Abstract
A new root-lesion nematode was isolated from sugarcane (Saccharum sinensis Roxb.) in Guangxi Zhuang Autonomous Region, China. Morphology and molecular analyses of rRNA small subunit (SSU), D2D3 expansion domains of large subunit (LSU D2D3) and internal transcribed spacer (ITS) confirmed that this nematode is different from other previously described root-lesion nematodes. Herein it is described, illustrated and named as Pratylenchus parazeae n. sp. This new species is characterised by plain and smooth En face, three lip annuli, stylet with anteriorly rounded to indented knobs, 17.0–19.0 μm long, lateral field composed of four lines with areolated outer bands at vulval region, vulva position 68.9–74.9 %, small, oval and empty spermatheca, and a subhemispherical tail usually with smooth terminus and sometimes with a more or less pronounced dorsally indented terminus. Phylogenetic trees inferred from BI analysis based on SSU, LSU D2D3 and ITS revealed that P. parazeae n. sp. can be distinguished from all described root-lesion nematodes, especially from the most closely related species P. zeae. A species-specific duplex PCR was also developed to separate the new species from other nematode species.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Sugarcane (Saccharum sinensis Roxb.) is one of the world’s largest crops and can be used as food, fiber and fuel. In 2010, FAO estimated it was cultivated in more than 90 countries, and China was one of the three major producers (FOOD AND AGRICULTURE ORGANIZATION OF THE UNITED NATIONS statistical database FAOSTAT 2010). Sugarcane is the principal source of sugar in China and about 80 % sugar is produced from sugarcane. In China, about 80 % sugarcane is grown in the south and southwest regions, including Guangxi Zhuang Autonomous Region, Guangdong and Yunnan provinces (FAO 1997).
The sugarcane production is threatened by various diseases including nematodes worldwide. Nematodes in sugarcane are more diverse than in most other cultivated crops. Over 310 species belonging to 48 genera have been reported from roots or rhizosphere of sugarcane (Cadet and Spaull 2005). Pratylenchus is one of the most common plant-parasitic nematodes infecting sugarcane. In China, two Pratylencus species, P. zeae Graham 1951 and P. thornei Sher and Allen 1953, had been recorded from sugarcane. However, in some other countries, at least ten species were recorded (Castillo and Vovlas 2007). P. zeae and P. brachyurus (Godfrey 1929) Filipjev and Schuurmans Stekhoven 1941 are the most frequently discovered species worldwide and are able to damage sugarcane. Moreover, P. zeae was frequently regarded as highly pathogenic to sugarcane (Cadet and Spaull 2005).
In China, sugarcane production in Guangxi accounts for about 40 % of the national total, which is the leading supplying province (FAO 1997). The subtropical climate in Guangxi is very suitable for sugarcane growth as well as for the reproduction of plant-parasitic nematodes. Therefore, plant-parasitic nematodes became one of the major contributing factors for low yield and quality of sugarcane. It is estimated that plant-parasitic nematodes could reduce yield of sugarcane by 16 ~ 23 % in Guangxi (Li 2000; Wei et al. 2012). More recently, 16 plant nematode genera were found in sugarcane from Guangxi and Pratylenchus was one of the most widespread nematodes (Wei et al. 2012). In a recent survey of plant-parasitic nematodes, one Pratylenchus population from damaged sugarcane field in Baise, Guangxi comprised of two species with significant different size in female. Therefore, single females of each were cultured on carrot disc separately to obtain two purified populations. Comparative morphological, morphometric and molecular studies of the two populations revealed they are two separate species, namely P. zeae and a new species. In addition, two more populations of this new species were collected from sugarcane fields in Laibin and Hechi from Guangxi. The new species is herein described as Pratylenchus parazeae n. sp. through extensive morphological and molecular studies on ribosomal DNA small subunit (SSU), D2D3 expansion domains of large subunit (LSU D2D3) and internal transcribed spacer (ITS) sequences. The host-parasite relationship of P. zeae on sugarcane was widely recognized (Cadet and Spaull 2005). In order to know the effect of P. parazeae sp. n. on sugarcane, the host-parasite relationship was studied in artificially infected sugarcane to determine the histopathology of the new species on sugarcane.
Materials and methods
Nematode populations
Soil and root samples of sugarcane were collected from Baise city, Guangxi Zhuang Autonomous Region, China. Root-lesion nematodes were isolated from the infected roots and soils by Baermann funnel method (Feng 2001). Single females were picked and transferred to carrot discs at 25 °C (Castillo et al. 1995). After 8 weeks, purified nematodes reared on carrot discs were used for morphological and molecular analysis. As a result, two species were identified, P. zeae (A29) and a new species (A90). In addition, two other populations (A6L and GX1043) of the new species were purified from samples collected from sugarcane fields in Laibin and Hechi from Guangxi.
Morphological studies
For light microscopy (LM) studies, nematodes were relaxed by gentle heat, fixed in a solution of 4 % formaldehyde + 1 % glycerin and processed by the glycerin-ethanol method (Feng 2001). Nematodes were measured and photographed with the aid of a Nikon ECLIPSE Ni microscope equipped with a Nikon Digital Sight Camera and exclusive NIS-Elements BR software (Nikon, Tokyo, Japan).
Specimens for scanning electron microscopy (SEM) were fixed in 2.5 % glutaraldehyde buffered with 0.1 M phosphate buffer (pH 7.2) at 4 °C, dehydrated in a graded series of ethanol, critical point dried with CO2 and sputter coated with gold palladium (Feng 2001). Nematodes were observed by using a XL-30-ESEM microscope (Philips, Netherlands).
DNA extraction, amplification and sequencing
DNA was extracted from individuals of female according to the protocol described in detail by Mundo-Ocampo et al. (2008). Three individuals from each population were used for DNA extraction. Three rDNA fragments, i.e., partial SSU, LSU D2D3 and ITS, of P. parazeae n. sp. (A90) and P. zeae (A29) purified from the mixed population and two other populations of P. parazeae n. sp. (A6L and GX1043), were amplified respectively. Primers for partial SSU amplification were G18SU (5′-GCTTGTCTCAAAGATTAAGCC-3′) and R18Tyl1 (5′-GGTCCAAGAATTTCACCTCTC-3′) (Chizhov et al. 2006). Primers for LSU D2D3 amplification were D2A (5′-ACAAGTACCGTGGGGAAAGTTG-3′) and D3B (5′-TCGGAAGGAACCAGCTACTA-3′) (Subbotin et al. 2006). Primers for ITS amplification were 18S (5′-TTGATTACGTCCCTGCCCTTT-3′) and 26S (5′-TTTCACTCGCCGTTACTAAGG-3′) (Vrain et al. 1992). Detailed protocols of PCR amplification were as described by Tanha Maafi et al. (2003). DNA sequencing was performed as described in Zhuo et al. (2010). The obtained sequences were deposited in the GenBank and the accession numbers are listed in Table 1.
Phylogenetic analysis
The sequences of P. parazeae n. sp. were compared with GenBank nematode sequences using the BLAST homology search program. The closest and published sequences were selected for phylogenetic analyses. Outgroup taxa for each dataset were chosen according to previous molecular phylogenetic analyses for root-lesion nematodes (Subbotin et al. 2008; Palomares-Rius et al. 2010; De Luca et al. 2011). DNA sequences were aligned in MEGA4.0 (Tamura et al. 2003) using default parameters. Models of base substitution were evaluated using MODELTEST3.7 (Posada and Crandall 1998; Huelsenbeck and Ronquist 2001) combined with PAUP4.0 (Swofford 1998). The Akaike-supported model, the base frequencies, the proportion of invariable sites, and the gamma distribution shape parameters and substitution rates were used in phylogenetic analyses. Bayesian analysis for SSU, LSU and ITS under the TrNef + I + G, TVM + I + G and TVM + I + G models, respectively, was performed to confirm the tree topology for each gene separately using MrBayes 3.2 (Huelsenbeck and Ronquist 2001) running the chain for 1 × 106 generations and setting the ‘burn-in’ at 2500. The MCMC (Markov Chain Monte Carlo) method was used within a Bayesian framework to estimate the posterior probabilities of the phylogenetic trees (Larget and Simon 1999) and generate a 50 % majority-rule consensus tree.
Duplex PCR with species specific primers
The ITS sequences of P. parazeae n. sp. were aligned with all available Pratylenchus sequences obtained from the GenBank and then the sequences specific to P. parazeae n. sp. was designed with the aid of the SP-Designer (Villard and Malausa 2013). The P. parazeae n. sp. specific primers PpzF (5′-CTGCTGCTGGATCATTACATT-3′) and PpzR (5′-TCAAATAGACATGCCCCAAT-3′) were selected. Six other Pratylenchus, three other Pratylenchidae and one free-living nematode samples (Table 2) were employed to test the specificity of the newly designed primers. DNA was extracted from single nematode as mentioned before. 2 μl DNA crude exact was used as template. The DNA quality was checked using the PCR amplification of the 28S rDNA universal primers D3A (5′-GACCCGTCTTGAAACACGGA-3′) and D3B (Al-Banna et al. 1997).
The duplex PCR system contained two sets of primers: the species-specific primers PpzF/PpzR and the universal primers D3A/D3B. To find the optimum annealing temperature and primers concentration, the reactions were performed in 54–62 °C, respectively. Species specific primers: universal primers concentrations were 0.2: 0.2, 0.2: 0.08, 0.2: 0.04, 0.2: 0.02, 0.4: 0.08 or 0.4: 0.04 μM, respectively. One optional combination was obtained based on the criteria mentioned by Waeyenberge et al. 2009. Finally, the optimized duplex PCR included 25 μl of sterile water, 1× PCR reaction buffer, 2 mM MgCl2, 200 μM of each dNTP, 0.2 μM of primers PpzF and PpzR, 0.04 μM of primers D3A and D3B, 1 U Ex Taq DNA Polymerase (TAKARA, Dalian, China) and 2 μl template DNA, was applied using PCR programme of initial denaturation at 94 °C for 5 min, 35 cycles at 94 °C for 30 s, 58 °C for 30 s and 72 °C for 40 s, followed by 72 °C for 7 min.
Histopathology
Two-weeks-old Yuetang-00236 sugarcane seedlings were transplanted into the four-liter pot containing autoclaved soil and maintained under greenhouse conditions at 28 °C. About 2000 nematodes were inoculated onto the plant roots. The sugarcane roots inoculated with water were used as a negative control. Two months after inoculation, infected sugarcane roots were selected for tissue staining and histopathological studies. Tissue staining was processed as described by Feng (2001). Necrotic roots were cut into ca 1 cm long. These samples were fixed, dehydrated and embedded according to the descriptions of Tepper (1992, 1993). Then, the necrotic root segments were sliced and the paraffin was removed following the description of Bachand and Castello (2001). Sections were stained with safranin and fast green (Sigma, USA) (Castillo et al. 1998), mounted in pure glycerol, examined under the Nikon ECLIPSE Ni microscope and photographed.
Results
Disease symptoms and histopathology
A mixed root-lesion nematode population comprised of P. parazeae n. sp. and P. zeae was detected in rhizosphere and roots of sugarcane in Baise, Guangxi, China. Sugarcanes in the field showed lesions on the roots, decline in stalks and yellowing in leaves. At 60 days post-inoculation of sugarcane with P. parazeae n. p. in glasshouse, the new species caused reddish, brown to dark lesions along the entire roots except for the root tips, single necrotic root lesions were ca 0.25 to 1.5 mm in length, some lesions fused to a large necrotic lesion (Fig. 1a, b). The above ground part of the plant became stunted with a yellowing of leaves, showing symptoms of water and nutrient deficiencies.
Symptoms and histology of sugarcane roots infected by Pratylenchus parazeae n. sp. a–b Necrotic areas on roots; c–e P. parazeae n. sp. and its eggs in the cortex; f–g longitudinal root section showing hypertrophied nucleus, granular cytoplasm, cavities and thicken walls of cortical cells adjacent to nematodes; h–i cross sections showing necrotic cells adjacent to nematodes. n, nematode; hn, hypertrophied nucleus; ca, cavity. (Scale bars: a = 2 cm; b = 2 mm; c–e = 200 μm; g–i = 50 μm)
Histopathological observations on P. parazeae n. sp.-infected sugarcane roots showed that the nematodes are migratory endoparasitic and move longitudinally in the root cortex, but do not invade stele (Fig. 1c, e, f–i). Massive necrosis, granular cytoplasm and hypertrophied nucleus of cortical cells were induced after nematode infection. Cavities and cells with thicken walls widely distributed in cortex (Fig. 2f–i). Each lesion usually contained more than one nematode, about 30 nematodes were observed maximum. Eggs were laid in the cortical tissue (Fig. 1d, e).
Description of the root-lesion nematode Pratylenchus parazeae n. sp.
Measurements of holotype and 60 paratype females are listed in Table 3. Illustrations and photos are in Figs. 2, 3 and 4.
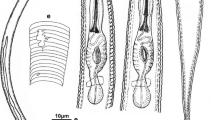
Females of Pratylenchus parazeae n. sp under the light microscope. a entire body; b pharyngeal region; c–e anterior region; f–h ovary; i vulval region; j post-vuval uterine sac; k lateral field at vuval region; l lateral field near mid-body; m lateral field at tail region; n–s tails. (Arrows point to phasmids; scale bars: a = 100 μm; b–s = 10 μm)
Female
Body almost straight or ventrally curved after heat-killing (Figs. 2a and 3a). Body annuli ca 1.0–1.5 μm wide at mid-body. Lip region with three annuli, continuous to body (Figs. 2b and 3b). Labial framework heavily sclerotised, outer margin extending two annuli into body. Under SEM, some with three annuli on one side and four annuli on the other side (Fig. 4b). En face plain and smooth, fused with all labial sectors and oral disc (Fig. 4a, b). Stylet long and robust, conus shorter than shaft, 44–48 % of entire stylet long (Figs. 2c–e and 3c–e). Stylet knobs developed, shape varies from rounded to indented anteriorly (Figs. 2b–e and 3b–e), based on the observation on 102 specimens according to knob indices of Mizukubo et al. (1990), their frequencies are: rounded = 55.9 % (Figs. 2b and 3b), lateral directed or flattened anteriorly = 32.3 % (Figs. 2c and 3c) and indented anteriorly = 11.8 % (Figs. 2d–e and 3d–e), respectively. Dorsal pharyngeal gland opening at 2.5–4.0 μm posterior to the stylet base (Figs. 2b–e and 3b–e). Median bulb well developed, oval (Figs. 2b and 3b). Isthmus slender, encircled by the anterior half of the nerve ring (Figs. 2b and 3b). Long pharyngeal gland lobe ventrally overlapping intestine, three gland nuclei present (Figs. 2b and 3b). Excretory pore near pharyngeal-intestinal junction (Figs. 2b and 3b). Hemizonid just anterior to excretory pore and two body annuli long (Figs. 2b and 3b). Lateral field occupies ca one third of corresponding body diameter with four incisures at vulval region, areolated in outer bands (Figs. 2g and 3k), and areolation occasionally extended into central band under SEM (Fig. 4c, d). Additional one or two oblique broken striae present in central band giving appearance of five or six lines near middle body in half of specimens observed by LM (Figs. 2h and 3l). Reproductive tract mono-prodelphic, germinative zone outstretched (Figs. 2a and 3f–g) [reflex in one specimen (Fig. 3h)], anterior part always posterior to pharyngeal glands. Growth zone distinct, with one row of oocytes (Fig. 3f), occasionally two rows in the middle region (Fig. 3g). Spermatheca small, oval and empty (Figs. 2f and 3i). Vulval lips usually slightly protruding (Figs. 2f and 3g). Postvulval uterine sac ca one-fourth to two-fifths of the vulva-anus distance, mostly differentiated with some cellular material in distal part (Figs. 2f and 3i–j). Phasmids pore like (Figs. 2i, 3m–s and 4f), at 35–61 % of tail length (Fig. 2j–o). Tail subcylindrical with rounded to bluntly pointed terminus, tapering toward tip, bearing 22–36 annuli (Figs. 2i–o, 3m–s and 4f). In 81 specimens observed, most tail (86.4 %) with smooth terminus (Figs. 2i–n, 3m–n; Figs. 2p–r and 4f), and less frequently (16.6 %) with a cleft (Figs. 2o and 3o). In 33.7 % specimens with tail in the lateral view (89 specimens observed), a more or less pronounced dorsally indented tail terminus present (Figs. 2l–o, 3m–p and 4g).
Male
Not found.
Type host and locality
Holotype and paratypes from a population extracted from roots and rhizosphere of sugarcane (Saccharum sinensis Roxb.) collected in Baise, Guangxi Zhuang Autonomous Region, China, in July, 2011.
Other localities
Besides the type locality, Pratylenchus parazeae n. sp. has been found in sugarcane fields of other two cities, i.e., Laibin and Hechi in Guangxi Zhuang Autonomous Region. A sample identified by us as P. parazeae n. sp., based on published sequences (Chen et al. 2009), came from maize roots collected in Taiwan.
Type specimens
Holotype and paratypes on slide were deposited in the USDA Nematode Collection, Beltsville, Maryland, USA and the Department of Nematology, University of California, Riverside, California, USA. Additional paratypes on slides and cultures on carrot disc were deposited in the Plant Nematode Research Laboratory, College of Natural Resources and Environment, South China Agricultural University, Guangzhou, China.
Etymology
The species name refers to its similarity to Pratylenchus zeae.
Diagnosis
Pratylenchus parazeae n. sp. can be differed from all other Pratylenchus spp. by morphological and molecular characteristics. It is characterized by a lip region bearing three annuli, SEM face plain, smooth and undivided, a 17.0–19.0 μm long stylet with rounded to indented anteriorly knobs, lateral field composed of four lines with areolated outer bands at vulval region and often additional one or two oblique striae in central band near middle body, vulva at 69–75 % of the body, a small, oval and empty spermatheca, and a subhemispherical tail usually with smooth terminus and sometimes with a more or less pronounced dorsally indented terminus. The matrix code of the new species, according to Castillo and Vovlas (2007) is: A2, B1, C3 (3,4), D3, E1, F6 (4,6), G2, H1, I3 (2,4), J1, K2 (range given between brackets).
Relationships
Pratylenchus parazeae n. sp. has a plain, smooth and undivided face with no division between the submedian and lateral segments, as shown in SEM (Group 1 according to Corbett and Clark 1983), three lip annuli, absence of male, and unique SSU, LSU D2D3 and rRNA-ITS sequences. Closely related species similar to P. parazeae n. sp. by three lip annuli, undivided and plain face, as seen with SEM, unfunctional spermatheca and absent or rare males include P. elamini Zeidan and Geraert 1991, P. japonicus Ryss 1988, P. mulchandi Nandakumar and Khera 1970, P. subranjani Mizukubo, Toida, Keereewan and Yoshida 1990, P. yassini Zeidan and Geraert 1991 and P. zeae Graham 1951.
P. parazeae n. sp. differs from P. zeae by the tail shape (subcylindrical vs conoid), tail terminus (rounded to bluntly pointed vs generally almost pointed, narrowly rounded to subacute), lateral field at vulva region [areolated vs smooth (code K2 vs K1)], relatively longer body (528–705 vs 360–580 μm), longer pharyngeal gland overlap (code I3 vs I2) and longer PUS (code F6 vs F5); from P. subranjani by the shape of stylet knobs (55.9 % of rounded and 11.8 % of indented anteriorly vs mostly in indented conditon and by no means rounded), more often smooth tail terminus (86.4 % smooth and 16.6 % with a cleft vs 71 % annulated, 18 % smooth and 11 % cleft), oval and empty spermatheca vs rarely observed, slightly lower ‘V’ value (69–75 vs 73–77), slightly longer body and longer vulva-anus distance (528–705 μm and 107–176 μm vs 386–572 and 77–118 μm, respectively); from P. yassini by more often smooth tail terminus (86.4 % smooth and 16.6 % with a cleft vs annulated), dorsally indented tail terminus (33.7 % vs none), different ornamentation of the lateral field (additional one or two oblique striae vs diagonally interrupted lines in central band), slightly longer PUS (33–61 μm vs 20–42 μm), slightly greater vulval body diameter (19–27 μm vs 15–20 μm) and shorter pharyngeal gland overlap (code I3 vs I4).
The new species differs from P. elamini mainly by the longer stylet, body and tail (17.0–19.0 μm, 528–705 μm and 31.5–44.0 μm vs 13–14.5 μm, 340–510 μm and 18–27 μm, respectively); from P. mulchandi by the relatively longer body (528–705 μm vs 440–580 μm), longer pharyngeal gland overlap (code I3 vs I2), lower ‘V’ value (69–75 vs 75–78), lateral field at vulva region [areolated vs smooth (code K2 vs K1)], slight lower ‘c’ value (14–20 vs 17–27), more tail annuli (22–36 vs 16–22) and dorsally indented tail terminus (33.7 % vs none); from P. japonicus by the shorter stylet length (17.0–19.0 μm vs 18.5–21.5 μm), lower ‘V’ value (69–75 vs 84–88), smaller stylet knob heigh (2.0–2.7 μm vs 3.3–4.6 μm) and longer distance between phasmid to tail terminus (15.0–23.9 μm vs 9.2–12.4 μm).
Other Pratylenchus species sharing with P. parazeae n. sp. by the combination of three lip annuli, unfunctional spermatheca and absent or rare males have been described without examining their lip patterns, such as P. arlingtoni Handoo et al. 2001, P. crenatus Loof 1960 P. cruciferus Bajaj and Bhatti 1984, P. curvicauda Siddiqi et al. 1991, P. delattrei Luc 1958, P. kumaoensis Lal and Khan 1990 and P. microstylus Bajaj and Bhatti 1984. The new species differs from all the species mentioned above by the dorsally indented tail terminus (33.7 % vs none) and the lateral field at vulva region [areolated vs smooth (code K2 vs K1)]. It can also be distinguished from P. arlingtoni and P. crenatus mainly by the lower ‘V’ value (69–75 vs 81–86 and 79–84, respectively) and more often smooth tail terminus (86.4 % smooth and 16.6 % with a cleft vs coarsely annulated); from P. curvicauda by longer stylet (17.0–19.0 μm vs 15–16.5 μm), longer body (528–705 μm vs 450–550 μm), more often smooth tail terminus (86.4 % smooth and 16.6 % with a cleft vs striated), relatively shorter pharyngeal gland overlap (code I3 vs I4), greater b’ value (3.7–5.3 vs 3.0–3.9); from P. delattrei by the longer pharyngeal gland overlap (code I3 vs I1), longer PUS (code F6 vs F3), longer vulva-anus distance (107–176 vs 77–116 μm), more tail annuli (22–36 vs 17–23) and longer tail (31.5–44.0 μm vs 23–29 μm); from P. cruciferus, P. kumaoensis and P. microstylus mainly by the longer stylet (17.0–19.0 μm vs 15–16 μm, 14–15 μm and 11–12 μm, respectively) and lower ‘V’ value (69–75 vs 76–81, 81–83, and 75–77, respectively).
Molecular characterization
Partial sequence of SSU rDNA
Nine 895-bp partial SSU products were sequenced, respectively, based on DNA template of three single females from three geographic populations from Baise, Laibin and Hechi from Guangxi (Table 1). Intraspecific variations for P. parazeae n. sp. were 0.3–1.9 % (3–17 bp). A Blastn search of all P. parazeae n. sp. sequences revealed the most similar being with three P. zeae isolates (GenBank accession number EU130832-EU130834). The interspecific variations between P. parazeae n. sp. and P. zeae (including P. zeae A29 obtained in this study) ranged from 4.0 to 5.2 % (36–47 bp) with 3–5 gaps. Intraspecific variations for P. zeae isolates were 0.7–1.6 % (6–14 bp). Fig. 5 represents a phylogenetic tree based on partial SSU from a multiple alignment of 707 total characters with 413 constant characters (58.4 %). The average nucleotide composition was equal. P. parazeae n. sp. was a sister taxon to P. zeae with 100 % support, and both species were monophyletic with 100 % support.
Bayesian consensus tree inferred from SSU of Pratylenchus parazeae n. sp. under TrNef + I + G model (lnL = 5974.769; AIC = 11957.5381; freqA = 0.25; freqC = 0.25; freqG = 0.25; freqT = 0.25; R(a) = 1; R(b) = 2.0923; R(c) = 1; R(d) = 1; R(e) = 4.1192; R(f) = 1; Pinva = 0.2377; Shape = 0.4131). Posterior probability values exceeding 50 % are given on appropriate clades. Newly obtained sequences are indicated in bold
D2D3 expansion domains of LSU rDNA
Nine LSU D2D3 sequences were obtained as shown in Table 1. Intraspecific variations for P. parazeae n. sp. were 0.5–1.8 % (4–14 bp) with 0–3 gaps. A Blastn search of P. parazeae n. sp. on the LSU D2D3 revealed high matches with some Pratylenchus species. The sequences from the new species differ from P. bhatti Siddiqi et al. 1991 (JN244269- JN244270) by 67–75 bp (8.5–9.5 %) with 7–18 gaps; from P. delattrei (JX261948-JX261949) by 65–70 bp (8.3–9.0 %) with 10–17 gaps; from Pratylenchus sp. ZTM-2013 (JX261959) by 58–63 bp (7.6–8.2 %) with 9–14 gaps. Compared with P. zeae (EU130889-EU130896 in GenBank and P. zeae A29 obtained in this study), one of the most closely related species in morphology, P. parazeae n. sp. had 72–95 variable sites (9.2–12.1 %) with 8–17 gaps. Intraspecific variations for P. zeae isolates were 0–8.3 % (0–65 bp) with 0–6 gaps. A phylogenetic tree (Fig. 6) based on LSU D2D3 was from a multiple alignment of 659 total characters with 229 constant characters (34.7 %). The average nucleotide composition was as follows: 16.6 % A, 24.44 % C, 32.11 % G and 26.85 % T. Using Boleodorus sp. (DQ328718) as outgroup taxon, all sequences of P. parazeae n. sp. isolates formed a strongly supported clade which was phylogentically related to P. zeae, Pratylenchus sp. ZTM-2013, P. delattrei and. P. bhatti.
Bayesian consensus tree inferred from LSU D2D3 of Pratylenchus parazeae n. sp. under TVM + I + G model (lnL = 9541.0137; AIC = 19100.0273; freqA = 0.166; freqC = 0.2444; freqG = 0.3211; freqT = 0.2685; R(a) = 1.143; R(b) = 5.5072; R(c) = 2.7122; R(d) = 0.4144; R(e) = 5.5072; R(f) = 1; Pinva = 0.2512; Shape = 0.8882). Posterior probability values exceeding 50 % are given on appropriate clades. Newly obtained sequences are indicated in bold
rDNA ITS
Nine rDNA-ITS sequences were obtained as shown in Table 1. Intraspecific variations for P. parazeae n. sp. were 0.1–5.4 % (1–53 bp) with 0–21 gaps. A Blastn search of P. parazeae n. sp. on the ITS region revealed the highest matches with P. zeae from Taiwan (FJ643590). The dissimilarity of the sequences from the new species and this sequence were 0–5.4 % (0–53 bp) with 0–23 gaps. However, interspecific variations of ITS region between P. parazeae n. sp. and other populations of P. zeae (JN020933-JN020935, JQ966893 and P. zeae A29 obtained in this study) were 19.2–22.4 % (190–222 bp) with 109–146 gaps. Moreover, the sequence variations between P. zeae from Taiwan (FJ643590) and other populations of P. zeae were 19.4–21.3 % (191–210 bp) with 109–132 gaps. However, intraspecific variations for P. zeae isolates (excluded P. zeae from Taiwan) were 4.4–9.5 % (39–84 bp) with 6–36 gaps. Figure 7 represented a phylogenetic tree based on ITS region from a multiple alignment of 850 total characters with 244 constant characters (28.7 %). The average nucleotide composition was as follows: 27.69 % A, 19.56 % C, 22.89 % G and 29.86 % T. When using Nacobbus aberrans as outgroup taxon, P. parazeae n. sp. was in a 100 %-supported monophyletic clade with P. zeae from Taiwan (FJ643590). This clade was sister to a 100 %-supported monophyletic clade of 7 sequences of P. zeae. P. bhatti and P. goodeyi were in a monophyletic clade with P. parazeae n. sp. and P. zeae with 100 % supported.
Bayesian consensus tree inferred from ITS of Pratylenchus parazeae n. sp. under TVM + I + G model (lnL = 10931.3662; AIC = 21880.7324; freqA = 0.2769; freqC = 0.1956; freqG = 0.2289; freqT = 0.2986; R(a) = 1.3414; R(b) = 3.1644; R(c) = 1.5217; R(d) = 1.1079; R(e) = 3.1644; R(f) = 1; Pinva = 0.2055; Shape = 1.3628). Posterior probability values exceeding 50 % are given on appropriate clades. Newly obtained sequences are indicated in bold. * Preliminary identified as P. zeae
Molecular diagnostic of Pratylenchus parazeae n. sp. using species-specific duplex PCR
Results of PCR with the species-specific primers and universal primers were given in Fig. 8a and b, respectively. The species specific PCR resulted in a single band of ca 570 bp in all studied populations of P. parazeae sp. n. All other samples gave negative results. Optimal duplex PCR revealed that a universal band of ca 340 bp for all samples tested, and a specific band of ca 570 bp appeared only for P. parazeae sp. n. populations (Fig. 8c).
The Gel with PCR results (a) PCR amplicons obtained with species specific primers for Pratylenchus parazeae n. sp.; b PCR products obtained with universal primer D3A/D3B; c Duplex PCR with the P. parazeae n. sp. specific primers and universal primer D3A/D3B. Lane M: DL2000 (Dongsheng, Guangzhou, China); 1–3: P. parazeae n. sp. (A90, A6L and GX1043); 4: P. zeae (A29); 5: P. brachyurus (HN336); 6: P. coffeae (JS43); 7: P. loosi (Pl); 8: P. neglectus (SDWG); 9: P. scribneri (SD12862); 10: P. thornei (XJDM); 11: P. vulnus (JXP); 12: Radophulus similis (Rs); 13: Hirschmanniella mucronata (Hm); 14: Pratylenchoides ritteri (NMG); 15: Caenorhabditis elegans (Ce); 16: negative control with sterile water
Discussion
Species identification on the genus Pratylenchus is very challenging because of the high interspecific overlapping and the high intraspecific variability of some morphological characters. Therefore, molecular techniques including comparative analysis of regions of rDNA and ITS-RFLP are of great assistance to the identification of Pratylenchus spp. (Castillo and Vovlas 2007). Species-specific PCR assay is a popular method for detecting root-lesion nematodes (Setterquist et al. 1996; Uehara et al. 1998a, b; Al-Banna et al. 2004; Waeyenberge et al. 2009; De Luca et al. 2012; Palomares-Rius et al. 2014). The species-specific duplex PCR system designed for P. parazeae n. sp. also showed stable and reliable results in our study.
In this study, a root-lesion nematode that is morphologically similar to P. zeae and P. subranjani was described as P. parazeae n. sp. based on morphological characters combined with molecular data. P. zeae and P. subranjani were classified as ‘subranjani’ group because of their similarity in morphology (Ryss 2002). They share the following characters with the new species: three lip annuli, undivided and plain En face, unfunctional spermatheca and absence of male. Therefore, P. parazeae n. sp. should be placed in this group based on morphological characters. Traditionally, identification of plant-parasitic nematode species mainly depends on morphology. However, species complexes or cryptic species exist in all groups of organisms including plant-parasitic nematodes. Usually, a species complex is a group of closely related but distinct species (Brown et al. 1995). A cryptic species is typically considered as two or more species with almost indistinguishable morphology, but distinct molecular characters (Bickford et al. 2007). In general, the definition of cryptic species is narrower. Some plant-parasitic nematodes cryptic species have been recognized, e.g., Ditylenchus dipsaci (Kühn 1857) Filipjev 1936 and D. gigas Vovlas et al. 2011 (Vovlas et al. 2011), P. floridensis De Luca et al. 2010 and P. parafloridensis De Luca et al. 2010 (De Luca et al. 2010), P. coffeae (Zimmermann 1898)Filipjev and Schuurmans Stekhoven 1941 and P. speijeri De Luca et al. 2012 (De Luca et al. 2012). In this study, P. parazeae n. sp. and P. zeae are very similar in appearance, but they could be separated from each other not only by molecular characters, but also by some morphological differences, including shape of tail and tail terminus, areolated lateral field at vulva region, body length, pharyngeal gland overlap and PUS length. Therefore, it may be more precise to consider P. parazeae n. sp. and P. zeae as a species complex but not cryptic species.
According to phylogenetic trees constructed from partial SSU and LSU D2D3 sequences, P. zeae and P. goodeyi were clustered in clade VI (Subbotin et al. 2008). Recently, P. bhatti, P. delattrei and two unknown species (Pratylenchus sp. ZTM-2013 and Pratylenchus sp. DP-2010) were added to the clade VI based on the LSU D2D3 tree (Taheri et al. 2013; Palomares-Rius et al. 2014). The trees obtained from molecular analysis of different rDNA sequences placed P. parazeae n. sp. in a high supported monophyletic clade with P. zeae, but still showed significant divergence. Obviously, the new species also affiliated to the clade VI. Therefore, the clade VI contained seven species (P. parazeae n. sp., P. zeae, P. bhatti, P. delattrei, P. goodeyi, Pratylenchus sp. ZTM-2013 and Pratylenchus sp. DP-2010).
ITS sequence variation was widely reported among plant-parasitic nematodes (Cherry et al. 1997; Powers et al. 1997; Blok et al. 1998; Hugall et al. 1999; Subbotin et al. 2001; Reid et al. 2003). Inter- and intraspecific variability in ITS region of root-lesion nematodes was also demonstrated (Waeyenberge et al. 2000, 2009; De Luca et al. 2011). Usually, intraspecific sequence variation was 1–3 % in the genus Pratylenchus. However, for several species, such as P. agilis (Thorne and Malek 1968) Hernández et al. 2000, P. brachyurus and P. penetrans (Cobb 1917) Filipjev and Schuurmans Stekhoven 1941, the variations were up to 8 % (Waeyenberge et al. 2009). In this study, intraspecific variations for P. parazeae n. sp. were 0.1–5.4 %. But, for P. zeae isolates, the variations were 4.4–9.5 %. Chen et al. (2009) reported P. zeae from rhizosphere of maize in Taiwan and provided an ITS sequence (FJ643590). The sequence divergences were 0–5.4 % between P. parazeae n. sp. and P. zeae from Taiwan, but interspecific ITS sequence divergences between P. parazeae n. sp. and other P. zeae were 19.2–22.4 %. Interestingly, the sequence variations were 19.4–21.3 % between P. zeae from Taiwan and other P. zeae. Moreover, P. zeae from Taiwan was in the same clade with P. parazeae n. sp. with 100 % support and short branch differences. Consequently, it is suggested that P. zeae from Taiwan should be reclassified as P. parazeae n. sp. In fact, P. parazeae n. sp. and P. zeae were found in the same sugarcane field in this study. Therefore, previously reported P. zeae from sugarcane field in China should be reexamined.
Pratylenchus is usually considered as a pathogen to sugarcane. It was reported that P. zeae causes lesions to sugarcane roots and results in a yellowing of the leaves and reductions in shoot and root mass and stalk length (Cadet and Spaull 2005). Pratylenchus brachyurus was reported to affect the length and mass of stalks, but did not affect root or shoot mass (Onapitan and Amosu 1982). Inoculation experiments showed the new species can invade sugarcane roots and caused similar disease symptom as P. zeae. Results from histopathological study showed P. parazeae n. sp. feed on cortex cells and doesn’t enter stele. Histopathological responses of sugarcane roots infected by P. parazeae n. sp. were different from that in P. brachyurus-sugarcane interactions. It is reported that P. brachyurus partly buried in the cortex and partly in the vascular bundle (Onapitan and Amosu 1982). In this study, P. parazeae n. sp. was isolated from sugarcane together with P. zeae in the same field, further evaluation on its pathogenicity and economic damage is needed.
References
Al-Banna, L., Williamson, V., & Gardner, S. L. (1997). Phylogenetic analysis of nematodes of the genus Pratylenchus using nuclear 26S rDNA. Molecular Phylogenetics and Evolution, 7(1), 94–102.
Al-Banna, L., Ploeg, A. T., Williamson, V. M., & Kaloshian, I. (2004). Discrimination of six Pratylenchus species using PCR and species-specific primers. Journal of Nematology, 36(2), 142–146.
Bachand, G. D., & Castello, J. D. (2001). Immunolocalization of tomato mosaic tobamovirus in roots of red spruce seedlings. Journal of Phytopathology, 149, 415–419.
Bajaj, H. K., & Bhatti, D. S. (1984). New and known species of Pratylenchus Filipjev, 1936 (Nematoda: Pratylenchidae) from Haryana, India, with remarks on intraspecific variations. Journal of Nematology, 16(4), 360–367.
Bickford, D., Lohman, D. J., Sodhi, N. S., Ng, P. K., Meier, R., Winker, K., Ingram K. K., & Das, I. (2007). Cryptic species as a window on diversity and conservation. Trends in Ecology & Evolution, 22(3), 148–155.
Blok, V. C., Malloch, G., Harrower, B., Phillips, M. S., & Vrain, T. C. (1998). Intraspecific variation in ribosomal DNA in populations of the potato cyst nematode Globodera pallida. Journal of Nematology, 30(2), 262.
Brown, J. K., Frohlich, D. R., & Rosell, R. C. (1995). The sweetpotato or silverleaf whiteflies: biotypes of Bemisia tabaci or a species complex? Annual Review of Entomology, 40(1), 511–534.
Cadet, P., & Spaull, V. W. (2005). Nematode parasites of sugarcane. In M. Luc, R. A. Sikora, & J. Bridge (Eds.), Plant-parasitic nematodes in subtropical and tropical agriculture (2nd ed., pp. 645–674). Wallingford: CAB International.
Castillo, P., & Vovlas, N. (2007). Pratylenchus (Nematoda: Pratylenchidae): Diagnosis, Biology, Pathogenicity and Management. In D. J. Hunt & R. N. Perry (Eds.), Nematology monographs and perspectives 6 (p. 529). Leiden: Brill.
Castillo, P., Trapero-Casas, J. L., & Jiménez-Díaz, R. M. (1995). Effect of time, temperature, and inoculum density on reproduction of Pratylenchus thornei in carrot disk cultures. Journal of Nematology, 27(1), 120–124.
Castillo, P., Vovlas, N., & Jiménez-Díaz, R. M. (1998). Pathogenicity and histopathology of Pratylenchus thornei populations on selected chickpea genotypes. Plant Pathology, 47(3), 370–376.
Chen, D. Y., Ni, H. F., Yen, J. H., & Tsay, T. T. (2009). Identification of a new recorded root-lesion nematode Pratylenchus zeae (Nematoda: Tylenchoidea, Pratylenchidae) from corn plantations in Taiwan. Plant Pathology Bulletin, 18(2), 111–118.
Cherry, T., Szalanski, A. L., Todd, T. C., & Powers, T. O. (1997). The internal transcribed spacer region of Belonolaimus (Nemata: Belonolaimidae). Journal of Nematology, 29(1), 23.
Chizhov, V. N., Chumakova, O. A., Subbotin, S. A., & Baldwin, J. G. (2006). Morphological and molecular characterization of foliar nematodes of the genus Aphelencholdes: A. fragariae and A. ritzemabosi (Nematoda: Aphelenchoididae) from the Main Botanical Garden of the Russian Academy of Sciences. Russian Journal of Nematology, 14(2), 179–184.
Cobb, N. A. (1917). A new parasitic nema found infesting cotton and potatoes. Journal of Agricultural Research, 11, 27–33.
Corbett, D. C. M., & Clark, S. A. (1983). Surface features in the taxonomy of Pratylenchus species. Revue de Nématologie, 6(1), 85–98.
De Luca, F., Troccoli, A., Duncan, L. W., Subbotin, S. A., Waeyenberge, L., Moens, M., & Inserra, R. N. (2010). Characterisation of a population of Pratylenchus hippeastri from bromeliads and description of two related new species, P. floridensis n. sp. and P. parafloridensis n. sp. from grasses in Florida. Nematology, 12(6), 847–868.
De Luca, F., Reyes, A., Troccoli, A., & Castillo, P. (2011). Molecular variability and phylogenetic relationships among different species and populations of Pratylenchus (Nematoda: Pratylenchidae) as inferred from the analysis of the ITS rDNA. European Journal of Plant Pathology, 130(3), 415–426.
De Luca, F., Troccoli, A., Duncan, L. W., Subbotin, S. A., Waeyenberge, L., Coyne, D. L., Brentu, F. C., & Inserra, R. N. (2012). Pratylenchus speijeri n. sp. (Nematoda: Pratylenchidae), a new root-lesion nematode pest of plantain in West Africa. Nematology, 14(8), 987–1004.
FAO (1997). Fiji/FAO Asia Pacific Sugar Conference. http://www.fao.org/docrep/005/x0513e/x0513e18.htm. Accessed 1 June 2014.
Feng, Z. X. (2001). Plant nematology (p. 208). Beijing: Chinese Agricultural Publishing.
Filipjev, I. N. (1936). On the classification of the Tylenchinae. Proceedings of the Helminthological Society of Washington, 3(2), 80–82.
Filipjev, I. N., & Schuurmans Stekhoven, J. H. (1941). A Manual of agricultural helminthology (p. 878). Leiden: Brill.
FOOD AND AGRICULTURE ORGANIZATION OF THE UNITED NATIONS statistical database (FAOSTAT) (2010). FAOSTAT production statistics of Sugarcane. Online database. http://faostat.fao.org/site/567/DesktopDefault.aspx?PageID=567. Accessed 1 June 2014.
Godfrey, G. H. (1929). A destructive root disease of pineapples and other plants due to Tylenchus brachyurus n. sp. Phytopathology, 19, 611–629.
Graham, T. W. (1951). Nematode root rot of tobacco and other plants. In South Carolina Agricultural Experiment Station Bulletin, 390 (p. 25). South Carolina Agricultural Experiment Station, Clemson Agricultural College.
Handoo, Z. A., Carta, L. K., & Skantar, A. M. (2001). Morphological and molecular characterisation of Pratylenchus arlingtoni n. sp., P. convallariae and P. fallax (Nematoda: Pratylenchidae). Nematology, 3(6), 607–618.
Hernández, M., Jordana, R., Goldaracena, A., & Pinochet, J. (2000). SEM observations of nine species of the genus Pratylenchus Filipjev, 1936 (Nematoda: Pratylenchidae). Journal of Nematode Morphology and Systematics, 3(2), 165–174.
Huelsenbeck, J. P., & Ronquist, F. (2001). MR BAYES: bayesian inference of phylogenetic tress. Bioinformatics, 17(8), 1754–1755.
Hugall, A., Stanton, J., & Moritz, C. (1999). Reticulate evolution and the origins of ribosomal internal transcribed spacer diversity in apomictic Meloidogyne. Molecular Biology and Evolution, 16(2), 157–164.
Kühn, J. (1857). Über das Vorkommen von Anguillulen in erkrankten Blüthenköpfen von Dipsacus fullonum L. Zeitschrift für wissenschaftliche Zoologie, 9, 129–137.
Lal, A., & Khan, E. (1990). Species of Pratylenchus Filipjev, 1936 and Helicotylenchus Steiner, 1945 (Nematoda:Tylenchida) found associated with forest trees in northern India. Indian Journal of Nematology, 19(1), 44–50.
Larget, B., & Simon, D. L. (1999). Markov chain Monte Carlo algorithms for the Bayesian analysis of phylogenetic trees. Molecular Biology and Evolution, 16, 750–759.
Li, Q. W. (2000). Improved technology of modern sugarcane (p. 340). Guangzhou: South China University of Technology Press.
Loof, P. A. A. (1960). Taxonomic studies on the genus Pratylenchus (Nematoda). Tijdschrift over Plantenziekten, 66(2), 29–90.
Luc, M. (1958). Les nématodes et le flétrissement des cotonniers dans le Sud-Ouest de Madagascar. Coton et Fibres Tropicales, 13(2), 1–18.
Mizukubo, T., Toida, Y., Keereewan, S., & Yoshida, M. (1990). Pratylenchus subranjani n. sp. (Nematoda: Pratylenchidae) from maize in Thailand. Applied Entomology and Zoology, 25(2), 311–318.
Mundo-Ocampo, M., Troccoli, A., Subbotin, S. A., Cid, J., Baldwin, J. G., & Inserra, R. N. (2008). Synonymy of Afenestrata with Heterodera supported by phylogenetics with molecular and morphological characterisation of H. koreana comb. n. and H. orientalis comb. n. (Tylenchida: Heteroderidae). Nematology, 10(5), 611–632.
Nandakumar, C., & Khera, S. (1970). A new nematode species, Pratylenchus mulchandi from millets of Rajasthan. Indian Phytopathology, 22(3), 359–363.
Onapitan, J. A., & Amosu, J. O. (1982). Pathogenicity and histopathology of Pratylenchus brachyurus and Helicotylenchus pseudorobustus on sugarcane. Nematropica, 12(1), 51–60.
Palomares-Rius, J. E., Castillo, P., Liebanas, G., Vovlas, N., Landa, B. B., Navas-Cortes, J. A., & Subbotin, S. A. (2010). Description of Pratylenchus hispaniensis n. sp. from Spain and considerations on the phylogenetic relationship among selected genera in the family Pratylenchidae. Nematology, 12(3), 429–451.
Palomares-Rius, J. E., Guesmi, I., Horrigue-Raouani, N., Cantalapiedra-Navarrete, C., Liébanas, G., & Castillo, P. (2014). Morphological and molecular characterisation of Pratylenchus oleae n. sp. (Nematoda: Pratylenchidae) parasitizing wild and cultivated olives in Spain and Tunisia. European Journal of Plant Pathology, 140(1), 53–67.
Posada, D., & Crandall, K. A. (1998). Modeltest: testing the model of DNA substitution. Bioinformatics, 14(9), 817–818.
Powers, T. O., Todd, T. C., Burnell, A. M., Murray, P. C. B., Fleming, C. C., Szalanski, A. L., Adams, B. A., & Harris, T. S. (1997). The rDNA internal transcribed spacer region as a taxonomic marker for nematodes. Journal of Nematology, 29(4), 441.
Reid, A., Manzanilla-López, R. H., & Hunt, D. J. (2003). Nacobbus aberrans (Thorne, 1935) Thorne & Allen, 1944 (Nematoda: Pratylenchidae); a nascent species complex revealed by RFLP analysis and sequencing of the ITS-rDNA region. Nematology, 5(3), 441–451.
Ryss, A. Y. (1988). Root parasitic nematodes of the family Pratylenchidae (Tylenchida) of the World Fauna (p. 367). Leningrad: USSR.
Ryss, A. Y. (2002). Phylogeny and evolution of the genus Pratylenchus according to morphological data (Nematoda: Tylenchida). Zoosystematica Rossica, 10(2), 257–273.
Setterquist, R. A., Smith, G. K., Jones, R., & Fox, G. E. (1996). Diagnostic probes targeting the major sperm protein gene that may be useful in the molecular identification of nematodes. Journal of Nematology, 28(4), 414–421.
Sher, S. A., & Allen, M. W. (1953). Revision of the genus Pratylenchus (Nematoda: Tylenchidae). University of California Publications in Zoology, 57(6), 441–470.
Siddiqi, M. R. (2000). Tylenchida parasites of plants and insects (2nd ed., p. 864). Wallingford: CABI Publishing.
Siddiqi, M. R., Dabur, K. R., & Bajaj, H. K. (1991). Descriptions of three new species of Pratylenchus Filipjev, 1936 (Nematoda: Pratylenchidae). Nematologia Mediterranea, 19(1), 1–7.
Subbotin, S. A., Vierstraete, A., De Ley, P., Rowe, J., Waeyenberge, L., Moens, M., & Vanfleteren, J. R. (2001). Phylogenetic relationships within the cyst-forming nematodes (Nematoda, Heteroderidae) based on analysis of sequences from the ITS regions of ribosomal DNA. Molecular Phylogenetics and Evolution, 21(1), 1–16.
Subbotin, S. A., Sturhan, D., Chizhov, V. N., Vovlas, N., & Baldwin, J. G. (2006). Phylogenetic analysis of Tylenchida Thorne, 1949 as inferred from D2 and D3 expansion fragments of the 28S rRNA gene sequences. Nematology, 8(3), 455–474.
Subbotin, S. A., Ragsdale, E. J., Mullens, T., Roberts, P. A., Mundo-Ocampo, M., & Baldwin, J. G. (2008). A phylogenetic framework for root-lesion nematodes of the genus Pratylenchus (Nematoda): evidence from 18S and D2-D3 expansion segments of 28S ribosomal RNA genes and morphological characters. Molecular Phylogenetics and Evolution, 48(2), 491–505.
Swofford, D. L. (1998). PAUP*-Phylogenetic Analyses Using Parsimony (* and other Methods). Version 4 b10 (p. 128). Sunderland: Sinauer Associates.
Taheri, Z. M., Maafi, Z. T., Subbotin, S. A., Pourjam, E., & Eskandari, A. (2013). Molecular and phylogenetic studies on Pratylenchidae from Iran with additional data on Pratylenchus delattrei, Pratylenchoides alkani and two unknown species of Hirschmanniella and Pratylenchus. Nematology, 15(6), 633–651.
Tamura, K., Dudley, J., Nei, M., & Kumar, S. (2003). MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Molecular Biology and Evolution, 24(8), 1596–1599.
Tanha Maafi, Z., Subbotin, S. A., & Moens, M. (2003). Molecular identification of cyst-forming nematodes (Heteroderidae) from Iran and a phylogeny based on the ITS sequences of rDNA. Nematology, 5(1), 99–111.
Tepper, H. B. (1992). Benzyladenine promotes shoot initiation in empty leaf axils of Stellaria media L. Journal of Plant Physiology, 140(2), 241–243.
Tepper, H. B. (1993). Developmental features accompanying the imposition and release of apical dominance in pea. Journal of Plant Physiology, 142(6), 722–729.
Thorne, G., & Malek, R. B. (1968). Nematodes of the northern Great Plains. Part I. Tylenchida (Nemata: Secernentea) South Dakota: South Dakota Agricultural Experiment Station, Technical Bulletin, 31 p. 111.
Uehara, T., Mizukubo, T., Kushida, A., & Momota, Y. (1998a). Identification of Pratylenchus penetrans (Cobb) by PCR using ITS-based species-specific primers. Japanese Journal of Nematology, 28(1), 1–7.
Uehara, T., Mizukubo, T., Kushida, A., & Momota, Y. (1998b). Identification of Pratylenchus coffeae and P. loosi using specific primers for PCR amplification of ribosomal DNA. Nematologica, 44(4), 357–368.
Villard, P., & Malausa, T. (2013). SP-Designer: a user-friendly program for designing species-specific primer pairs from DNA sequence alignments. Molecular Ecology Resources, 13(4), 755–758.
Vovlas, N., Troccoli, A., Palomares-Rius, J. E., De Luca, F., Liébanas, G., Landa, B. B., Subbotin, S. A., & Castillo, P. (2011). Ditylenchus gigas n. sp. parasitizing broad bean: a new stem nematode singled out from the Ditylenchus dipsaci species complex using a polyphasic approach with molecular phylogeny. Plant Pathology, 60(4), 762–775.
Vrain, T. C., Wakarchuk, D. A., Levesque, A. C., & Hamilton, R. I. (1992). Intraspecific rDNA restriction fragment length polymorphism in the Xiphinema americanum group. Fundamental and Applied Nematology, 15(6), 563–573.
Waeyenberge, L., Ryss, A., Moens, M., Pinochet, J., & Vrain, T. C. (2000). Molecular characterisation of 18 Pratylenchus species using rDNA restriction fragment length polymorphism. Nematology, 2(2), 135–142.
Waeyenberge, L., Viaene, N., & Moens, M. (2009). Species-specific duplex PCR for the detection of Pratylenchus penetrans. Nematology, 11(6), 847–857.
Wei, J. L., Huang, C. H., Shang, X. K., Pan, X. H., Huang, D. F., & Wang, B. H. (2012). Nematode species and their distribution in sugarcane fields in Guangxi. Journal of Southern Agriculture, 43(2), 184–186.
Zeidan, A. B., & Geraert, E. (1991). Pratylenchus from Sudan, with the description of two new species (Nemata: Tylenchida). Revue de Nematologie, 14, 221–229.
Zhuo, K., Cui, R. Q., Ye, W. M., Luo, M., Wang, H. H., Hu, X. N., & Liao, J. L. (2010). Morphological and molecular characterization of Aphelenchoides fujianensis n. sp. (Nematoda: Aphelenchoididae) from Pinus massoniana in China. Zootaxa, 2509, 39–52.
Zimmermann, A. (1898). De Nematoden der koffiewortels. Deel I. Mededeelingen uit's Lands Plantentuin, 27, 1–64.
Acknowledgments
This research was supported by National Key Basic Research Program of China (973 Program, grant number 2013CB127501), the Planning Project for Science and Technology in Guangzhou City (grant number 11A62100574) and the special fund for agro-scientific research in the public interest of China (grant numbers 201103018 and 200903040).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Wang, H., Zhuo, K., Ye, W. et al. Morphological and molecular charaterisation of Pratylenchus parazeae n. sp. (Nematoda: Pratylenchidae) parasitizing sugarcane in China. Eur J Plant Pathol 143, 173–191 (2015). https://doi.org/10.1007/s10658-015-0674-z
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10658-015-0674-z