Abstract
Streptomyces scabiei is largely accepted as the causal organism of common scab on potato in South Africa, and other Streptomyces species associated with common scab are not often considered. This study therefore aims to determine the diversity and prevalence of Streptomycetes associated with common scab on potatoes in South Africa. Isolates from 11 of the 16 potato producing regions in South Africa were characterized morphologically, physiologically and genetically. Most isolates resembled S. scabiei based on morphology and physiology. Most pathogenic isolates were S. scabiei and S. stelliscabiei, and no S. acidiscabies or S. turgidiscabies isolates were found. All three pathogenicity/virulence genes (txtAB, nec1, tomA) were found in South African isolates. Pathogenicity could not be linked to the presence of a single one or any combination of two of the three genes. These results represent the most comprehensive published survey of Streptomycetes isolated from common scab lesions on potatoes in South Africa.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In South Africa, common scab on potato was reported in 1906 by Pole Evans (Doidge 1950). This disease causes downgrading of tubers on the fresh produce market and decreases processing potential of infected tubers (Gouws and van der Waals 2012). South Africa is the fourth largest potato producer in Africa, producing over two million tons a year (FAOSTAT 2013). Production is done on about 66,000 ha, making South Africa the largest producer in tons per hectare in Africa (FAOSTAT 2013). Potatoes are produced in 16 regions throughout the country. Most of South African potato production is done under irrigation but some regions still have about 15 % of their production under dry land conditions. In South Africa, the potato industry is one that is, in most part, based on individual small to large scale farmers (Potatoes South Africa 2013). Thus, it is important for South African potato growers to optimize their disease management strategies and avoid downgrading of tubers on the fresh produce market.
The interactions between the Streptomyces species, environment and host are complex and result in unpredictable disease incidences, severity and symptomatology between regions and years, making this disease difficult to understand (Wanner 2009). Common scab is not caused by a single species, but rather a species complex within the Streptomyces genus (Loria et al. 1997; Wanner 2009). Reports of various scab-causing species found within the same field and within the same lesion have led to investigations on species relatedness to specific symptoms and on the sensitivity of species to different environmental conditions (Lindholm et al. 1997; Bouchek-Mechiche et al. 2000; Krištůfek et al. 2000; Aittamaa et al. 2008; Wiechel and Crump 2010; Khodakaramian and Khodakaramian 2012; Tashiro et al. 2012). S. scabiei and S. turgidiscabies often co-occur in the same lesions and it has been found that when isolating directly after sampling more S. scabiei than S. turgidiscabies is isolated than when samples are left for a couple of days in storage (Aittamaa et al. 2008). It is thus unclear if the Streptomycetes isolated at the end of the season represent the causal pathogen species complex or if the complex of species changes during the season from the time of the first infection. Another aspect which requires more research is the role of the non-pathogenic Streptomyces spp. in pathogen establishment and disease development.
There are four well known causal species of potato common scab; Streptomyces scabiei (Lambert and Loria), Streptomyces europaeiscabiei (Bouchek-Mechiche), Streptomyces turgidiscabies (Miyajima) and Streptomyces acidiscabies (Lambert & Loria) (Loria et al. 2006). These are not the only scab-causing species and reports of other species are geographically distinct. S. scabiei is found worldwide but is not common in Europe; while S. europaeiscabiei is more often found in Europe, Korea and North America; and S. turgidiscabies is frequently found in Japan and Finland (Lehtonen et al. 2004; Wanner 2009; Dees et al. 2012). A multitude of other, poorly described Streptomyces spp. have been isolated from lesions (Song et al. 2004; Taddei et al. 2006; Pánková et al. 2012). Their function in the disease complex is not understood and some are closely related (Song et al. 2004; Taddei et al. 2006). For example the diastochromogenes group contains S. scabiei, S. europaeiscabiei, S. stelliscabiei, Streptomyces ipomoeae (Waksman and Henrici) and Streptomyces diastatochromogenes ((Krainsky) Waksman and Henrici). Streptomyces stelliscabiei (Bouchek-Mechiche) and Streptomyces bottropensis (Waksman) are 98.4 % related based on their genetic makeup. S. scabiei and S. acidiscabies differ in morphology but their DNA is approximately 90 % similar (Pánková et al. 2012).
Morphological, physiological and molecular characterization remains an integral part of species identification (Leiminger et al. 2012). Morphological characterization is based on the spore and colony colours when grown on yeast malt extract agar (Tashiro et al. 2012). The structure of the spore chain and sometimes the spores are examined under a microscope. Physiological testing involves growth at different pH levels, production of melanin in the presence of tyrosine, utilization of different sugars and resistance to various antibiotics (Loria et al. 1995; Lindholm et al. 1997; Bouchek-Mechiche et al. 1998). Genetically the isolates are screened for the presence of the pathogenicity island (PAI) genes and identified using different PCR based techniques, such as 16S rDNA sequencing, DNA-DNA hybridization and the use of species-specific primers (Leiminger et al. 2012; Pánková et al. 2012; Dees et al. 2013). Only about 10 species harbour all or part of this PAI (Loria et al. 2006). It is also widely accepted that if the txtAB gene is present, the isolate will be pathogenic (St-Onge et al. 2008; Dees et al. 2013).
Pathogenicity of Streptomyces spp. is said to be transferred through horizontal gene transfer of pathogenicity and virulence genes (such as txtAB, tomA and nec1) located on a PAI, and there has been an increasing abundance of pathogenic Streptomyces species identified over time (Kers et al. 2005; Loria et al. 2006; Wanner 2009; Dees et al. 2013). For instance S. scabiei and S. europaeiscabiei grow at pH above 5 (Lindholm et al. 1997), however, in Japan isolates such as S. acidiscabies that grow at pH 4 are said to cause acid scab (Tashiro et al. 2012). Conditions that normally cause common scab are dry and warm soils (Wanner 2006). However, scab was also found to occur under irrigated conditions in Northern Europe, Israel and Canada (Doering-Saad et al. 1992; Goyer et al. 1996). Different cultural practices in managing the disease have led to the selection of species capable of surviving under altered cropping conditions.
Cultural practices remain the primary method for disease management of common scab (Dees and Wanner 2012). Currently there are no resistant cultivars in South Africa (Gouws and van der Waals 2012) and with the limitations on the use of products for the control of bacteria it is becoming difficult to control this disease (Agrios 2005). Tubers with common scab have been harvested from virgin soils and from soils with no history of common scab (isolations made during this study). Identifying the different species within a disease complex and under unique environmental conditions may give insight into better management or even preventative action for common scab. For example, decreasing the soil pH will be ineffective if the majority of pathogenic Streptomycestes are S. acidiscabies that causes common scab at a lowered soil pH. Although the inheritance of resistance or the mechanism of resistance in potato cultivars against common scab is unclear, knowledge of the pathogenic Streptomyces species present would be important when screening for resistance, especially if the species differ in conditions that are favourable for disease development.
Streptomyces scabiei was thought to be the only causal organism of common scab in South Africa until about 2000 (Bouchek-Mechiche et al. 2000; Gouws and van der Waals 2012). However in 2000 Bouchek-Mechiche et al. (2000) isolated S. stelliscabiei from a common scab lesion in South Africa. Up until now the species associated with common scab in South Africa have not been characterized. The presence of the txtAB, tomA and nec1 genes in South African Streptomyces isolates has not yet been investigated, but they are presumed to play a role in pathogenicity and virulence of Streptomyces species associated with common scab of potato. The aims of this study were therefore to determine the diversity of Streptomycetes associated with common scab lesions on potato tubers in South Africa; to investigate the presence of the pathogenicity and virulence related genes in combination with pathogenicity tests and to determine which gene best corresponds to pathogenicity in Streptomyces species on potatoes in South Africa.
Materials and methods
Sample collection and bacterial isolation
Tubers showing common scab symptoms were collected from 11 of the 16 potato production regions in South Africa. Lesion type and disease severity score were recorded prior to isolation (Table 1). Disease score was recorded as 1 for no lesions; 2 if lesions covered <6.25 % of the tuber surface; 3 for 6.25–12.4 %; 4 for 12.5–24 %; 5 for 25–49 %; 6 for 50–74 % and 7 for 75 % - 100 % coverage. Tubers were surface sterilized with 70 % ethanol. From each lesion a 100mm3 piece of the edge of the lesion was excised, including the straw coloured tissue directly underneath. Each piece was placed in a Bioreba macerating bag (Labretoria) and macerated with a rubber mallet. Nine millilitres of sterile distilled water was added to each bag containing the macerated tissue. A serial dilution was made to 10−5 dilution and 100 μl thereof plated in triplicate onto water agar (WA) (Loria et al. 2001). Plates were incubated at 28 °C for 12 days, in the dark. Plates were examined at regular intervals during the incubation period for typical Streptomyces colonies and three colonies per plate were transferred to yeast malt extract agar (YME). Pure cultures were stored in 20 % glycerol at −80 °C. DNA was extracted using the ZR Soil Microbe DNA kit from Zymo Research (Inqaba) according to the manufacturer protocol and stored at −20 °C until use.
Morphological and physiological characterisation
Spore chain morphology on water agar (WA) was noted with the use of a light microscope at 400× magnification, prior to re-plating onto YME. Following seven days of incubation at 28 °C on YME, colony colour and spore colour for each isolate were noted. The presence or absence of diffusible pigments in the media was noted. Melanin production was determined after growth on tyrosine agar (TA) at 28 °C after four, eight and 12 days of incubation. Isolates altering the clear colour of the medium with a dark diffusible pigment were considered as positive for melanin production (Lindholm et al. 1997; Park et al. 2003). Growth at pH 4, 5, 6 and 7 was determined by growing the isolates on YME medium with adjusted pH levels for 14 days at 28 °C. The pH was altered by adding 1 M hydrochloric acid or 1 M sodium hydroxide to the media until the desired pH was reached. A tuber slice assay as described by Loria et al. (1995) was used to give an indication of the pathogenicity of the isolates. Briefly isolates were grown on oatmeal agar (OA), and tuber slices of the susceptible cultivar BP1 were used. Agar plugs from each isolate were placed inverted onto the tuber slice; this was done in triplicate in separate Petri dishes. Moist filter paper discs were placed in Petri dishes to ensure the tuber slices did not dry out. A sterile agar plug was used as control. Petri dishes were sealed and placed in the incubator in the dark for five days at 28 °C. Tuber slices were examined for a necrotic area surrounding the agar plug and a collapse of the cells directly underneath the agar plug.
Molecular characterisation
Conventional PCR was used to determine the presence of three genes known to be related to pathogenicity within the Streptomyces genus (Wanner 2006). Primers Nf and Nr were used to amplify the 700 bp product of the nec1 gene (Bukhalid et al. 1998); Tom3 and Tom4 to amplify tomA that is 392 bp in size; and TxtAtB1 and TxtAtB2 were used to amplify the 385 bp thaxtominAB gene product (Wanner 2006). The PCR analysis was carried out in 25 μl reactions containing 5 u/μl Taq DNA polymerase (recombinant) from Fermentas Life Science, 10xNH4 reaction buffer (Bioline), 2.5 mM dNTP mix (Bioline), 50 mM MgCl2 (Bioline) and 0.5 μl template DNA. Amplification was carried out in an MJ Mini Personal Thermal Cycler (Bio-Rad) with an initial denaturation step at 95 °C for 5 min; followed by 35 cycles of denaturation at 95 °C for 20 s, annealing for 30 s and extension at 72 °C for 1 min, followed by a final extension step at 72 °C for 7 min and holding at 4 °C. The annealing temperature for TxtAtB1/TxtAtB2 and Tom3/Tom4 was 50 °C; and for Nr/Nf the annealing temperature was 64 °C. Amplified DNA products were separated on 1 % agarose gels (stained with ethidium bromide) and viewed under UV light. Primers 16S-1F and 16S-1R (Bukhalid et al. 2002) as described by Wanner (2006) were used to amplify the 16S rDNA gene. Twenty-five isolates were selected at random to include pathogenic and non-pathogenic isolates, with and without pathogenicity/virulence genes, for identification by sequencing of the 16S rDNA gene. Products were viewed by agarose gel electrophoresis and purification of the amplicon was done with the Bioline Isolate PCR and gel kit (Celtic Diagnostic) as per manufacturer instructions. Sequencing was carried out on an Applied Biosystems ABI 3500xl (Life Technologies Carlsbad). Consensus sequences were obtained in Bio-edit and aligned with the NCBI sequencing database using the online Basic Local Alignment Tool (BLAST) against prokaryotes.
Results
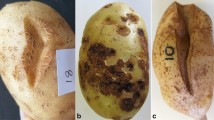
Scab symptoms from which isolations were made ranged from deep pitted and star shaped to shallow and raised. Tubers showing netted scab symptoms were only collected from the Western Cape region. Most isolates were yellow-brown with grey spores on YME (Table 1). Morphology was variable and the ability to produce pigments on YME media was lacking in most of the isolates. All the isolates grew at pH 7 while some could grow at pH 4 and above (Table 2). Melanin pigment production on tyrosine agar was only observed in half of the isolates; delayed or weak production of melanin was still noted as positive. Pathogenicity based on the tuber slice assay could not be linked to melanin production. Isolates with flexuous spore chains tended to not have all three of the pathogenicity/virulence genes. Only 29 % of the isolates were pathogenic in the tuber slice assay, which was expected as only a few Streptomycetes are pathogenic, and other non-pathogenic Streptomycetes are frequently isolated from lesions (Song et al. 2004; Taddei et al. 2006).
The three pathogenicity/virulence genes were found in the same frequency among isolates (Table 2). About 19 % of the pathogenic isolates did not contain any of the three pathogenicity/virulence genes tested for. All isolates containing all three pathogenicity/virulence genes were pathogenic. One isolate had txtAB only and was pathogenic. However none of the other isolates containing only txtAB, a combination of txtAB and one other pathogenicity/virulence gene or a combination of nec1 and tomA genes were pathogenic, which was surprising as txtAB is considered a determining factor for pathogenicity within the Streptomyces complex. One S. scabiei isolate did not test positive for nec1 and another did not test positive for any of the three pathogenicity/virulence genes. Analysis revealed that the presence of all three genes is a good indicator of pathogenicity based on the tuber slice assay.
Sequencing results revealed 22 % of the isolates selected for sequencing aligned to S. scabiei. Streptomyces padanus comprised 9 % of the sequenced isolates, Streptomyces flavofuscus 6 % and S. stelliscabiei 3 %. No S. acidiscabies or S. turgidiscabies were found. S. stelliscabiei was only isolated from the Western Cape region. GenBank accession numbers and genetic characterization for each isolate is shown in Table 2.
Discussion
Various common scab symptoms have been described in South Africa and symptoms cannot be linked to specific pathogenic Streptomyces spp. as found by Dees et al. (2013). The variability in morphology and physiology of the Streptomycetes isolated from the lesions is great, similar to a study in Israel (Doering-Saad et al. 1992). It is difficult to use morphological and physiological parameters to determine pathogenic Streptomycete species as some non-pathogenic isolates with grey spores and producing melanin, resemble pathogenic S. scabiei isolates (Faucher et al. 1992). The morphology of most isolates in this study resembled that of S. scabiei. Although Bukhalid et al. (2002) found a South African S. scabiei isolate that produced flexuous spore chains this characteristic was not observed in the isolates in this study. Morphological characteristics are said to be more constant than physiological characteristics (Park et al. 2003), however, the isolates tend to change in spore and colony colour after continuous re-culturing. On the other hand, Keinath and Loria (1989) estimated that the morphology and physiology of less than 10 % of pathogenic isolates resemble that of S. scabiei.
Leiminger et al. (2012) found most isolates in their study to produce melanin; this is contrary to what was observed in this study where only half of the isolates produced melanin. However, some of the isolates from this study had delayed or weak melanin production, which was also found by Taddei et al. (2006) in their study of Streptomyces spp. from Venezuelan soils. Isolates from netted scab in The Netherlands were positive for melanin production, and considered identical to S. scabiei (Bouchek-Mechiche et al. 2000). Netted scab isolates that did not produce melanin were however also reported from Denmark and Sweden. Pathogenicity and melanin production could not be linked, which was also found in a study by Leiminger et al. (2012).
Only a few of the isolated strains in this study were found to be pathogenic based on a tuber slice assay, which is consistent with previous findings in similar studies (Loria et al. 1986; Faucher et al. 1992; Lindholm et al. 1997). Although the tuber slice assay is not sensitive enough for virulence testing, it is an efficient screening method and correlates well with pot trials for pathogenicity evaluation (Park et al. 2003; Hao et al. 2009; Meng et al. 2012). Most studies base pathogenicity determination of isolates on the presence of txtAB and only conduct pathogenicity pot trials tests on selected isolates (Wanner 2009; Leiminger et al. 2012; Dees et al. 2013), although Park et al. (2003) reported two potato scab-causing isolates from Korea that did not produce thaxtomin. Leiminger et al. (2012) and Wanner (2009) also isolated pathogenic but txtAB-negative S. acidiscabies, S. stelliscabiei and S. bottropensis from Germany and North America respectively.
Lerat et al. (2010) pointed out that pathogenicity depends on the ability of the species to synthesize toxins. Little is known about the molecular mechanisms involved in Streptomyces pathogenicity. Most researchers believe that similar mechanisms are shared by Streptomycete pathogens (Bukhalid et al. 2002; Wanner 2004; Cullen and Lees 2007; Flores-González et al. 2008; Qu et al. 2008); however because of the rapid symptom development in the Streptomyces-potato pathosystem Loria et al. (2003) proposed multiple mechanisms of pathogenicity. Non-pathogenic S. scabiei, S. acidiscabies and S. turgidiscabies have been reported from Finland and America (Faucher et al. 1992; Lindholm et al. 1997; Wanner 2006), which makes species identification alone insufficient to classify an isolate as pathogenic.
Pathogenicity of common scab-causing isolates in South Africa, seems to be linked to the presence of all three pathogenicity/virulence genes (89 % of isolates tested), based on the results of the tuber slice assay. In this study we found at least one isolate of every haplotype, where haplotypes are depicted here in the order txtAB, nec1, tomA and capitals indicate the presence of the gene. The haplotype composition of the South African population was as follows: haplotype tnt comprised 67 % of the isolates, 56 in total; 11 TNT isolates (mostly deep pitted lesions but some shallow lesions were also associated with these isolates); 7 Tnt isolates; 1 tNt isolates (associated with various lesion types); 3 tnT isolates; 4 TnT isolates (mostly from shallow lesions); 1 TNt isolate (from a shallow lesion) and 1 tNT isolate. Pánková et al. (2012) did not find the haplotypes TNt, Tnt or tNT in their study focusing on plant pathogenic Streptomyces spp. in Central Europe, and the TNT haplotype was mostly associated with the deeper and more severe lesions. We also found that the TNT haplotype is associated with S. scabiei and S. stelliscabiei. Dees et al. (2013) found the haplotypes TNT, TnT, TNt and Tnt in Norway; 60 % of the population from this study in Norway did not have nec1 but did have txtAB, while 37 % did not have tomA but had txtAB. Pathogenic strains lacking the nec1 gene have been reported in the United States, Japan, Hungary, South Africa and Korea (Bukhalid et al. 1998; Kreuze et al. 1999; Park et al. 2003; Gouws 2006).
Genetic variation and evolution within species is not uncommon (Dees et al. 2013). The nec1 and tomA genes are at the opposite end of the chromosome to the txtAB gene and can be transferred and evolve independently (Aittamaa et al. 2010). A good example of the evolution of pathogens and their ability to acquire genes from other pathogens is the horizontal transfer of the nec1 gene in Streptomycetes. Wanner (2009) proposed that the presence of nec1 and tomA without the presence of txtAB might be due to non-pathogenic Streptomyces spp. acting as reservoirs for genes associated with pathogenicity. One of the S. scabiei isolates in the present study had an incomplete PAI; while another S. scabiei isolate had none of the PAI genes.
Bignell et al. (2010) discussed the multitude of other S. scabiei (strain 87–22) genes that were found by comparative genomics. These genes were revealed to be conserved within other sequenced microbial pathogens and play a role in disease development and virulence. Kinkel et al. (1998) stated that 40 % of variation in disease severity cannot be attributed to thaxtomin levels. Wanner (2004) stated that other pathogenicity determinants besides thaxtomin are involved in the disease caused by Streptomycetes on radish, and could be well be the case in pathogenic Streptomyces spp. that do not contain the known pathogenicity/virulence genes.
South African Streptomycete isolates may contain rearrangements or deletions in the pathogenicity island. Similar to results in this study, Gouws (2006) also found pathogenic Streptomyces isolates, which did not produce thaxtomin (14 %), and non-pathogenic isolates that did produce thaxtomin (6 %). Streptomyces padanus was frequently isolated from common scab lesions in South Africa and even though these isolates did not have any of the known pathogenicity genes, they caused necrosis and collapse of cells during the tuber slice assay. Streptomyces luridiscabiei and Streptomyces puniscabiei are speculated by Park et al. (2003) to have a pathogenicity factor other than txtAB, as they were also found to cause common scab on potatoes in Korea.
Acknowledging that 16S rDNA sequences of some Streptomyces spp. have high similarity in the 16S rDNA gene region; results from this and other studies (Bouchek-Mechiche et al. 2000; Gouws 2006; Gouws and van der Waals 2012) nonetheless show that S. scabiei is the most abundant species associated with common scab of potatoes in South Africa, with most isolates possessing all three genes associated with the PAI. It may be possible that some of the S. scabiei isolates are in fact S. europaeiscabiei as 16S rDNA sequencing cannot distinguish these two species. S. stelliscabiei was only isolated from the Western Cape production region. However the small sample sizes from some of the other regions could explain this discrepancy and further investigation is needed to confirm if the Western Cape ecological niche selects for S. stelliscabiei. The distribution of pathogenic Streptomycetes associated with common scab of potato in South Africa appears to be random, as no one species is restricted to a certain region. Dees et al. (2013) also found no correlation between species and geographical distribution within and between fields in Norway. Gouws (2009) stated that different soil types may select for different scab-causing species and symptoms, suggesting that netted scab prevalence is related to heavier soils found in Kwa-Zulu Natal. This is contrary to what we found; tubers showing netted scab symptoms mostly originated from the Western Cape which includes the Sandveld region, known for its sandy soils. It is possible that the lack of a specific geographical species distribution could be due to the use of infected seed tubers or that more than one species could naturally occur in a specific location (Lehtonen et al. 2004; Wanner 2009).
In a previous study in South Africa 56 % of isolates were S. scabiei and 0.01 % S. turgidiscaies; no S. acidiscabies was found (Gouws 2009). This study revealed a population composition of 22 % S. scabiei, 3 % S. stelliscabiei and 6 % S. flavofuscus. Streptomyces padanus comprised 9 % of the sequenced isolates. No S. turgidiscabies or S. acidiscabies were isolated in our study. Washing tubers and even storing tubers before isolation of the pathogen can result in more S. turgidiscabies than S. scabiei being isolated from the lesions (Lehtonen et al. 2004; Valkonen 2004). Dees et al. (2013) could only find S. turgidiscabies in a few countries but attributed this to the small sample size. Wanner (2009) found only two S. turgidiscabies isolates in a sample of 1074 txtAB positive isolates in North America. Climatic conditions may be a reason for low or no S. turgidiscabies isolations as the largest number of S. turgidiscabies isolates have been obtained from Norway (Dees et al. 2012), Japan and Finland (Miyajima et al. 1998; Kreuze et al. 1999; Lehtonen et al. 2004). There may also be ecological competition between S. scabiei and S. turgidiscabies (Hiltunen et al. 2009). Another explanation for the apparent absence of S. turgidiscabies and S. acidiscabies in South Africa could be that the association with potato is weak. Wanner (2009) stated that these two species are the furthest related to the most common species usually associated with common scab on potato and may have difficulty acquiring or maintaining the PAI. A larger survey is thus needed to determine if S. acidiscabies and S. turgidiscabies are indeed associated with common scab of potatoes in South Africa.
The unidentified Streptomycetes associated with the lesions in this study comprised 32 % of all isolates; similar to a study conducted by Bukhalid et al. (2002), in which 25 % of the isolates were unidentified. Doumbou et al. (2001) found two unusual isolates associated with common scab lesions, but which are possibly not pathogenic. In addition to this; 9 % of the isolates found in this study were identified as S. padanus; a species that is not regarded as a common scab pathogen. One of these isolates was capable of producing mild symptoms in a pot trial and all isolates were pathogenic in the tuber slice assay. However, none had any of the three pathogenicity/virulence genes usually associated with pathogenic Streptomycetes. Wanner (2007) identified non-pathogenic Streptomycetes associated with less severe common scab symptoms. The ratio of pathogenic to non-pathogenic Streptomycetes may play a role in disease development at different stages, but is not yet fully understood.
Conclusion
This study is an estimation of the true population of Streptomycetes associated with common scab of potatoes in South Africa. Future work could focus on a larger sample size to confirm these results. Some of the aspects that need re-evaluation are: the effect of the ratio of pathogenic to non-pathogenic Streptomycetes on potato common scab lesions; why pathogenicity is acquired or lost; how Streptomycetes are distributed and the interactions between host and pathogen in different ecological niches.
Knowledge of the multitude of genes involved in pathogenicity and comparative genome sequences will lead to a better understanding of pathogenicity within Streptomyces. In this study, not one single gene could be linked to all pathogenic Streptomyces isolates in South Africa, but instead it appears as if all three genes need to be present for the isolate to be pathogenic. Optimization of quantification techniques of Streptomyces species for predicting common scab in South African potato production should focus on txtAB, nec1 and tomA together. Quantification of the pathogen population based on the presence of one gene alone may lead to overestimation.
It is important to know the factors involved in pathogenicity as they can be used in in vitro screening of germplasm for potato breeding programs. Future disease management programs will rely strongly upon determination and characterization of pathogenic species within populations and selecting for tolerance in potato cultivars. This is the most comprehensive study of Streptomyces isolated from tubers post-harvest in South Africa. It still remains important to note that the soil microflora changes during the growing season and the Streptomyces responsible for the disease could thus also change during the season and post-harvest.
References
Agrios, G. N. (2005). Plant diseases caused by prokaryotes: bacteria and mollicutes. In Plant pathology (5th ed., pp. 674–675). USA: Elsevier Academic Press.
Aittamaa, M., Somervuo, P., Pirhonen, M., Mattinen, L., Nissinen, R., Auvinen, P., & Valkonen, J. P. T. (2008). Distinguishing bacterial pathogens of potato using a genome-wide microarray approach. Molecular Plant Pathology, 9, 705–717.
Aittamaa, M., Somervuo, P., Laakso, I., Auvinen, P., Valkonen, J. P. T. (2010). Microarray-based comparison of genetic differences between strains of Streptomyces turgidiscabies with focus on the pathogenicity island. Molecular Plant Pathology, 11, 733–746.
Bignell, D. R. D., Seipke, R. F., Huguet-Tapia, J. C., Chambers, A. H., Parry, R. J., & Loria, R. (2010). Streptomyces scabies 87–22 contains a coronafacic acid-like biosynthetic cluster that contributes to plant-microbe interactions. Molecular Plant-Microbe Interactions, 23, 161–175.
Bouchek-Mechiche, K., Guérin, C., Jouan, B., & Gardan, L. (1998). Streptomyces species isolated from potato scab in France: numerical analysis of “biotype-100” carbon source assimilation data. Research in Microbiology, 149, 653–663.
Bouchek-Mechiche, K., Pasco, C., Andrivon, D., & Jouan, B. (2000). Differences in host, pathogenicity to potato cultivars and response to soil temperature among Streptomyces species causing common and netted scab in France. Plant Pathology, 49, 3–10.
Bukhalid, R. A., Chung, S. Y., & Loria, R. (1998). Nec1, a gene conferring a necrogenic phenotype, is conserved in plant-pathogenic Streptomyces spp. and linked to a transposase pseudogene. Molecular Plant-Microbe Interactions, 11, 960–967.
Bukhalid, R. A., Takeuchi, T., Labeda, D., & Loria, R. (2002). Horizontal transfer of the plant virulence gene, nec1, and flanking sequences among genetically distinct Streptomyces strains in the diastatochromogenes cluster. Applied and Environmental Microbiology, 68, 738–744.
Cullen, D. W., & Lees, A. K. (2007). Detection of the nec1 virulence gene and its correlation with pathogenicity in Streptomyces species on potato tubers and in soil using conventional and real-time PCR. Journal of Applied Microbiology, 102, 1082–1094.
Dees, M. W., & Wanner, L. A. (2012). In search of better management of potato common scab. Potato Research, 55, 249–268.
Dees, M. W., Somervuo, P., LysØe, E., Aittamaa, M., & Valkonen, J. P. T. (2012). Species’ identification and microarray-based comparative genome analysis of Streptomyces species isolated from potato scab lesions in Norway. Molecular Plant Pathology, 13, 174–186.
Dees, M. W., Sletten, A., & Hermansen, A. (2013). Isolation and characterization of Streptomyces species from potato common scab lesions in Norway. Plant Pathology, 62, 217–225.
Doering-Saad, C., Kämpfer, P., Manulis, S., Kritzman, G., Schneider, J., Zakrzewska-Czerwinska, J., Schrempf, H., & Barash, I. (1992). Diversity among Streptomyces strains causing potato scab. Applied and Environmental Microbiology, 58, 3932–3940.
Doidge, E. M. (1950). The South African fungi and lichens to the end of 1945. Bothalia, 5, 1094.
Doumbou, C., Akimov, V., Côté, M., Charest, P,. & Beaulieu, C. (2001). Taxonomic study on nonpathogenic streptomycetes isolated from common scab lesions on potato tubers. Systematic and Applied Microbiology, 24, 451–456.
Faucher, E., Savard, T., & Beaulieu, C. (1992). Characterization of actinomycetes isolated from common scab lesions on potato tubers. Canadian Journal of Plant Pathology, 14, 197–202.
Flores-González, R., Velasco, I., & Montes, F. (2008). Detection and characterization of Streptomyces causing common scab in western Europe. Plant Pathology, 57, 162–169.
Food and Agriculture Organization of the United Nations. (2013) http://faostat.fao.org Accessed 7 July 2013.
Gouws, R. (2006). Etiology and integrated control of common scab on seed potatoes in South Africa. M.Sc. (Agric). Thesis, University of Pretoria, Pretoria, South Africa.
Gouws, R. (2009). Potatoes South Africa Congress 09: How do I manage a common scab problem? Ilanga estate, Bloemfontein 16–17 September 2009. Pretoria: ARC Rodeplaat.
Gouws, R., & van der Waals, J. E. (2012). Occurrence and Control of Bacterial Diseases. In F. D. N. Denner, S. L. Venter, & J. G. Niederwieser. Guide to Potato Production in South Africa (pp. 157–166). Pretoria, South Africa: Agricultural Research Council.
Goyer, C., Faucher, E., & Beaulieu, C. (1996). Streptomyces caviscabies sp. nov., from deep pitted lesions in potatoes in Quebec. International Journal of Systematic Bacteriology, 46, 635–639.
Hao, J. J., Meng, Q. X., Yin, J. F., & Kirk, W. W. (2009). Characterization of a new Streptomyces strain, DS3024, that causes potato common scab. Plant Disease, 93, 1329–1334.
Hiltunen, L. H., Ojanpera, T., Kortemaa, H., Richter, E., Lehtonen, M. J., & Valkonen, J. P. T. (2009). Interactions and biocontrol of pathogenic Streptomyces strains co-occuring in potato scab lesions. Journal of Applied Microbiology, 106, 199–212.
Keinath, A. P., & Loria, R. (1989). Population dynamics of Streptomyces scabies and other actinomycetes as related to common scab of potato. Phytopathology, 79, 681–687.
Kers, J. A., Cameron, K. D., Joshi, M. V., Bukhalid, R. A., Morello, J. E., Wach, M. J., Gibson, D. M., & Loria, R. (2005). A large, mobile pathogenicity island confers plant pathogenicity on Streptomyces species. Molecular Microbiology, 55, 1025–1033.
Khodakaramian, G., & Khodakaramian, N. (2012). Pattern of host range, phytotoxin production and pathogenicity related genes among Streptomyces complex inducing potato scab disease. International conference on Eco-systems and Biological Science, 19-20, 55–58.
Kinkel, L. L., Bowers, J. H., Shimizu, K., Neeno-Eckwall, E. C., & Schottel, J. L. (1998). Quantitative relationships among thaxtomin a production, potato scab severity and fatty acid composition in Streptomyces. Canadian Journal of Microbiology, 44, 768–776.
Kreuze, J. F., Suomalainen, S., Paulin, L., & Valkonen, J. P. T. (1999). Phylogenetic analysis of 16S rRNA genes and PCR analysis of the nec1 gene from Streptomyces spp. causing common scab, pitted scab, and netted scab in Finland. Phytopathology, 89, 462–469.
Krištůfek, V., Didiš, J., Dostálková, I., & Kalčík, J. (2000). Accumulation of mineral elements in tuber periderm of potato cultivars differing in susceptibility to common scab. Potato Research, 43, 107–114.
Lehtonen, M. J., Rantala, H., Kreuze, J. F., Bång, H., Kuisma, L., Koski, P., Virtanen, E., Vihlman, K., & Valkonen, J. P. T. (2004). Occurrence and survival of potato scab pathogens (Streptomyces species) on tuber lesions: quick diagnosis based on a PCR-based assay. Plant Pathology, 53, 280–287.
Leiminger, J., Frank, M., Wenk, C., Poschenrieder, G., Kellermann, A., & Schwarzfischer, A. (2012). Distribution and characterization of Streptomyces species causing potato common scab in Germany. Plant Pathology, 62, 611–623.
Lerat, S., Simao-Beaunoir, A., Wu, R., Beaudoin, N., & Beaulieu, C. (2010). Involvement of the plant polymer suberin and the disaccharide cellobiose in triggering thaxtomin a biosynthesis, a phytotoxin produced by the pathogenic agent Streptomyces scabies. Phytopathology, 100, 91–96.
Lindholm, P., Kortemaa, H., Kokkola, M., Haahtela, K., Salkinoja-Salonen, M., & Valkonen, J. P. T. (1997). Streptomyces spp. isolated from potato scab lesions under Nordic conditions in Finland. Plant Disease, 81, 1317–1322.
Loria, R., Kempter, A., & Jamieson, A. A. (1986). Characterization of Streptomyces spp. Causing potato Scab in the Northeast. American Potato Journal, 63, 440.
Loria, R., Bukhalid, R. A., Creath, R. A., Leiner, R. H., Olivier, M., & Steffens, J. C. (1995). Differential production of thaxtomins by pathogenic Streptomyces species in vitro. Phytopathology, 85, 537–541.
Loria, R., Bukhalid, R. A., Fry, B. A., & King, R. R. (1997). Plant pathogenicity in the genus Streptomyces. Plant Disease, 81, 836–846.
Loria, R., Clark, C. A., Bukhalid, R. A., & Fry, B. A. (2001). Gram-positive bacteria Streptomyces. In N. W. Schaad, J. B. Jones, & W. Chun (Eds.), Laboratory guide for identification of plant pathogenic bacteria (3rd ed., pp. 236–249). Minnesota: APS Press.
Loria, R., Coombs, J., Yoshida, M., Kers, J., & Bukhalid, R. (2003). A paucity of bacterial root diseases: Streptomyces succeeds where others fail. Physiological and Molecular Plant Pathology, 62, 65–72.
Loria, R., Kers, J., & Joshi, M. (2006). Evolution of plant pathogenicity in Streptomyces. Annual Review of Plant Pathology, 44, 469–487.
Meng, Q. X., Yin, J. F., Rosenzweig, N., Douches, D., & Hao, J. J. (2012). Culture-based assessment of microbial communities in soil suppressive to potato common scab. Plant Disease, 96, 712–717.
Miyajima, K., Tanaka, F., Takeuchi, T., & Kuninaga, S. (1998). Streptomyces turgidiscabies sp. nov. International Journal of Systematic Bacteriology, 48, 495–502.
Pánková, I., Sedláková, V., Sedlák, P., & Krejzar, V. (2012). The occurrence of plant pathogenic Streptomyces spp. in potato-growing regions in Central Europe. American Journal of Potato Research, 89, 207–215.
Park, D. H., Yu, Y. M., Kim, J. S., Cho, J. M., Hur, J. H., & Lim, C. K. (2003). Characterization of streptomycetes causing potato common scab in Korea. Plant Disease, 87, 1290–1296.
Qu, X., Wanner, L. A., & Christ, B. J. (2008). Using the TxtAB operon to quantify pathogenic Streptomyces in potato tubers and soil. Phytopathology, 98, 405–412.
Song, J., Lee, S., Kang, J., Baek, H., & Suh, J. (2004). Phylogenetic analysis of Streptomyces spp. isolated from potato scab lesions in Korea on the basis of 16S rRNA gene and 16S-23S rDNA internally transcribed spacer sequences. International Journal of Systematic and Evolutionary Microbiology, 54, 203–209.
St-Onge, R., Goyer, C., Coffin, R., & Filion, M. (2008). Genetic diversity of Streptomyces spp. causing common scab of potato in eastern Canada. Systematic and Applied Microbiology, 31, 474–484.
Taddei, A., Rodríguez, M. J., Márquez-Vilchez, E., & Castelli, C. (2006). Isolation and identification of Streptomyces spp. from Venezuelan soils: morphological and biochemical studies. Microbiological Research, 161, 222–231.
Tashiro, N., Manabe, K., Saito, A., & Miyashita, K. (2012). Identification of potato scab-causing Streptomyces sp. occurring in strongly acidic soil conditions in saga prefecture in Japan. Journal of General Plant Pathology, 78, 353–359.
Valkonen, J. P. T. (2004). International Potato Scab Symposium: Novel approaches to the control of potato scab (ISPP 2004). Sapporo, Japan 6–7 September 2004. Sapporo: Naito, S.
Wanner, L. A. (2004). Field isolates of Streptomyces differ in pathogenicity and virulence on radish. Plant Disease, 88, 785–796.
Wanner, L. A. (2006). A survey of genetic variation in Streptomyces isolates causing potato common scab in the United States. Phytopathology, 96, 1363–1371.
Wanner, L. A. (2007). High proportions of nonpathogenic Streptomyces are associated with common scab-resistant potato lines and less sever disease. Canadian Journal of Microbiology, 53, 1062–1075.
Wanner, L. A. (2009). A patchwork of Streptomyces species isolated from potato common scab lesions in North America. American Journal of Potato Research, 86, 247–264.
Wiechel, T. J., & Crump, N. S. (2010). Soil nutrition and common scab disease of potato in Australia. 19th World Congress of Soil Science, Soil Solutions for a Changing World, August 1-6, 237–240.
Acknowledgments
This work is based on the research supported in part by a number of grants from the National Research Foundation of South Africa (UID: 78566 (NRF RISP grant for the ABI3500)). The grant holders acknowledge that opinions, findings and conclusions or recommendations expressed in any publication generated by NRF supported research are that of the author(s), and that the NRF accepts no liability whatsoever in this regard.
The University of Pretoria and Potatoes South Africa are acknowledged for funding this work which formed part of an MSc degree at the University of Pretoria.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Jordaan, E., van der Waals, J.E. Streptomyces species associated with common scab lesions of potatoes in South Africa. Eur J Plant Pathol 144, 631–643 (2016). https://doi.org/10.1007/s10658-015-0801-x
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10658-015-0801-x