analysis of the FGSC associated with wheat, barley and maize in South Africa identified six FGSC species. Fusarium acaciae-mearnsii, F. cortaderiae and F. meridionale possess the NIV chemotype, the single F. brasilicum isolate a 3-ADON chemotype and F. boothii the 15-ADON chemotype. Isolates of F. graminearum predominately possesses the 15-ADON chemotype, with only two of the 391 isolates characterized representing a 3-ADON chemotype (Boutigny et al. 2011). The study by Boutigny et al. (2011) provided an understanding of the prevalence, geographic distribution and host range (of the host species included) of FGSC species in South Africa. Fusarium graminearum was found to be the dominant FGSC member associated with FHB of wheat and barley, accounting for more than 85% of the isolates collected. However, F. boothii was exclusively associated with GER of maize ears.
Wheat, barley and maize are often grown in rotation in South Africa, especially in the North West and Northern Cape provinces. Differences in FGSC species composition observed on these crops suggest a competitive advantage for F. boothii to colonise maize ears and F. graminearum to infect wheat heads (Boutigny et al. 2011). Evidence of the predilection of F. boothii towards maize as host was also demonstrated in China. Zhang et al. (2016) concluded that F. boothii is selected when maize is cultivated without rotation and the rotation of maize with wheat might restrict the infection cycle of F. boothii. Further indications of potential host preference among species of the FGSC was also observed for rice in Korea, where F. asiaticum was the dominant FGSC species causing FHB rather than F. graminearum or F. boothii (Lee et al. 2009). Likewise, F. asiaticum was mainly found in areas where rice is grown in rotation with wheat in China (Zhang et al. 2012), whereas F. graminearum was more common in wheat-maize rotation systems. In Brazil, F. graminearum was found to be dominant in wheat (Del Ponte et al. 2015), F. asiaticum in rice (Gomes et al. 2015), and F. meridionale was determined as the predominant FGSC species on maize ears and stalks (Kuhnem et al. 2016).
Understanding host-specific differences in pathogen composition is of critical importance in the development of disease and mycotoxin control strategies. Additionally, such information can provide clarity about the value of crop rotation systems as part of an integrated management strategy. It could also lead to novel approaches to achieve improved resistance in commercial cultivars, by using isolates with high virulence towards the host crop in inoculation studies when screening for resistance. In this study, the disease development and mycotoxin production by isolates representing FGSC species in maize, grown under field and greenhouse conditions, were investigated to better understand host preferences. These isolates were also inoculated onto wheat, under field conditions, to investigate differential disease development and mycotoxin production between maize and wheat.
Materials and methods
Fungal isolates and inoculum preparation
Eighteen FGSC isolates representing five species, collected from diseased wheat and maize in South Africa, were used in this study (Boutigny et al. 2011) (Table 1). The isolates are maintained in the culture collection of the Department of Plant Pathology at Stellenbosch University, South Africa. Conidial suspensions for greenhouse maize inoculations were prepared by cultivating FGSC isolates in 100 mL mung bean broth (Evans et al. 2000) at 25 °C on a rotary shaker at 100 rpm. After 14 days the fungal spores were collected by filtration through double layer sterile cheesecloth. The suspensions were then centrifuged at 4000 rpm for 10 min at 4 °C, the supernatant discarded and the conidia washed twice with de-ionised, autoclaved water. The conidial pellet was suspended in sterile de-ionised water and the spore concentration determined using a haemocytometer. The spore suspension was adjusted to a final concentration of 4 × 104 conidia mL−1.
Little to no macroconidia production by some FGSC isolates necessitated the use of mycelia for inoculations of maize and wheat field trials. The FGSC isolates were, therefore, inoculated in 100 mL potato dextrose broth (PDB), incubated at 25 °C and shaken at 100 rpm, and mycelia harvested by filtration through four layers of cheesecloth after 1 week. The harvested mycelia was rinsed with sterile de-ionised water and allowed to dry for 30 min. For the inoculation of maize and wheat, 3.0 g and 4.0 g mycelia, respectively, was weighed into 100 mL sterile water. The mycelia was then pulverised (VirTis® model no 6303–0002, The Virtis Company, USA) to form a homogenous suspension. The viability of the inoculum was confirmed by plating 1 mL suspension onto potato dextrose agar (PDA) after the completion of inoculations. Isolates were kept separate for all inoculations and were not co-inoculated onto the same plant.
Plant material and artificial inoculation
Maize: A white maize cultivar, PHB30D07B (DuPont Pioneer®, South Africa) characterised as susceptible to GER, was planted at the Welgevallen experimental farm at Stellenbosch University, Stellenbosch, Western Cape, South Africa. In the greenhouse, seeds were planted directly into 15-L plant bags filled with organic coconut coir growth medium (Vegtech 2000, South Africa). Plants were irrigated twice daily with approximately 500 mL of non-fertilised water until the seedlings were 2 weeks old. Thereafter, plants were irrigated twice daily with 1 L of elemental nutrient solution per day. For field trials, the seeds were planted manually in 5-m long rows with an intra-row spacing of 0.3-m and an inter-row spacing of 1-m. The field was fertilised 3 weeks after planting with 2:3:2 N:P:K (22) + ZN at a rate of 200 kg ha−1, and top-dressed with LAN 28 at the eighth leaf stage (Small et al. 2012). Water was supplied using a Tritan 2 L hr.−1 drip-irrigation pipe (Loxton Irrigation, South Africa) for 90 min per day for the first 2 months, after which irrigation time was reduced to 60 min every second day.
Silk channel inoculation was performed 6 days after silk emergence (Reid et al. 2002) according to Small et al. (2012). In the greenhouse, 2 mL of the liquid spore suspension was injected down the silk channel of primary ears, with three plants per treatment, repeated three times in a randomised complete block design (RCBD). Mock inoculations with sterile de-ionised water served as controls. In the field trial, experimental plots of +/−15 plants represented a replicate with each treatment replicated three times (RCBD). Sterile de-ionised water served as the control treatment. After inoculation, overhead irrigation was applied daily to ensure sufficient humidity for disease development. Inoculated maize ears were handpicked 8 weeks after inoculation, de-husked and artificially dried in a forced-air oven at 50 °C to 12–14% moisture measured with a Delmhorst G-7 Grain Moisture Meter (Delmhorst Instrument Co., USA).
Wheat: A wheat cultivar, SST047 (Sensako (Pty) Ltd., South Africa) characterised as susceptible to FHB, was planted under dryland conditions in a winter rainfall area at the Sensako Research Farm, Napier, Western Cape, South Africa. The trial was fertilized using standard fertilization practices. Experimental units (plots) were each 5-m long, and plots were inoculated in triplicate (RCBD) with the 18 FGSC isolates. For inoculations, wheat plants were tied into bunches containing six heads, and ten bunches per 5-m row served as a replicate. Bunches were spray-inoculated with 10 mL of freshly prepared inoculum (Engle et al. 2003) during full anthesis, and sterile water was used for mock-inoculations. The inoculated wheat heads were then covered with plastic bags to maintain a humid micro-environment (Teich and Michelutti 1993), and brown paper bags to prevent ultra-violet (UV) damage of the inoculated fungus. The bags were removed 3 days after inoculations and inoculated bunches were handpicked 10 weeks after inoculations.
Disease evaluation and grain handling
At harvest, maize ears (between 12 and 18% kernel moisture content) were handpicked, de-husked and inspected for visual disease symptoms. Disease severity was rated as a percentage of each ear surface showing Gibberella ear rot (GER) symptoms, which include dark purple or pink mold that usually starts at the tip of the ear. Kernels exhibiting ‘starburst’ symptoms (white streaking radiating from the cap of the kernels) were also considered as diseased (Clements et al. 2004; Picot et al. 2012). Maize ears were hand-shelled and bulked according to each replicate. The maize kernels were crushed using a Husqvarna cast-iron corn mill hand grinder (Reliance, Sweden), with the grinder being cleaned thoroughly between batches with high-pressure air to avoid cross contamination. Disease incidence, expressed as the percentage of infected wheat heads per inoculated bunch (Hilton et al. 1999; Wegulo et al. 2011), was determined 6 weeks post-inoculation. The presence of the disease was characterised by symptoms of premature bleaching. Wheat heads were excised 1 cm below the basal spikelet at harvest and the grain was mechanically threshed and winnowed. A sub-sample (~25 g) of the coarsely-milled maize and wheat kernels of each replicate, respectively, were further milled into a fine flour with a Philips 400 W blender (South Africa) and stored at −20 °C. Flour sub-samples (2 g and 5 g) from each replicate was stored at −20 °C until DNA and mycotoxin extractions were performed.
Fungal colonisation determination: DNA isolation for real-time PCR
A reference isolate of F. graminearum (NRRL 28439), provided by Dr. Kerry O’Donnell (USDA-ARS, Peoria, IL., USA), was used to produce DNA standards for the quantification of FGSC species in maize and wheat. The fungus was grown on PDA for 7 days and genomic DNA was extracted from freeze-dried mycelia using the commercial Qiagen DNeasy® Plant Mini Kit (Whitehead Scientific (Pty) Ltd., South Africa), with some modifications in the cell lysis and protein removal steps (Fredlund et al. 2008; Boutigny et al. 2012). DNA from the inoculated maize and wheat samples, as well as additional DNA extractions from FGSC-free, water-inoculated maize and wheat was performed according to Boutigny et al. (2012). The fungal and plant DNA was quantified using the Nanodrop ND-1000 Spectrophotometer (Inqaba Biotechnical Industries, South Africa). The DNA purity was evaluated by comparing the absorbance ratios A260/280 (pure DNA ~1.8) and A260/230 (ideally 1.8 to 2.2), and the plant DNA was diluted to 10 ng μL−1.
Quantification of FGSC fungal DNA
Primers ZEA-F (5′-CTGAGAAATATCGCTACACTACCGAC-3′) and ZEA-R (5′-CCCACTCAGGTTGATTTTCGTC-3′) (Atoui et al. 2012) were used to quantify FGSC DNA. The qPCR assay was carried out in a total volume of 20 μL comprising 1X KAPA SYBR® FAST, 2 μL DNA (10 ng μL−1), 200 nM of each primer for maize and 250 nM for wheat. Real-time PCR was performed on a Rotor-GeneTM 6000 (Corbett Life Science, Whitehead Scientific (Pty) Ltd., South Africa) using the cycling conditions: 95 °C for 5 min, 35 cycles of 95 °C for 15 s, 65 °C for 35 s and 72 °C for 1 s, followed by melt curve analyses from 72 °C to 95 °C, rising by 1 °C each step.
The analytical specificity of the primer set to amplify the target sequences of the 18 FGSC isolates (Table 1), was determined. The isolates were grown on PDA plates for 5 days and genomic DNA was then extracted from fresh aerial mycelia using the Promega Wizard® SV Genomic DNA Purification System (Anatech Instruments (Pty) Ltd., South Africa). Fungal DNA was quantified using the Nanodrop ND-1000 Spectrophotometer and diluted to ~10 ng μL−1 for use in the qPCR assays. The qPCR reactions were prepared using the optimised cycling conditions as described.
For direct comparisons with inoculated grain samples, a matrix matched mixed DNA standard curve was prepared by diluting the F. graminearum (NRRL 28439) DNA (~20 ng μL−1) 41–46-fold in FGSC-free, water-inoculated maize or wheat DNA (10 ng μL−1). Real-time PCR reactions for each dilution standard were performed in triplicate. Standard curves were analysed using the Rotor-GeneTM 2.0.2.4 software (Corbett Life Science, Whitehead Scientific (Pty) Ltd., South Africa), which plotted the cycle threshold (Ct) values of the diluted standards against the logarithm of the known DNA concentration (Nanodrop Spectrophotometer measurement). Dynamic tube normalisation, starting at Cycle 1 and noise slope correction for standard curve analysis was selected on the Rotor-GeneTM 2.0.2.4 software programme for all runs performed.
To assess the linearity and presence of inhibitors, the correlation coefficient (R2), slope (M-value) and reaction efficiency (E-value) were noted after analysis of each standard curve. Stringent parameters were set to ensure accurate, inhibition-free qPCR assays with R2 > 0.99, M-values between −3.2 and −3.4, and an efficiency of 0.98 to 1.05 (Corbett Life Science 2006; European Commission 2008; Bustin et al. 2009). The melting temperatures (Tm) of qPCR products were evaluated to confirm analytical specificity in planta.
To quantify FGSC target DNA in the inoculated maize and wheat samples, the qPCR assays contained a duplicate of each sample and a triplicate of the standard (F. graminearum DNA diluted 42-fold in target free host DNA). Using the matrix specific standard curves, the Ct values were transformed into DNA concentrations using the Rotor-Gene™ 2.0.2.4 software.
Mycotoxin quantification
Mycotoxin extractions were performed on 5-g maize and wheat flour samples according to Rose et al. (2016). The analytes, together with the freshly prepared standard dilution series of known concentration, were sent to the Central Analytical Facility (CAF), Stellenbosch University for the simultaneous quantification of DON, NIV and ZEA using liquid chromatography tandem mass spectrometry (LC-MS/MS). The analytical standards, used to prepare the standard dilution series of DON, NIV and ZEA, were obtained from Sigma-Aldrich (South Africa), and were all guaranteed >98% pure.
Statistical analyses
All data obtained was subjected to univariate analysis of variance (ANOVA) using the generalised linear model (GLM) procedure of SAS statistical software version 9.2 (SAS Institute Inc., Cary, NC, USA). The replicates of each isolate were used to determine differences between isolates. To determine the most aggressive FGSC species, isolates were considered as replicates within a FGSC species (means of replicates for each isolate). The Shapiro-Wilk test was performed to test for normality (Shapiro and Wilk 1965) and variables (fungal colonisation, DON, NIV and ZEA content) were ln transformed [Ln(x + 1)] to stabilise the variance and improve normality for all measurements (Snedecor and Cochran 1980). Transformation did not improve normality, thus the untransformed data was reported on. Student’s t-test least significant difference (LSD) was calculated at the 5% level to compare isolates and species means with respect to the variable tested (Ott 1998), and a probability level of 5% (P < 0.05) was considered significant for all significance tests.
Pearson correlation coefficients (R) were used to determine the relationship between fungal colonisation and mycotoxin contamination (DON or NIV and ZEA) using the CORR procedure in SAS. The relationships were classified as very strong (R > 0.7), strong (40 < R < 69), moderate (30 < R < 39), weak (20 < R < 29) and negligible (1 < R < 19) (Quinnipiac University 2015), and were only considered significant when P < 0.05.
Results
Standard curves for qPCR assays
The standard curves obtained for the quantification of FGSC target DNA in both maize and wheat DNA matrices indicated that the correlation coefficient (R2) for linearity and inhibition tests were 0.999 for each. The slopes were also within the acceptance criterion with M values of −3.32 for maize and −3.31 for wheat. The reaction efficiencies within both matrices were 1.00 (Table 2). The analytical specificity of primer pair ZEA-F/ZEA-R was demonstrated as all the FGSC isolates were successfully amplified. Melt curve analysis of the amplicons revealed a melt temperature (Tm) of 82.5–82.7 °C for all the isolates. The NTC reaction, which generated a fluorescent signal, produced a different PCR product with a Tm of 75 °C.
Disease severity, fungal colonisation and mycotoxin contamination in maize
Greenhouse trial: Significant differences between FGSC isolates were determined in their ability to cause disease on maize grain. All F. boothii isolates caused significantly (P < 0.05) more disease than the water-inoculated control, with three isolates being most virulent based on the variation in the degree of disease severity (Burlakoti et al. 2012) (Table 3). The F. boothii isolates STE-U 8009 (51.9%) and STE-U 8013 (51.1%) caused significantly more disease (P < 0.05) then the other FGSC isolates evaluated, with the exception of the third most virulent isolate, F. boothii STE-U 8010 (42.6%). Isolates of F. acaciae-mearnsii and F. cortaderiae did not cause significantly (P > 0.05) more disease than the water-inoculated control, and only one isolate each of F. graminearum, STE-U 8018 (20.0%), and F. meridionale, STE-U 8021 (23.6%), displayed significant more disease symptoms (P < 0.05) than the water-inoculated control (Table 3).
FGSC isolates differed significantly (P < 0.05) in their ability to colonise and produce their associated mycotoxins in maize (Table 3). Fusarium boothii isolates were the most successful maize colonisers, with significantly (P < 0.05) more fungal DNA of isolate STE-U 8013 measured in inoculated grain (15.46 ng μL−1) than in any of the other FGSC isolates used (Table 3). The F. boothii isolates STE-U 8010 (8.54 ng μL−1) and STE-U 8011 (7.41 ng μL−1) also resulted in significantly (P < 0.05) more fungal DNA compared to the water control (0.17 ng μL−1). The highest DON concentration (40.12 μg mL−1) in maize grain was produced by F. graminearum isolate STE-U 8018, followed by three F. boothii isolates; STE-U 8013 (30.63 μg mL−1), STE-U 8010 (27.63 μg mL−1) and STE-U 8012 (23.19 μg mL−1). These four isolates differed significantly (P < 0.05) from the water-inoculated control, but not from each other (Table 3). Two F. meridionale isolates, STE-U 8021 (5.93 μg mL−1) and STE-U 8022 (4.93 μg mL−1), were the only treatments which resulted in significantly higher (P < 0.05) NIV concentrations than measured in the water control, although they did not differ significantly (P > 0.05) from the other isolates (Table 3). Only three isolates, the two F. boothii isolates STE-U 8013 (7.91 μg mL−1) and STE-U 8009 (7.12 μg mL−1), as well as the F. graminearum isolate STE-U 8018 (7.48 μg mL−1), resulted in significantly (P < 0.05) higher ZEA levels than measured in the water inoculated control (Table 3).
The most virulent FGSC species on maize, based on disease severity, was F. boothii (37.44%), as it was the only species to cause significantly (P < 0.05) more disease than all the FGSC species (Table 4). Fusarium boothii was also the only species to result in significantly (P < 0.05) higher fungal DNA levels (7.57 ng μL−1) than the water-inoculated control, although it did not differ significantly (P > 0.05) from F. graminearum (1.9 ng μL−1) (Table 4). Neither F. boothii (21.53 μg mL−1) nor F. graminearum (22.31 μg mL−1) -inoculated grain contained significantly more DON than the water-inoculated control (0.49 μg mL−1), and only F. meridionale-inoculated maize contained significantly (P < 0.05) higher NIV levels (4.21 μg mL−1) than the water control (not detected), although it did not differ significantly (P > 0.05) from the other two NIV-producing species (Table 4). No significant differences (P > 0.05) were observed between any of the species and the water control when evaluating ZEA contamination (Table 4).
Significant (P < 0.05) correlations were obtained among disease severity, target DNA concentrations and mycotoxin contamination in maize grain (Fig. 1 a-i)). Strong positive correlations were obtained between disease severity and fungal colonisation (R = 0.64; P < 0.0001) (Fig. 1 a) as well as between disease severity and DON (R = 0.52; P = 0.005) (Fig. 1 b) and disease severity and NIV contamination (R = 0.67; P = 0.0001) (Fig. 1 g). A very strong relationship was found between disease severity and ZEA contamination (R = 0.70; P < 0.0001) (Fig. 1 c). Strong positive correlations were observed between fungal colonisation and DON (R = 0.54; P = 0.0046) (Fig. 1 d), NIV (R = 0.61; P = 0.0008) (Fig. 1 h) and ZEA contamination (R = 0.51; P = 0.0002) (Fig. 1 f). Additionally, a strong, positive correlation was found between ZEA and DON contamination (R = 0.69; P < 0.0001) (Fig. 1 e), and a very strong correlation between ZEA and NIV contamination of maize grain (R = 0.70; P < 0.0001) (Fig. 1 i).
Pearson correlation coefficients (R) demonstrating the relationship between (a) disease severity and fungal colonisation, (b) disease severity and deoxynivalenol (DON) contamination, (c) disease severity and zearalenone (ZEA) contamination, (d) DON contamination and fungal colonisation, (e) DON and ZEA contamination levels, (f) ZEA contamination and fungal colonisation, (g) disease severity and nivalenol (NIV) contamination, (h) fungal colonisation and NIV contamination, and (i) ZEA and NIV contamination levels on greenhouse grown maize grain inoculated with 18 isolates representing five Fusarium graminearum species complex species
Field maize: The disease severity of GER symptoms on maize ears ranged from 0.4 to 24.5% (Table 5). Significant differences (P < 0.05) in disease severity were determined amongst some of the 18 FGSC isolates (Table 5). The highest disease severity was caused by F. boothii isolates STE-U 8009 (10.8%), STE-U 8008 (13.3%), STE-U 8010 (24.5%) and STE-U 8011 (10.3%). Isolate STE-U 8009 was the most virulent, causing significantly more (P < 0.05) disease than all isolates inoculated. Notably, an isolate of F. acaciae-mearnsii (STE-U 8006) and F. meridionale (STE-U 8021) also produced 9.4 and 9.8% disease severity, respectively. However, only F. boothii isolates STE-U 8008 and STE-U 8010 produced significantly more GER symptoms than the water-inoculated control (0.4%).
FGSC isolates differed significantly (P < 0.05) in their ability to colonise and produce their associated mycotoxins in maize (Table 5). The F. boothii isolate, STE-U 8010, was the most successful maize coloniser (1.62 ng μL−1) which was significantly more (P < 0.05) than all other isolates, except for isolates STE-U 8006 (F. acaciae-mearnsii) (0.93 ng μL−1) and STE-U 8013 (F. boothii) (0.92 ng μL−1) (Table 5). Not all isolates belonging to F. acaciae-mearnsii, F. cortaderiae and F. meridionale, classified as NIV producers based on chemotype, were able to produce detectable levels of NIV in maize grain. However, F. boothii and F. graminearum isolates, classified as DON producers, all produced detectable levels of DON.
The F. boothii isolate, STE-U 8010 was the highest DON-producing isolate and did not differ significantly from F. boothii isolate STE-U 8011 (20.3 μg mL−1). Both, however, differed significantly from the water-inoculated control (P < 0.05), but not from the remaining F. boothii isolates. The highest DON-producing F. graminearum isolate, STE-U 8017 (5.6 μg mL −1), did not differ significantly (P > 0.05) from the water control or from the remaining F. graminearum isolates. The NIV content of grain inoculated with the highest NIV-producing isolates for F. acaciae-mearnsii (STE-U 8005) (0.68 μg mL−1), F. cortaderiae (STE-U 8015) (0.54 μg mL−1) and F. meridionale (STE-U 8022) (0.11 μg mL−1) did not differ significantly (P > 0.05) from the water-inoculated controls. The highest ZEA levels were detected in maize grain inoculated with F. boothii isolate STE-U 8013 (2.98 μg mL−1) and was the only isolate to differ from the water inoculated control (P < 0.05) (Table 5).
All FGSC species caused disease, colonised maize ears and produced detectable TCT-B levels, but the most aggressive FGSC species on maize grain could not be conclusively established (Table 6). Fusarium boothii caused the highest disease severity (11.9%) and was the only species to differ significantly (P < 0.05) from the water control, but did not differ significantly from the other FGSC species. This was also observed for fungal colonisation, in which Fusarium boothii was the most successful grain coloniser with the highest target DNA level (0.77 ng μL−1) but did not differ significantly (P > 0.05) from the other FGSC species. It was, however, the only FGSC species that differed significantly (P < 0.05) from the water-inoculated control (Table 6). The DON levels produced by F. boothii (14.1 μg mL−1) and F. graminearum (3.8 μg mL−1) did not differ significantly from each other, but F. boothii differed significantly (P < 0.05) from the water-inoculated control (not detected). Fusarium acaciae-mearnsii was the highest NIV-producing FGSC species (0.42 μg mL−1), followed by F. cortaderiae (0.22 μg mL−1) and F. meridionale (0.04 μg mL−1). None of their NIV concentrations measured, however, differed significantly (P > 0.05) from the water-inoculated control (not detected). The highest ZEA levels were produced by F. boothii (1.11 μg mL−1), followed by F. acaciae-mearnsii (0.26 μg mL−1)- and F. meridionale (0.25 μg mL−1)-inoculated maize grain, which did not differ significantly from one another, or the water control (0.01 μg mL−1) (Table 6).
A highly significant positive relationship was recorded between disease severity and fungal colonisation (R = 0.805; P < 0.0001) (Fig. 2 a), as well as between disease severity and DON accumulation (R = 0.845; P < 0.0001) Fig. 2 b). A significant, strong positive relationship (R = 0.441; P = 0.0006) was also determined between disease severity and ZEA accumulation (Fig. 2 c). A significant, very strong positive relationship was recorded between fungal colonisation and DON accumulation (R = 0.895; P < 0.0001) (Fig. 2 d). No significant correlation could be established between NIV accumulation and fungal colonisation (P = 0.947). Significant, strong positive relationships were observed between ZEA and DON accumulation (R = 0.59; P < 0.0001) (Fig. 2 e), and between fungal colonisation and ZEA accumulation (R = 0.68; P < 0.0001) (Fig. 2 f). An insignificant correlation (P = 0.981) was observed between NIV and ZEA accumulation.
Pearson correlation coefficients (R) to demonstrate the relationship between (a) disease severity and fungal colonisation, (b) disease severity and deoxynivalenol (DON) contamination, (c) disease severity and zearalenone (ZEA) contamination, (d) DON contamination and fungal colonisation, (e) ZEA contamination and DON contamination, (f) fungal colonisation and ZEA contamination levels on field grown maize grain inoculated with 18 isolates representing five Fusarium graminearum species complex species
Disease incidence, fungal colonisation and mycotoxin contamination in wheat
Wheat: FGSC isolates differed significantly (P < 0.05) in their ability to cause FHB to wheat (Table 7). Fusarium boothii isolate STE-U 8013 (68.0%) caused the highest disease incidence, followed by F. cortaderiae isolate STE-U 8015, (58.0%) and F. meridionale isolate STE-U 8022 (55.9%). The disease incidence determined by these isolates did not differ significantly (P < 0.05), but differed significantly from the water-inoculated control (21 .4%).
FGSC isolates also differed significantly (P < 0.05) in their ability to colonise and produce their associated mycotoxins in wheat (Table 7). Two F. graminearum isolates, STE-U 8017 and STE-U 8018, were the most successful grain colonisers, resulting in significantly higher (P < 0.05) fungal target DNA levels than the remaining isolates and the water inoculated control, at 0.96 and 0.73 ng μL−1, respectively. The highest DON-producing isolate, F. graminearum isolate STE-U 8018 (0.24 μg mL−1), produced significantly more DON (P < 0.05) than the other DON-producing isolates and the water control (Table 7). The highest DON-producing F. boothii isolate (STE-U 8010) (0.05 μg mL−1) did not produce significantly more DON (P > 0.05) than the levels detected in the other F. boothii isolates or in the water-inoculated control (Table 7). The F. acaciae-mearnsii isolate STE-U 8005 was the only isolate to produce significantly more NIV (0.05 μg mL−1) (P < 0.05) than the water-inoculated control. No significant differences (P > 0.05) in the NIV content of wheat grain inoculated with the highest NIV-producing isolates of F. cortaderiae (STE-U 8015) (0.01 μg mL−1) and F. meridionale (STE-U 8021) (0.01 μg mL−1) was observed when compared to the water-inoculated controls (Table 7).
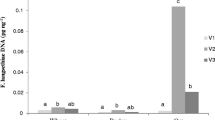
Evaluation of the five FGSC species revealed that the highest visual disease incidence on inoculated wheat grain was caused by F. cortaderiae (54.4%), followed by F. meridionale (49.2%), F. graminearum (47.3%) and F. acaciae-mearnsii (44.4%). These FGSC species did not differ significantly from one another or Fusarium boothii (43.6%). However, F. boothii was the only species that did not cause significantly more disease (P > 0.05) than the water-inoculated control plants (21.4%) (Table 8). Fusarium graminearum was the most successful coloniser of wheat kernels accumulating significantly more (P < 0.05) fungal target DNA (0.68 ng μL−1) than the other FGSC species. It also produced significantly (P < 0.05) more DON (0.16 μg mL−1) than the only other DON-producing species, F. boothii (0.04 μg mL−1), and the water control. The NIV content in wheat grain inoculated with the highest NIV-producing species, F. acaciae-mearnsii (0.038 μg mL−1), differed significantly (P < 0.05) from the other NIV-producing species which failed to produce significantly more NIV (P > 0.05) than the water control (Table 8). No ZEA was detected in inoculated wheat grain.
No significant relationships were determined between disease incidence and fungal accumulation or mycotoxin contamination in wheat grain. Fungal colonisation correlated significantly with DON (P < 0.0001) and NIV (P = 0.003) accumulation, with a stronger relationship between fungal colonisation and DON (R = 0.72) (Fig. 3 a) than between fungal colonisation and NIV (R = 0.52) (Fig. 3 b). ZEA was not detected in any of the samples evaluated.
Discussion
In this study, significant differences in disease expression, fungal colonisation and mycotoxin contamination of maize and wheat grain by individual isolates of FGSC were observed. Inoculation with F. boothii isolates resulted in the highest fungal target DNA in greenhouse and field-grown maize. Isolates of F. graminearum were the most successful kernel colonisers and highest DON producers in wheat grain. These findings are in accordance with a study by Boutigny et al. (2011), who also found F. boothii and F. graminearum to be the predominant FGSC species associated with South African maize and wheat, respectively. Furthermore, Lamprecht et al. (2011) found significant differences in pathogenicity between F. boothii, F. graminearum and F. meridionale in causing maize root rot. Hao et al. (2017), however, found no differences between FGSC isolates from wheat and maize when determining their pathogenicity on both maize and wheat using a Petri plate assay.
Significant differences in disease expression, fungal colonisation among the five FGSC species present in South Africa could not be definitively established in field grown maize. However, F. boothii was the most successful FGSC species in colonising maize ears when compared to the four other species evaluated under greenhouse conditions. For maize, FGSC species associated with GER differ according to geographical region and cropping systems. In Nepal, GER of maize was predominantly associated with F. meridionale and F. asiaticum. In South Korea and France, GER infected maize was mostly contaminated with F. graminearum, while F. boothii was the only FGSC species found on maize in South Africa (Lee et al. 2012; Desjardins and Proctor 2011; Boutigny et al. 2011, 2014). Noticeably, F. boothii was also commonly detected in regions in South Korea where only maize was grown (Lee et al. 2012), while the occurrence of F. boothii on maize decreased in areas in China where maize and wheat was grown in rotation (Zhang et al. 2016).
In wheat grain, however, F. graminearum was the most aggressive species, resulting in the highest fungal colonisation and DON contamination levels. This suggest an infection-specific adaptation of F. graminearum towards wheat, which was not surprising, as this species is the FGSC species also most commonly associated with wheat world-wide (Gale et al. 2007; Boutigny et al. 2011, 2014; Qiu and Shi 2014). However, it must be noted that F. graminearum is not the only FGSC species affecting wheat.
Host-specificity and host-preference within the FGSC have been implied (Lee et al. 2009; Boutigny et al. 2011). Host-specificity, referring to an exclusive FGSC species-host relationship, is not unlikely, as various FGSC species have only been found on a single host plant (Aoki et al. 2012). The frequency of occurrence of FGSC species on specific host crops, and a selective advantage to infect these, suggests host-preference. The value of TCT-Bs to act as virulence factors in maize and wheat, but not in barley, suggests a selective advantage for F. graminearum to infect maize and wheat rather than barley (Proctor et al. 1995; Maier et al. 2006). Differences in the development of FHB in wheat and barley caused by F. graminearum are also known to exist (Kazan et al. 2012). This was demonstrated by the differential gene expression in F. graminearum observed during infection of the two grain crops. Genes expressed exclusively in wheat were enriched for genes involved in allantoin and allantoate transport, which could contribute to increased pathogen fitness (Lysøe et al. 2011). In another study, 69 F. graminearum genes were preferentially expressed in developing maize kernels compared to inoculated wheat and barley spikes (Harris et al. 2016). The genomic flexibility of F. graminearum to adapt to a range of hosts was demonstrated by these host-specific differences. Studying transcriptional differences between FGSC species during the infection of different host crops could contribute to understanding host specificity and/or preference.
A strong correlation was observed in this study between disease severity and fungal colonisation in maize, and an insignificant correlation between disease incidence and fungal colonisation in wheat. This demonstrated that disease severity, rather than incidence, can be used as an indicator of colonisation by FGSC species. Disease severity is an informative measurement of aggressiveness, and can allow for improved distinction when comparing species and isolates (Burlakoti et al. 2007). Disease incidence is defined as the number of infected plants and is, therefore, not an indicator of the degree to which plants are infected (Hilton et al. 1999; Wegulo et al. 2011). A strong positive correlation between disease severity and DON accumulation in maize, while no meaningful correlation between DON and disease incidence in wheat could be established, and further demonstrates the limitations of disease incidence to serve as an indicator of the aggressiveness of FGSC isolates.
The strong, positive correlations between DON accumulation and fungal colonisation in maize and wheat confirmed results of previous studies on F. graminearum infection of these crops (Burlakoti et al. 2007; Fredlund et al. 2008; Boutigny et al. 2012; Malbrán et al. 2014). The findings of this and other studies supported the role of DON as a virulence factor, allowing the fungus to colonise infected grain more successfully (Proctor et al. 1995, 2002). The most prolific DON-producing isolates on maize and wheat in the current study, irrespective of species annotation, were also the best grain colonisers. Weaker correlations were observed between NIV and fungal colonisation. This may be due to the higher phytotoxicity of DON when compared to NIV (Desjardins 2006), indicating DON’s potential as a better contributor to virulence. The understanding that TCT-B mycotoxins act as virulence factors in host plants could be misleading as only DON, and not NIV, was measured in most studies (Proctor et al. 1995, 2002; Jansen et al. 2005; Menke et al. 2012). However, Purahong et al. (2014) found no significant difference in aggressiveness among isolates representing the 3-ADON, 15-ADON and NIV chemotypes in a single-floret wheat inoculation study.
The absence of significant correlations between NIV concentrations and fungal colonisation could be attributed to the lack of sensitivity of the LC-MS/MS method to detect NIV at low concentrations, thereby reducing the ability to determine significant differences and correlations. Weaker correlations were observed between ZEA and fungal colonisation in maize than between DON and fungal colonisation, with no measurable levels of ZEA in wheat. ZEA is not known as a virulence factor and does not contribute to the establishment and colonisation of host tissue by the pathogen. Rather, high concentrations of ZEA have been shown to have a stimulatory effect on maize embryo development (McLean 1995).
The determination of fungal colonisation by qPCR for predicting mycotoxin contamination in grains have previously been proposed, as strong positive correlations between fungal biomass and mycotoxin contamination have been reported (Sarlin et al. 2006; Nicolaisen et al. 2009; Atoui et al. 2012). However, the identity of mycotoxigenic fungi in the target grain should be known in order to select qPCR primers with sufficient sensitivity to amplify these species. This was made apparent by the weak correlations between F. graminearum DNA and DON levels in wheat and barley, due to the co-occurrence of F. culmorum, compared to the better correlation between DON and TCT-producing Fusarium DNA levels in barley (Sarlin et al. 2006). Therefore, employing primers that target genes responsible for mycotoxin production (Sarlin et al. 2006; Atoui et al. 2012), rather than species-specific primers (Sarlin et al. 2006; Nicolaisen et al. 2009), should be considered when attempting to predict mycotoxin contamination risks. Furthermore, fluctuating climatic conditions, the effect of fungicide applications and cultivar resistance could also influence mycotoxin production, irrespective of the extent of fungal colonisation (Wegulo et al. 2011; Boutigny et al. 2012; Covarelli et al. 2015).
Plant-pathogen interactions are influenced by environmental conditions that could influence disease development, pathogen colonisation and mycotoxin production (Proctor et al. 2002; Malbrán et al. 2012). A higher aggressiveness of F. boothii isolates on maize, and F. graminearum isolates on wheat, was observed in this study. Climatic conditions play an intricate role in host-pathogen interactions, which could further complicate host specificity studies. Host preference within the FGSC, supported by the prevalence of FGSC species in specific host plants (Lee et al. 2009; Boutigny et al. 2011; Qiu and Shi 2014), were also strongly correlated with the temperature range of the surveyed regions. Fusarium graminearum has been recorded at a higher frequency in the cooler regions of China, Japan and Korea, both on rice and maize, while F. asiaticum incidence increased in the warmer regions (Qu et al. 2008; Suga et al. 2008; Lee et al. 2009). Differences in FGSC species distribution, linked to variation in temperature, could account for the differences in FGSC composition observed by Boutigny et al. (2011) on South African grains. Maize is grown during the hot summer months, and wheat during the cooler winter months in South Africa. The higher colonisation and TCT-B production by F. boothii isolates in maize, planted in summer, and F. graminearum in wheat, planted in winter, might therefore be influenced by temperatures as much as by their preference of the host plant. However, Zhang et al. (2012) observed differences in FGSC species composition in different regions in China, which could not be attributed to differences in average temperatures between the regions studied. In contrast to the studies which found F. graminearum associated with cooler regions of (Qu et al. 2008; Suga et al. 2008; Lee et al. 2009), the study of Zhang et al. (2012) revealed no significant difference between the average temperatures in the areas where either F. graminearum or F. asiaticum were mostly found. A thorough review on the biogeography of the FGSC by van der Lee et al. (2015) further demonstrated host preference and concluded preferential infection of rice by F. asiaticum and maize by F. boothii and F. meridionale, while F. graminearum is found in association with wheat, barley and maize.
Host specificity, or host preference, remains to be elucidated within the FGSC. The degree of fungal colonisation, following fungal infection, and the subsequent accumulation of harmful mycotoxins, depend on an interaction between the pathogen, the host and the environment. The findings of this study supports differential aggressiveness, host preference and mycotoxin production between FGSC species. Fusarium boothii isolates were the best colonisers and mycotoxin producers in maize, and F. graminearum isolates in wheat. The selective advantage of F. boothii to cause disease on maize was supported in this study. The high level of variability among isolates of the FGSC seen in this and other studies (Goswami and Kistler 2005; Moradi et al. 2010; Malbrán et al. 2012), however, highlights the complexities associated with determining host preference by FGSC species.
References
Alexander, N. J., McCormick, S. P., Waalwijk, C., van der Lee, T., & Proctor, R. H. (2011). The genetic basis for 3-ADON and 15-ADON trichothecene chemotypes in Fusarium. Fungal Genetics and Biology, 48, 485–495.
Aoki, T., Ward, T. J., Kistler, H. B., & O’Donnell, K. (2012). Systematics, phylogeny and Trichothecene mycotoxin potential of fusarium head blight cereal pathogens. Mycotoxins, 62(2), 91–102.
Atoui, A., El Khoury, A., Kallassy, M., & Lebrihi, A. (2012). Quantification of Fusarium graminearum and Fusarium culmorum by real-time PCR system and zearalenone assessment in maize. International Journal of Food Microbiology, 154, 59–65.
Beukes, I., Rose, L.J., Shephard, G.S., Flett, B.C. & Viljoen, A. (2017). Mycotoxigenic Fusarium species associated with grain crops in South Africa – A review. South African Journal of Science, 113 (3/4), art. #2016-0121, 12 pages.
Boutigny, A.-L., Ward, T. J., Van Coller, G. J., Flett, B., Lamprecht, S. C., O’Donnell, K., et al. (2011). Analysis of the Fusarium graminearum species complex from wheat, barley and maize in South Africa provides evidence of specie-specific differences in host preference. Fungal Genetics and Biology, 48(9), 914–920.
Boutigny, A.-L., Beukes, I., Small, I., Zuhlke, S., Spiteller, M., Van Rensburg, B. J., Flett, B., & Viljoen, A. (2012). Quantitative detection of Fusarium pathogens and their mycotoxins in south African maize. Plant Pathology, 61(3), 522–531.
Boutigny, A.-L., Ward, T. J., Ballois, N., Iancu, G., & Ioos, R. (2014). Diversity of the Fusarium graminearum species complex on French cereals. European Journal of Plant Pathology, 138, 133–148.
Burlakoti, R. R., Estrada Jr., R., Rivera, V. V., Boddeda, A., Secor, G. A., et al. (2007). Real-time PCR quantification and mycotoxin production of Fusarium graminearum in wheat inoculated with isolates collected from potato, sugar beet, and wheat. Phytopathology, 97(7), 835–841.
Burlakoti, P., Rivera, V., Secor, G. A., Qi, A., Del Rio-Mendoza, L. E., & Khan, M. F. R. (2012). Comparative pathogenicity and virulence of Fusarium species on sugar beet. Plant Disease, 96, 1291–1296.
Bustin, S. A., Benes, V., Garson, J. A., Hellemans, J., Huggett, J., Kubista, M., et al. (2009). The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clinical Chemistry, 55(4), 611–622.
Clements, M. J., Maragos, C. A., Pataky, J. K., & White, D. G. (2004). Sources of resistance to fumonisin accumulation in grain and fusarium ear and kernel rot of corn. Phytopathology, 94, 251–260.
Corbett Life Science (2006). Rotor-gene™ 6000 operator manual. Corbett Robotics, Queensland, Australia. 152 pp.
Covarelli, L., Beccari, G., Prodi, A., Generotti, S., Etruschi, F., Juan, C., et al. (2015). Fusarium species, chemotype characterisation and trichothecene contamination of durum and soft wheat in an area of central Italy. Journal of the Science of Food and Agriculture, 95, 540–551.
Del Ponte, E. M., Spolti, P., Ward, T. J., Gomes, L. B., Nicolli, C. P., Kuhnem, P. R., Silva, C. N., & Tessmann, D. J. (2015). Regional and field-specific factors affect the composition of fusarium head blight pathogens in subtropical no-till wheat agroecosystem of Brazil. Phytopathology, 105, 246–254.
Desjardins, A. E. (2006). Fusarium mycotoxins: chemistry, genetics, and biology. APS press, St. Paul, MN, USA 203 pp.
Desjardins, A. E., & Proctor, R. H. (2011). Genetic diversity and trichothecene chemotypes of the Fusarium graminearum clade isolated from maize in Nepal and identification of a putative new lineage. Fungal Biology, 115, 38–48.
Engle, J. S., Madden, L. V., & Lipps, P. E. (2003). Evaluation of inoculation methods to determine resistance reactions of wheat to Fusarium graminearum. Plant Disease, 87(12), 1530–1535.
European Commission: Institute for Health and Consumer Protection (2008). Report of the verification of the performance of a method for the detection of DAS-59132-8 (event 32) in maize using real-time PCR. CRL-EM-01⁄08. Ispra, Italy: Community Reference Laboratory for GM Food and Feed. http://gmo-crl.jrc.ec.europa.eu/doc/Maize_E32_verification_report.pdf. Accessed 23 Jan 2015.
Evans, C. K., Xie, W., Dill-Macky, R., & Mirocha, C. J. (2000). Biosynthesis of deoxynivalenol in spikelets of barley inoculated with macroconidia of Fusarium graminearum. Plant Disease, 84(6), 654–660.
Fredlund, E., Gidlund, A., Olsen, M., Börjesson, T., Spliid, N. H. H., & Simonsson, M. (2008). Method evaluation of Fusarium DNA extraction from mycelia and wheat for down-stream real-time PCR quantification and correlation to mycotoxin levels. Journal of Microbiological Methods, 73, 33–40.
Gale, L. R., Ward, T. J., Balmas, V., & Kistler, H. C. (2007). Population subdivision of Fusarium graminearum sensu stricto in the upper midwestern United States. Phytopathology, 97(11), 1434–1439.
Gomes, L. B., Ward, T. J., Badiale-Furlong, E., & Del Ponte, E. M. (2015). Species composition, toxigenic potential and pathogenicity of Fusarium graminearum species complex isolates from southern Brazilian rice. Plant Pathology, 64, 980–987.
Goswami, R. S., & Kistler, H. C. (2005). Pathogenicity and in planta mycotoxin accumulation among members of the Fusarium graminearum species complex on wheat and rice. Phytopathology, 95(12), 1397–1404.
Hao, J. J., Xie, S. N., Sun, J., Yang, G. Q., Liu, J. Z., Xu, F., Ru, Y. Y., & Song, Y. L. (2017). Analysis of Fusarium graminearum species complex from wheat–maize rotation regions in Henan (China). Plant Disease, 101(5), 720–725.
Harris, L. J., Balcerzak, M., Johnston, A., Schneiderman, D., & Ouellet, T. (2016). Host-preferential Fusarium graminearum gene expression during infection of wheat, barley, and maize. Fungal Biology, 120, 111–123.
Hilton, A., Jenkinson, P., Hollins, T. W., & Parry, D. W. (1999). Relationship between cultivar height and severity of fusarium ear blight in wheat. Plant Pathology, 48, 202–208.
Jansen, C., von Wettstein, D., Schäfer, W., Kogel, K.-H., Felk, A., & Maier, F. J. (2005). Infection patterns in barley and wheat spikes inoculated with wild-type and trichodiene synthase gene disrupted Fusarium graminearum. Proceedings of the National Academy of Sciences of the United States of America, 102(46), 16892–16897.
Kazan, K., Gardiner, D. M., & Manners, J. M. (2012). On the trail of a cereal killer: Recent advances in Fusarium graminearum pathogenomics and host resistance. Molecular Plant Pathology, 13(4), 399–413.
Kuhnem, P. R., Ward, T. J., Silva, C. N., Spolti, P., Ciliato, M. L., Tessmann, D. J., & Del Ponte, E. M. (2016). Composition and toxigenic potential of the Fusarium graminearum species complex from maize ears, stalks and stubble in Brazil. Plant Pathology, 65, 1185–1191.
Lamprecht, S. C., Tewoldemedhin, Y. T., Botha, W. J., & Calitz, F. J. (2011). Fusarium graminearum species complex associated with maize crowns and roots in the KwaZulu-Natal province of South Africa. Plant Disease, 95, 1153–1158.
Lee, J., Chang, I. Y., Kim, H., Yun, S.-H., Leslie, J. F., & Lee, Y. -W. (2009). Genetic diversity and fitness of Fusarium graminearum populations from rice in Korea. Applied and Environmental Microbiology, 75(10), 3289–3295.
Lee, J., Kim, H., Jeon, J.-J., Kim, H.-S., Zeller, K. A., Carter, L. L. A., et al. (2012). Population structure of and mycotoxin production by Fusarium graminearum from maize in South Korea. Applied and Environmental Microbiology, 78(7), 2161–2167.
Lysøe, E., Seong, K. Y., & Kistler, H. C. (2011). The transcriptome of Fusarium graminearum during the infection of wheat. Molecular Plant-Microbe Interactions, 24(9), 995–1000.
Maier, F. J., Miedaner, T., Hadeler, B., Felk, A., Salomon, S., Lemmens, M., et al. (2006). Involvement of trichothecenes in fusarioses of wheat, barley and maize evaluated by gene disruption of the trichodiene synthase (Tri5) gene in three field isolates of different chemotype and virulence. Molecular Plant Pathology, 7(6), 449–461.
Malbrán, I., Mourelos, C. A., Girotti, J. R., Aulicino, M. B., Balatti, P. A., & Lori, G. A. (2012). Aggressiveness variation of Fusarium graminearum isolates from Argentina following point inoculation of field grown wheat spikes. Crop Protection, 42, 234–243.
Malbrán, I., Mourelos, C. A., Girotti, J. R., Balatti, P. A., & Lori, G. A. (2014). Toxigenic capacity and trichothecene production by Fusarium graminearum isolates from Argentina and their relationship with aggressiveness and fungal expansion in the wheat spike. Phytopathology, 104(4), 357–364.
McLean, M. (1995). The phytotoxicity of selected mycotoxins on mature, germinating Zea mays embryos. Mycopathologia, 132(3), 173–183.
Menke, J., Dong, Y., & Kistler, H. C. (2012). Fusarium graminearum Tri12p influences virulence to wheat and Trichothecene accumulation. Molecular Plant-Microbe Interactions, 25(11), 1408–1418.
Moradi, M., Oerke, E.-C., Steiner, U., Tesfaye, D., Schellander, K., & Dehne, H. -W. (2010). Microbiological and Sybr® green real-time PCR detection of major fusarium head blight pathogens on wheat ears. Microbiology, 79(5), 646–654.
Munkvold, G. P. (2003). Epidemiology of Fusarium diseases and their mycotoxins in maize ears. European Journal of Plant Pathology, 109, 705–713.
Nicolaisen, M., Suproniene, S., Nielsen, L. K., Lazzaro, I., Spliid, N. H., & Justesen, A. F. (2009). Real-time PCR for quantification of eleven individual Fusarium species in cereals. Journal of Microbiological Methods, 76, 234–240.
O’Donnell, K., Kistler, H. C., Tacke, B. K., & Casper, H. H. (2000). Gene genealogies reveal global phylogeographic structure and reproductive isolation among lineages of Fusarium graminearum, the fungus causing wheat scab. Proceedings of the National Academy of Sciences of the United States of America, 97(14), 7905–7910.
O’Donnell, K., Ward, T. J., Geiser, D. M., Kistler, H. C., & Aoki, T. (2004). Genealogical concordance between the mating type locus and seven other nuclear genes supports formal recognition of nine phylogenetically distinct species within the Fusarium graminearum clade. Fungal Genetics and Biology, 41, 600–623.
O’Donnell, K., Ward, T. J., Aberra, D., Kistler, H. C., Aoki, T., Orwig, N., et al. (2008). Multilocus genotyping and molecular phylogenetics resolve a novel head blight pathogen within the Fusarium graminearum species complex from Ethiopia. Fungal Genetics and Biology, 45, 1514–1522.
Ott, R. L. (1998). An introduction to statistical methods and data analysis (p. 837). Belmont: Duxbury Press.
Picot, A., Hourcade-Marcolla, D., Barreau, C., Pinson-Gadais, L., Caron, D., Richard-Forget, F., & Lannou, C. (2012). Interactions between Fusarium verticillioides and Fusarium graminearum in maize ears and consequences for fungal development and mycotoxin accumulation. Plant Pathology, 61, 140–151.
Proctor, R. H., Hohn, T. M., & McCormick, S. P. (1995). Reduced virulence of Gibberella zeae caused by disruption of a trichothecene toxin biosynthetic gene. Molecular Plant-Microbe Interactions, 8(4), 593–601.
Proctor, R. H., Desjardins, A. E., McCormick, S. P., Plattner, R. D., Alexander, N. J., & Brown, D. W. (2002). Genetic analysis of the role of trichothecene and fumonisin mycotoxins in the virulence of Fusarium. European Journal of Plant Pathology, 108, 691–698.
Purahong, W., Nipoti, P., Pisi, A., Lemmens, M., & Prodi, A. (2014). Aggressiveness of different Fusarium graminearum chemotypes within a population from northern-Central Italy. Mycoscience, 55(1), 63–69.
Qiu, J., & Shi, J. (2014). Genetic relationships, Carbendazim sensitivity and mycotoxin production of the Fusarium graminearum populations from maize, wheat and Rice in eastern China. Toxins, 6, 2291–2309.
Qu, B., Li, H. P., Zhang, J. B., Huang, T., Carter, J., Liao, Y. C., et al. (2008). Comparison of genetic diversity and pathogenicity of fusarium head blight pathogens from china and Europe by SSCP and seedling assays on wheat. Plant Pathology, 57, 642–651.
Quinnipiac University (2015). http://faculty.quinnipiac.edu/libarts/polsci/Statistics.html . Accessed on 14 June 2015.
Reid, L. M., Woldemariam, T., Zhu, X., Stewart, D. W., & Schaafsma, A. W. (2002). Effect of inoculation time and point of entry on disease severity in Fusarium graminearum, Fusarium verticillioides, or Fusarium subglutinans inoculated maize ears. Canadian Journal of Plant Pathology, 24, 162–167.
Rose, L. J., Mouton, M., Beukes, I., Flett, B. C., van der Vyver, C., & Viljoen, A. (2016). Multi-environment evaluation of maize inbred lines for resistance to fusarium ear rot and fumonisins. Plant Disease, 100, 2134–2144.
Sarlin, T., Yli-Mattila, T., Jestoi, M., Rizzo, A., Paavanen-Huhtala, S., & Haikara, A. (2006). Real-time PCR for quantification of toxigenic Fusarium species in barley and malt. European Journal of Plant Pathology, 114, 371–380.
Sarver, B. A., Ward, T. J., Gale, L. R., Broz, K., Kistler, H. C., Aoki, T., et al. (2011). Novel fusarium head blight pathogens from Nepal and Louisiana revealed by multilocus genealogical concordance. Fungal Genetics and Biology, 48, 1096–1107.
Shapiro, S. S., & Wilk, M. B. (1965). An analysis of variance test for normality (complete samples). Biometrika, 52(3–4), 591–611.
Small, I. M., Flett, B. C., Marasas, W. F. O., McLeod, A., Stander, M. A., & Viljoen, A. (2012). Resistance in maize inbred lines to Fusarium verticillioides and fumonisin accumulation in South Africa. Plant Disease, 96(6), 881–888.
Snedecor, G. W., & Cochran, W. G. (1980). Statistical methods (7th ed.p. 478). Ames: The Iowa State University Press.
Starkey, D. E., Ward, T. J., Aoki, T., Gale, L. H., Kistler, H. C., Geiser, D. M., et al. (2007). Global molecular surveillance reveals novel Fusarium head blight species and trichothecene toxin diversity. Fungal Genetics and Biology, 44, 1191–1204.
Suga, H., Karugia, G. W., Ward, T., Gale, L. R., Tomimura, K., Nakajima, T., et al. (2008). Molecular characterization of the Fusarium graminearum species complex in Japan. Phytopathology, 98(2), 159–166.
Taylor, J. W., Jacobson, D. J., Kroken, S., Kasuga, T., Geiser, D. M., Hibbett, D. S., et al. (2000). Phylogenetic species recognition and species concepts in fungi. Fungal Genetics and Biology, 31, 21–32.
Teich, A. H., & Michelutti, R. (1993). Determining resistance to wheat scab by covering field inoculated heads with plastic bags. Cereal Research Communications, 21(1), 69–73.
van der Lee, T., Zhang, H., van Diepeningen, A., & Waalwijk, C. (2015). Biogeography of Fusarium graminearum species complex and chemotypes: A review. Food Additives & Contaminants: Part A, 32(4), 453–460.
Ward, T. J., Bielawski, J. P., Kistler, H. C., Sullivan, E., & O'Donnell, K. (2002). Ancestral polymorphism and adaptive evolution in the trichothecene mycotoxin gene cluster of phytopathogenic Fusarium. Proceedings of the National Academy of Sciences of the United States of America, 99(14), 9278–9283.
Wegulo, S. N., Bockus, W. W., Hernandez Nopsa, J., De Wolf, E. D., Eskridge, K. M., Peiris, K. H. S., et al. (2011). Effects of integrating cultivar resistance and fungicide application on fusarium head blight and deoxynivalenol in winter wheat. Plant Disease, 95(5), 554–560.
Windels, C. E. (2000). Economic and social impacts of fusarium head blight: Changing farms and rural communities in the northern Great Plains. Phytopathology, 90(1), 17–21.
Yli-Mattila, T., Gagkaeva, T., Ward, T. J., Aoki, T., Kistler, H. C., & O'Donnell, K. (2009). A novel Asian clade within the Fusarium graminearum species complex includes a newly discovered cereal head blight pathogen from the Russian far east. Mycologia, 101(6), 841–852.
Zhang, H., van der Lee, T., Waalwijk, C., Chen, W., Xu, J., Xu, J., Zhang, Y., & Feng, J. (2012). Population analysis of the Fusarium graminearum species complex from wheat in China show a shift to more aggressive isolates. PloS One, 7, e31722.
Zhang, H., Brankovics, B., Luo, W., Xu, J., Xu, J. S., Guo, C., et al. (2016). Crops are a main driver for species diversity and the toxigenic potential of Fusarium isolates in maize ears in China. World Mycotoxin Journal, 9(5), 701–715.
Acknowledgements
This research was financially supported by the South African Winter Cereal Trust and the National Research Foundation: Technology and Human Resources for Industry Programme (THRIP) of South Africa. We are grateful to Dr. Emmanuel Terrasson, Karlien van Zyl, Ilana Visser and Dewald Gouws for excellent technical assistance. Thank you to Ms. M. van der Rijst for statistical analyses and Mr. D. Lesch for management of the wheat field trials. The mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by Stellenbosch University.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors hereby declare that they have no financial or non-financial conflict of interest.
Rights and permissions
About this article
Cite this article
Beukes, I., Rose, L.J., van Coller, G.J. et al. Disease development and mycotoxin production by the Fusarium graminearum species complex associated with South African maize and wheat. Eur J Plant Pathol 150, 893–910 (2018). https://doi.org/10.1007/s10658-017-1331-5
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10658-017-1331-5