Abstract
Bois noir (BN), which is induced by ‘Candidatus Phytoplasma solani’ (‘Ca. P. solani’), is an important grapevine yellows disease that causes severe damage in viticultural regions throughout the Euro-Mediterranean basin. An epidemiological survey to determine potential insect vectors and the primary reservoir plants of BN phytoplasma in Macedonian vineyards was undertaken between 2012 and 2013 in the southeastern part of the country. A study on the species diversity from the suborder Auchenorrhycha revealed the prevalence of the principal vector of ‘Ca. P. solani’, which is the planthopper Hyalesthes obsoletus. Reptalus panzeri, which is the second-most documented BN vector, was not recorded in Macedonian vineyards. Three leafhopper species, namely Psammotettix alienus, Artianus manderstjernii and Euscelis incisus, were also widespread in the BN-affected vineyards, but only H. obsoletus tested positive for ‘Ca. P. solani’. Molecular characterizations were performed by the sequencing and/or RFLP typing of tuf, vmp1 and stamp genes, and the results were used to gain detailed insight into the molecular diversity of the ‘Ca. P. solani’ strains associated with grapevines, tentative reservoir plants (Urtica dioica and Convolvulus arvensis) and the H. obsoletus associated with these plants. Among the 91 ‘Ca. P. solani’ strains detected in diverse plant and insect hosts, three tuf, five vmp1 and 11 distinct stamp genotypes were identified. Twelve comprehensive genotypes of ‘Ca. P. solani’ were detected according to the tuf/vmp1/stamp genotyping. The highest diversity of genotypes was detected among the strains from H. obsoletus individuals associated with U. dioica, of which the most frequent genotype was tuf-ab/V18/M1 (43 %). The tuf-b/V2-TA/STOL comprehensive genotype was found in 33 % of naturally infected grapevines. Two ‘Ca. P. solani’ genotypes were associated with U. dioica, namely (i) tuf-ab/V18/M1 (60 %) and tuf-a/V3/M4 (40 %), and only one genotype (tuf-b/V2-TA/Rqg50) was associated with C. arvensis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Bois noir (BN) is an important grapevine yellows disease, and it is induced by the stolbur phytoplasma from the 16SrXII-A subgroup that was recently described as ‘Candidatus Phytoplasma solani’ (‘Ca. P. solani’) (Quaglino et al. 2013). The disease is widespread all over Europe, the Mediterranean area and in the Middle East and causes serious economic losses in grapevine production (Johannesen et al. 2012; Aryan et al. 2014; Cvrković et al. 2014).
Phytoplasmas are wall-less, non-helical prokaryotes, and they are members of the class Mollicutes that colonize plant phloem. These organisms are obligatorily transmitted by insects, grafting or parasitic plants (Weintraub and Beanland 2006). The insect vectors of phytoplasmas include leafhoppers, planthoppers and psyllids, and these insects belong to the suborders Auchenorrhyncha and Sternorrhyncha, order Hemiptera.
‘Ca. P. solani’ is an emerging plant pathogen that causes yellows diseases in grapevines (bois noir) and various cultivated plants of economic importance including potatoes, maize, sugar beets and others (Gatineau et al. 2002; Jović et al. 2009, 2011). The documented vectors that transmit ‘Ca. P. solani’ to grapevines are polyphagous planthoppers of xerothermic habitats, namely Hyalesthes obsoletus and Reptalus panzeri, both of which belong to the Cixiidae family (Maixner 1994; Cvrković et al. 2014). Nevertheless, other species such as R. quinquecostatus and Anaceratagallia ribauti are also known to harbor ‘Ca. P. solani’ infections and were demonstrably able to transmit the pathogen to experimental plants or artificial feeding medium; however, their ability to vector the pathogen to grapevines has not yet been demonstrated (Pinzauti et al. 2008; Riedle-Bauer et al. 2008; Aryan et al. 2014).
The epidemiology of phytoplasma-induced diseases is determined by the interaction between the vector and pathogen and their common natural hosts. In the case of BN, grapevines are an erroneous food substrate for H. obsoletus and, consequently, a dead-end host for ‘Ca. P. solani’ (Johannesen et al. 2012). The epidemiology of BN is coupled to the infection of herbaceous host plants, which are the primary food sources for H. obsoletus nymphs and pathogen reservoirs. According to observations of the elongation factor Tu (tuf) gene, the ‘Ca. P. solani’ consists of two genetically divergent strain types, tuf-a and tuf-b, which are involved in the two diverse epidemiological cycles of BN (Langer and Maixner 2004). The tuf-b type is primarily associated with field bindweed (Convolvulus arvensis), the major host plant of H. obsoletus in the eastern and southeastern occurrences of the disease (Langer and Maixner 2004; Johannesen et al. 2012), although it infects a number of diverse weedy plants (Riedle-Bauer et al. 2008; Johannesen et al. 2012; Cvrković et al. 2014). Tuf-a strains of ‘Ca. Phytoplasma solani’ are spread via an epidemic cycle sourced in stinging nettle (Urtica dioica) which is the only documented herbaceous reservoir of this ‘Ca. P. solani’ genotype (Langer and Maixner 2004; Johannesen et al. 2012). The nettle-sourced epidemiological cycle is the most common in the northwestern disease range (Germany, Switzerland and northern France), and it was recently registered as the cause of an epidemic BN outbreak in Austria (Aryan et al. 2014).
In addition to the epidemiological significance of the tuf housekeeping gene, two stolbur-specific genes encoding putative membrane proteins that are involved in host recognition and interaction, namely vmp1 and stamp, are proposed for the characterization of genetic diversity in ‘Ca. Phytoplasma solani’ through a multilocus sequencing approach (Cimerman et al. 2009; Fabre et al. 2011; Pacifico et al. 2009). These two genes have higher sequence variability than the tuf gene, and thus they are more widely used in epidemiological studies of BN phytoplasmas (Johannesen et al. 2012; Aryan et al. 2014; Cvrković et al. 2014; Kostadinovska et al. 2014).
The grapevine is one of the most important cultivated plants and has a long tradition of cultivation and economic significance in the Republic of Macedonia. The presence of BN has been reported in different viticultural regions of Macedonia (Šeruga et al. 2003), infecting diverse grapevine varieties with a constantly increasing incidence of infection throughout the vineyards (Kostadinovska et al. 2014). However, there is a lack of information about the diversity of planthoppers and leafhoppers in affected vineyards and their surroundings, potential insect vectors and the putative reservoir plants that are involved in the epidemiological cycle(s) of BN phytoplasma in Macedonian vineyards. Therefore, the aim of this study was as follows: (i) to identify insect species from the suborder Auchenorrhyncha that occur in BN-affected vineyards and that harbor ‘Ca. P. solani’, and (ii) to examine the epidemiologically informative tuf, vmp1 and stamp genes of ‘Ca. P. solani’ strains in naturally infected grapevines, insects and reservoir plants to elucidate the epidemiological cycle(s) involved in BN transmission in Macedonia.
Material and methods
A survey of potential insect vectors
A survey of potential phytoplasma vectors in the suborder Auchenorrhyncha was performed in 2012 and 2013. The survey sites included three vineyards with symptoms of phytoplasma infection in the southeastern Macedonian viticulture region. Insects were collected every 15 days from May 15th until the end of September. Potential hemipteran vectors including leafhoppers, planthoppers and cixiids were collected from grapevines, along the inter-rows, in the rows and around the vineyard on different herbaceous and woody plants, with a focus on the major documented hosts of ‘Ca. P. solani’ - H. obsoletus, C. arvensis and U. dioica. Insects were collected with sweep nets and mouth aspirators. The insects collected for PCR analyses were placed in 2-ml plastic vials (Sarstedt) containing 96 % ethanol, and they were subsequently identified to the species level with taxonomic keys provided by Holzinger et al. (2003) and Biedermann and Niedringhaus (2004).
Plant sampling
Symptomatic grapevines and the predominant weeds inside and around the vineyards were collected for ‘Ca. P. solani’ detection and multilocus sequence typing.
In the beginning of September 2012, leaves with symptoms of phytoplasma infection, such as the rolling of leaf margins and partial discoloration, were sampled from vineyards. Fresh leaf veins and petioles were dissected, distributed into 1 g aliquots and stored at −20 °C prior to DNA extraction.
During August and at the beginning of September 2013, 118 samples of the two most abundant weeds, C. arvensis and U. dioica, were collected from the study vineyards. Because they were asymptomatic, the weeds were sampled randomly, dug out with roots that were later sliced, distributed into 0.5-1.0 g aliquots and stored at −20 °C until DNA extraction.
DNA extraction
Total nucleic acids were extracted from fresh grapevine leaf midribs and petioles and from weed roots by using the CTAB protocol described by Angelini et al. (2001).
To detect ‘Ca. P. solani’ in the insects, we analyzed species that were represented by more than a 100 individuals per collection year, along with all the specimens of cixiid species that were present in the vineyards. The insects were analyzed in pools of 3–5 adults, depending on the specimen size, or individually in the case of cixiids. DNA was isolated by applying a modified CTAB method according to Gatineau et al. (2001).
‘Ca. P. solani’ detection in plants and insects
The presence of ‘Ca. P. solani’ in field-collected plant and insect material was detected by using a modification of the stolbur-specific Stol11 protocol with an F2/R1 primer pair for direct PCR, followed by F3/R2 for nested PCR (Clair et al. 2003). DNA amplification was performed in a 20 μl reaction volume by following amplification conditions according to Radonjić et al. (2009). DNA extracts of BN-infected grapevines from Serbia (Cvrković et al. 2014) were used as a positive control in all amplification reactions. Amplified products were separated on a 1 % agarose gel by electrophoresis in TBE buffer (Tris-Borate 90 mM, EDTA 1 mM), stained with ethidium bromide and visualized under a UV transilluminator.
Characterization of ‘Ca. P. solani’ based on RFLP and sequence typing
The amplification of the following three phytoplasma genomic loci was performed for the molecular characterization of ‘Ca. P. solani’ strains detected in grapevine plants, insects and weedy reservoir plants: (i) the tuf gene encoding the translation elongation factor Tu, (ii) the vmp1 gene encoding a putative ‘Ca. P. solani’ membrane protein, and (iii) the stamp gene encoding the antigenic membrane protein in ‘Ca. P. solani’.
Tuf gene
Tuf gene amplification was performed by nested PCR with the primers Tuf1f/r, followed by TufAYf/r as described by Langer and Maixner (2004). The amplicons obtained by nested PCR were subjected to restriction analysis with HpaII endonuclease to obtain information about the tuf type present in the collected material. Restriction analyses were performed according to the manufacturer’s instructions (Fermentas, Lithuania). The restriction products were separated by automated capillary electrophoresis by using a QIAxcel advanced system (Qiagen) with a Screening Gel Cartridge (Qiagen) under the following parameters from the applied method: sample injection voltage 5 kV, sample injection time 8 s, separation voltage 6 kV and separation time 320 s. The QX alignment marker for 15 bp/5 kb (Qiagen) was used to align the resulting restriction fragments and the QX DNA size marker FX174/HaeIII (Qiagen) was used for fragment size comparisons. The DNA of ‘Ca. P. solani’ tuf-a and tuf-b types was isolated from naturally infected H. obsoletus from the Middle-Rhine and Mosel regions of Germany, respectively (as provided by M. Maixner, Bernkastel-Kues), and they were used as the reference controls to compare with the restriction profiles. Strains with the tuf-b restriction profile that were associated with stamp and vmp1 types and generally considered to be associated with the nettle were additionally subjected to a sequencing analysis of the tuf nested products. Sequencing was performed by Macrogen Inc. (Seoul, South Korea) and the sequences are deposited in the NCBI GenBank under the accession numbers KP337324-7. The tuf sequences were edited by using FinchTV v.1.4.0 (http://www.geospiza.com) and aligned with the reference strains (Aryan et al. 2014) by using Clustal W as integrated into MEGA5 software (Tamura et al. 2011) to compare the SNPs associated with each of the tuf-a/-b strains.
Vmp1 gene
The vmp1 gene amplification was performed as a nested PCR with the primer pair StolH10F1/R1 (Cimerman et al. 2009), followed by primer pair TYPH10F/R (Fialová et al. 2009), by using reaction conditions specified by Fialová et al. (2009). The TYPH10F/R amplicons of all the characterized strains were digested with RsaI restriction enzyme, and for some of the resulting profiles, an additional TaqI and AluI digestion was performed to distinguish between the V2 and V2-TA vmp1 profiles. The restriction fragments were separated by capillary electrophoresis as described above. The phytoplasma strains employed as references for vmp1 restriction pattern comparison were taken from Cvrković et al. (2014) or provided by X. Foissac (Bordeaux-France) (Fig. 1).
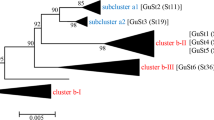
RsaI RFLP profiles of the vmp1 marker for ‘Candidatus Phytoplasma solani’ associated with different hosts in BN-diseased vineyards in Macedonia and reference strains. The restriction fragments were separated by automated capillary electrophoresis by using the QIAxcel advanced system (Qiagen). The following phytoplasma strains were employed as references for vmp1 restriction pattern comparisons: GGY, Sp-infected grapevine from Germany, V2 profile; Rpg39, Sp-infected R. panzeri from Serbia, V2-TA profile; Hr-Br (HR-BR18-09), Sp-infected grapevine from Croatia, V3 profile; Rqg50, Sp-infected R. quinquecostatus from Serbia, V4 profile; 19–25, Sp-infected grapevine from Germany, V5 profile; and Vv5, Sp-infected grapevine from Serbia, V14 profile. Reference strains were taken from Cvrković et al. (2014) or provided by X. Foissac (Bordeaux-France). FX174/HaeIII: QX DNA size marker (Qiagen). The fragment sizes (bp) of the marker (1353, 1078, 872, 603, 310, 281, 271, 234, 194, 118 and 72) and alignment marker QX 15 bp/5 kb (15 and 5000) are designated
Stamp gene
The stamp gene, which encodes the antigenic membrane protein in ‘Ca. P. solani’, was amplified by nested PCR with StampF/R0 followed by StampF1/R1 primers, with PCR conditions according to Fabre et al. (2011). The resulting amplicons were sequenced by Macrogen Inc. by using the forward primer only and the sequences are deposited in the GenBank database under accession numbers KP337309-23.
The stamp sequences were edited with FinchTV v.1.4.0 and compared with reference stamp strains (Fabre et al. 2011; Johannesen et al. 2012; Aryan et al. 2014; Cvrković et al. 2014; Kostadinovska et al. 2014).
All phylogenetic analyses were conducted under the GTR+I+G nucleotide substitution model as chosen by jModeltest 2.1.7 (Darriba et al. 2012) according to the Akaike information criterion (AIC). Bayesian and maximum parsimony (MP) approaches were applied. The Bayesian analysis was performed in MrBayes 3.1.2 (Huelsenbeck and Ronquist 2001) with the settings as follows: two simultaneous Markov Chain Monte Carlo (MCMC) runs for one million generations, with a sampling frequency of 100 generations and a relative burn-in of 25 %. The convergence of the MCMC chains and their stationarity were checked by using Tracer 1.5 (Rambaut and Drummond 2009). The MP analysis was conducted with PAUP* 4.0b10 (Swofford 2002). One hundred replicates of a heuristic search were performed with an initial random stepwise addition of sequences and tree bisection-reconnection (TBR) branch-swapping. The trees obtained from both analyses were visualized in FigTree 1.4 (Rambaut 2012).
Results
Auchenorrhyncha species in the vineyards of Macedonia
The surveys performed in and around vineyards in 2012 and 2013 led to the collection of 1180 Auchenorrhyncha specimens, which belonged to 29 species from the following six families: Cicadellidae (21), Aphrophoridae (3), Cixiidae (2), Delphacidae (1), Dictyopharidae (1) and Issidae (1) (Table 1). Despite the high diversity of Auchenorrhyncha, only three species were found in numbers greater than 200 over the 2 years survey. The predominant species was the primary BN vector H. obsoletus, followed by leafhoppers Psammotettix alienus, Artianus manderstjernii and Euscelis incisus; the species Dictyophara europaea, Cicadella viridis, Doratura impudica and Anaceratagallia ribauti were collected in numbers <50. The other cixiid vector of BN, R. panzeri, was not recorded in the inspected vineyards, and only four R. quinquecostatus individuals were collected as suspect vectors along the weed-covered borders adjacent to the vineyards. For 18 species, <10 total specimens were identified in all the inspected vineyards during the 2-year survey.
The majority of all H. obsoletus specimens (75 %) were collected from Urtica dioica, and approximately 25 % of individuals were associated with Convolvulus arvensis.
‘Ca. P. solani’ detection in plants and insects
The presence of ‘Ca. P. solani’ was detected in all 12 symptomatic grapevine samples that were subjected to analyses, and in 22 % (15 of 68) and 14 % (7 of 50) of asymptomatic U. dioica and C. arvensis plants, respectively (Table 2).
Because the amount of symptomatic grapevine plants that occurred in the studied vineyards was high (ca. 50 %), we analyzed the specimens of the most abundant leafhoppers and planthoppers species as tentative vectors, as well as R. quinquecostatus, which was the only cixiid species present in the vineyards in addition to H. obsoletus. PCR amplifications with stolbur-specific Stol11 primers indicated that out of the five analyzed insect species in BN-affected vineyards, only H. obsoletus harbored ‘Ca. P. solani’ (Table 1). ‘Ca. P. solani’ were detected in H. obsoletus individuals at a rate of approximately 18 % (Table 2). None of the four analyzed R. quinquecostatus specimens was positive for the ‘Ca. P. solani’ presence.
The molecular typing of the ‘Ca. P. solani’ strains
A molecular differentiation of the’Ca. P. solani’ strains that were infecting grapevines and tentative reservoir plants and were harbored by the insects, was performed by PCR-RFLP and sequence typing for three presumably epidemiologically informative genes, that is, tuf, vmp1 and stamp.
All 91 positive plant and insect samples yielded successful amplifications of the tuf gene with Tuf1f/r and TufAYf/r primers. The HpaII restriction profiles showed the presence of the so-called nettle-associated tuf-a type as defined by Langer and Maixner (2004) in 25 % of infected grapevines (3 of 12), 40 % of ‘Ca. P. solani’-infected nettle plants (6 of 15) and 49 % of ‘Ca. P. solani’-infected H. obsoletus that were collected from nettles (21 of 43). PCR-RFLP analysis revealed the presence of the so-called bindweed-associated tuf-b type in 75 % of ‘Ca. P. solani’-infected grapevines, all the infected bindweeds and H. obsoletus collected on bindweeds. However, 9 out of 15 (60 %) stinging nettles and 45 % of H. obsoletus collected on U. dioica surprisingly exhibited RFLP profiles corresponding to the tuf-b type. In order to confirm these results, the tuf fragments were sequenced for these samples, in addition to the grapevine samples. A sequence comparison determined the presence of the following three tuf types: tuf-a, tuf-b and a third type that was genealogically intermediate between tuf-a and tuf-b and was designated in this study as tuf-ab (Table 2) that clusters within the tuf-a type, which is the nettle-associated lineage tuf-b2 as defined by Aryan et al. (2014). The presence of an intermediate tuf type was revealed in all nettles and the nettle-associated H. obsoletus designated as the tuf-b type according to RFLP typing, in addition to 17 % of grapevine samples (2 of 12; Table 2).
Vmp1 gene amplicons of approximately 1450 bp in length were obtained from all 91 ‘Ca. P. solani’ strains. A restriction digestion with RsaI, TaqI and AluI enzymes allowed us to identify five diverse vmp1 profiles among the phytoplasmas infecting the grapevines, H. obsoletus, nettles and bindweeds called V2-TA, V3, V4, V14 and V18 (Fig. 1). All the detected vmp1 profiles were previously published and designated with these names (Murolo et al. 2010, 2013; Cvrković et al. 2014).
The V18 and V3 profiles were the most common with 36 and 33 % of all the strains assigned to these types, respectively. All V18 vmp1 profiles were associated with an intermediate tuf-ab type, and they were detected in grapevines, U. dioica and its corresponding H. obsoletus populations. The V3 profile was found to be uniquely associated with the tuf-a type in grapevines, stinging nettles and H. obsoletus specimens collected on nettles. The less frequently identified V2-TA, V4 and V14 profiles were detected in grapevines and bindweed-associated H. obsoletus. The V14 type was found only in grapevines, V4 was detected only in H. obsoletus associated with bindweed, and V2-TA was shared by grapevines, bindweeds and bindweed-associated H. obsoletus.
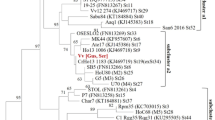
The stamp gene-based phylogeny that employed 91 sequences revealed the highest diversity of ‘Ca. P. solani’ strains and allowed us to identify 11 distinct genotypes with a maximum variability of 4.9 %. Among the identified stamp genotypes, six were identical to previously published reference strains: 19–25, SB5, Rqg50, Vv24, GGY and STOL (Fabre et al. 2011; Cvrković et al. 2014), and five were unique and designated as M1, M2, M3, M4 and M5 (Fig. 2, Table 2). Both Bayesian and MP phylogeny revealed two major phylogenetic groups, each of which was associated with bindweed and nettle (Fig. 2). The first one consisted of the three clusters b-I, b-II and b-III, among which the ‘Ca. P. solani’ strains from Macedonian vineyards clustered within the latter two. The second cluster associated with nettle consisted of two subclusters, which were designated a1 and a2, both of which encompassed the strains detected in this study. Four of the stamp genotypes that were detected for the first time in this study (M1-M4) were clustered within nettle-associated cluster a (three in a2 subcluster and one in a1) and the M5 genotype belonged to cluster b-III (Fig. 2). The majority of the characterized stamp sequences belonged to the newly identified M1 genotype.
The MP cladogram obtained from stamp sequences of ‘Candidatus Phytoplasma solani’ strains as detected in grapevines, weeds and H. obsoletus in the Republic of Macedonia and in reference strains (Fabre et al. 2011; Johannesen et al. 2012; Aryan et al. 2014; Cvrković et al. 2014; Kostadinovska et al. 2014). The strains of each stamp genotype detected in this study are designated in bold letters. Strains from the stamp genotype as previously detected in grapevines from Macedonia according to Kostadinovska et al. (2014) are designated in red. Maximum parsimony bootstrap values/Bayesian posterior probabilities are indicated for each cluster node
Four diverse stamp genotypes were identified in grapevines. A sequence comparison and a maximum parsimony phylogenetic analysis revealed that two of them clustered within the b group and had 100 % sequence similarity with reference strains STOL (cluster b-III) and Vv24 (cluster b-II) from Serbia. The third genotype clustered within the a1 subcluster, and it had sequence characteristics identical to those of reference strains 19–25. The fourth genotype was clustered within the a2 subcluster and has a unique sequence designated here as M3, which is present only in grapevines and H. obsoletus collected on nettles.
Five stamp genotypes were identified among the H. obsoletus specimens collected on nettles. Three of them, namely M1, M2 and M3, have unique sequences in comparison with the reference strains. The other two genotypes are identical to reference strains 19–25 from Germany and SB5 from Croatia, respectively. However, in U. dioica, only two different stamp genotypes were identified, that is, M1 and M4, and they belong to the a1 and a2 subclusters, respectively, and have unique sequences in comparison with the reference strains. The first one was also detected in H. obsoletus associated with nettle, and the second is characteristic only for the nettles that share a 99.6 % sequence similarity with the M3 genotype detected in grapevines and nettles (with a 2 nt difference).
All ‘Ca. P. solani’ strains from bindweed and bindweed-associated H. obsoletus belonged to cluster b-II. The majority of H. obsoletus and all the bindweed strains had sequences that were identical to one that was previously found in tentative insect vectors, H. obsoletus and grapevines in Serbia and Austria (Rqg50; Aryan et al. 2014; Cvrković et al. 2014). Four H. obsoletus strains have an identical sequence to that of H. obsoletus individuals collected from bindweed in Germany and Slovenia (GGY and NGA9; Fabre et al. 2011).
The comprehensive tuf/vmp1/stamp genotypes of ‘Ca. P. solani’ hosted by grapevines, reservoir plants and H. obsoletus
Overall, twelve tuf/vmp1/stamp comprehensive genotypes of ‘Ca. P. solani’ were detected in grapevines, nettles, bindweeds and associated populations of H. obsoletus in the vineyards of southeastern Macedonia.
Four ‘Ca. P. solani’ genotypes were detected among naturally infected grapevines, six among nettle and nettle-associated population of H. obsoletus and four in bindweed and its associated insects. According to the tuf/vmp1/stamp genotyping, the most frequent genotypes were those associated with nettle, i.e., tuf-ab/V18/M1 detected in 60 % of naturally infected nettles and in 44 % of its corresponding H. obsoletus populations, and tuf-a/V3/M3 detected in 25 % of analyzed grapevine and in 19 % of nettle-associated H. obsoletus. The majority of ‘Ca. P. solani’ genotypes were detected among the strains from H. obsoletus that were associated with U. dioica: (i) tuf-ab/V18/M1, (ii) tuf-a/V3/M2, infecting 23 % of analyzed samples, (iii) tuf-a/V3/M3, infecting 18 % of analyzed samples, (iv) tuf-ab/V18/19-25, and (v) tuf-a/V3/SB5, both of which infected approximately 7 % of the analyzed samples (Table 2). All ‘Ca. P. solani’ strains associated with C. arvensis and 35 % of strains associated with its corresponding H. obsoletus populations belonged to the tuf-b/V2-TA/Rqg50 genotype. However, tuf-b strains found in naturally infected grapevine were associated with V14/Vv24 and V2-TA/STOL vmp1/stamp types.
Discussion
Bois noir infections of diverse grapevine varieties have been recorded in the vineyards of the Republic of Macedonia (Šeruga et al. 2003), and a molecular characterization of ‘Ca. P. solani’ infecting BN-affected grapevines have recently been documented (Kostadinovska et al. 2014).
An increased incidence of BN throughout Macedonian vineyards initiated a study on genetic diversity of ‘Ca. P. solani’ in symptomatic grapevine samples, potential insect vectors and herbaceous host plants as the primary reservoirs of infection, in order to clarify the epidemiology of the BN. During the two-year survey, we detected the presence of ‘Ca. P. solani’ in all analyzed grapevine samples, the major ‘Ca. P. solani’ vector H. obsoletus and the two principal ‘Ca. P. solani’ reservoirs, bindweed and nettle. A qualitative analysis of the planthoppers and leafhoppers captured in the vineyards of southeastern Macedonia determined a high diversity with 29 species recorded. Of all the analyzed species, only H. obsoletus tested positive for the presence of ‘Ca. P. solani’ despite the high diversity of Auchenorrhyncha species.
The incidence and dispersal of vector-borne plant pathogens depends upon the abundance of the vector(s), their interplant movement and a high infection rate (Power 1992; Orenstein et al. 2003; Trivellone et al. 2005). The infection rate of H. obsoletus specimens collected from U. dioica and C. arvensis was approximately 20 %, and the abundance of their populations was high in southeastern Macedonia. Additionally, the other cixiid species R. panzeri, a documented vector of BN, was not present in the studied vineyards, and the potential vector R. quinquecostatus was present in a negligible number. This finding clearly indicates that H. obsoletus plays a major role in the BN epidemiology of the vineyards we studied, and diminishes the hypothesis made by Kostadinovska et al. (2014) that R. panzeri could be a vector of ‘Ca. P. solani’ in Macedonian vineyards. However, the G2 genotype (tuf-b/V2-TA/STOL) was detected in 33 % of the diseased grapevines but in none of the H. obsoletus found infected. A similar incidence (42 %) was observed in northeastern Serbia where this strain proved to be the only strain transmitted by R. panzeri (Cvrković et al. 2014).
A molecular characterization of the ‘Ca. P. solani’ strains indicates the presence of two epidemiological BN cycles in Macedonian vineyards, with one sourced by C. arvensis and the other by U. dioica as weedy phytoplasma reservoirs. Overall, 12 genotypes were detected according to the tuf/vmp1/stamp typing. Four genotypes were found to be associated with grapevines, with equally distributed tuf-a and tuf-b types. According to the comprehensive tuf/vmp1/stamp typing, one of the grapevine-associated BN genotypes that occurred in Macedonian vineyards corresponded to the most prevalent genotype in Serbian grapevines and planthoppers (STOLg), and another was identical to the Vv24g associated with BN-affected grapevines in Serbia (Cvrković et al. 2014). Regarding the tuf-a and intermediate tuf-ab ‘Ca. P. solani’ types, they were found in grapevines, nettles and nettle-associated H. obsoletus. One of them corresponded to the tuf-ab/V18/19-25 that was previously found in grapevines from Macedonia (Kostadinovska et al. 2014) and in grapevines, H. obsoletus and nettles from Austrian vineyards (genotype CPsM4_At1) where it was found to be a major genotype that induced the current BN epidemics (Aryan et al. 2014). The second is a new stamp genotype, known as comprehensive tuf-a/V3/M3, which was found to be restricted to grapevines and H. obsoletus associated with nettles. Two new genotypes were associated with nettles, called tuf-ab/V18/M1 and tuf-a/V3/M4; the former was also associated with the corresponding population of H. obsoletus, and the latter restricted solely to nettle. The high genotype diversity of ‘Ca. P. solani’ was detected in H. obsoletus collected from both bindweeds and stinging nettles. In addition to the above mentioned genotypes, five genotypes were associated exclusively with H. obsoletus. Two genotypes were found in nettle-associated H. obsoletus, with tuf-a/V3/SB5 as previously reported in Croatia (Fabre et al. 2011) and a new genotype called tuf-a/V3/M2. The following three genotypes were detected in bindweed-associated H. obsoletus: tuf-b/V4/GGY, tuf-b/V4/M5 and tuf-b/V14/Rqg50. The third genotype corresponds to the Rqg50g identified in Serbian vineyards and to the CPsM4_At12 genotype that occurs in Austrian vineyards. Only one genotype was associated with bindweed, called tuf-b/V2-TA/Rqg50, which was previously reported in grapevines from Montenegro (A. Kosovac, personal communication). Profile V3 was only found in association with type tuf-a, which is consistent with previous evidence (Foissac et al. 2013), and profile V18 was always found in association with the intermediate type tuf-ab.
H. obsoletus adults were found to feed on diverse plant species, but for nymphal development, during which the insects acquire phytoplasmas, only a few are preferred (Langer and Maixner 2004). Convolvulus arvensis is generally considered as both the primary host plant and ‘Ca. P. solani’ reservoir, but within the last decade, U. dioica has become an equally preferred host plant (Johannesen et al. 2012). Until recently, stinging nettle was considered a primary host only in Italy (Lessio et al. 2007). However, the latest studies emphasize that tuf-a type associated with stinging nettles is the most common ‘Ca. P. solani’ strain and U. dioica is the dominant host plant in Germany, northeastern France, Switzerland and Austria, where it is predominantly responsible for the mass occurrence of H. obsoletus and severe BN outbreaks (reviewed in Johannesen and Riedle‐Bauer 2014).
Our research indicates that BN epidemiological cycles in the Republic of Macedonia are correlated with both bindweed and stinging nettle as reservoir plants, with the prevalence of tuf type a and H. obsoletus associated with Urtica dioica, which is the preferred host plant for H. obsoletus in this country. The high incidence of nettle-associated tuf-a and tuf-ab types might be related to the agricultural praxis in southeastern Macedonia, which includes intensive irrigation during hot and dry summers and provides suitable habitats and environmental conditions for the growth of stinging nettles in vineyard surroundings. Because stinging nettles do not express the symptoms of phytoplasma infection and the plants were randomly selected for ‘Ca. P. solani’ identification, only two different genotypes were identified in infected reservoir plants. However, a high diversity of stamp genotypes was detected in ‘Ca. P. solani’ strains from H. obsoletus collected on U. dioica, in addition to strains from H. obsoletus associated with C. arvensis.
The increased BN incidence since the first report in 2003 in the Republic of Macedonia (Šeruga et al. 2003) and the high abundance of H. obsoletus on stinging nettles suggest the possible occurrence of sudden outbreaks and the existence of two host races for the vector, with one specialized in stinging nettle and one in bindweed. Further research should be focused on the host-race diversification of H. obsoletus among Macedonian host-plant populations and the design of adequate management strategies.
References
Angelini, E., Clair, D., Borgo, M., Bertaccini, A., & Boudon-Padieu, E. (2001). Flavescence dorée in France and Italy - occurence of closely related phytoplasma isolates and their near relationships to Palatinate grapevine yellows and an alder yellows phytoplasma. Vitis, 40, 79–86.
Aryan, A., Brader, G., Mörtel, J., Pastar, M., & Riedle-Bauer, M. (2014). An abundant ‘Candidatus Phytoplasma solani’ tuf b strain is associated with grapevine, stinging nettle and Hyalesthes obsoletus. European Journal of Plant Pathology, 140, 213–227.
Biedermann, R., & Niedringhaus, R. (2004). Die Zikaden Deutschlands - Bestimmungstafeln für alle Arten. Scheessel: WABV.
Cimerman, A., Pacifico, D., Salar, P., Marzachì, C., & Foissac, X. (2009). Striking diversity of vmp1, a variable gene encoding a putative membrane protein of the stolbur phytoplasma. Applied and Environmental Microbiology, 75, 2951–2957.
Clair, D., Larrue, J., Aubert, G., Gillet, J., Cloquemin, G., & Boudon-Padieu, E. (2003). A multiplex nested-PCR assay for sensitive and simultaneous detection and direct identification of phytoplasma in the Elm yellows group and Stolbur group and its use in survey of grapevine yellows in France. Vitis, 42, 151–157.
Cvrković, T., Jović, J., Mitrović, M., Krstić, O., & Toševski, I. (2014). Experimental and molecular evidence of Reptalus panzeri as a natural vector of bois noir. Plant Pathology, 63, 42–53.
Darriba, D., Taboada, G. L., Doallo, R., & Posada, D. (2012). jModelTest 2: more models, new heuristics and parallel computing. Nature Methods, 9, 772.
Fabre, A., Danet, J. L., & Foissac, X. (2011). The stolbur phytoplasma antigenic membrane protein gene stamp is submitted to diversifying positive selection. Gene, 472, 37–41.
Fialová, R., Válová, P., Balakishiyeva, G., Danet, J. L., Šafárová, D., Foissac, X., & Navrátil, M. (2009). Genetic variability of stolbur phytoplasma in annual crop and wild plant species in south Moravia. Journal of Plant Pathology, 91, 411–416.
Foissac, X., Carle, P., Fabre, A., Salar, P., Danet, J. L. & STOLBUR-EUROMED consortium. (2013). ‘Candidatus Phytoplasma solani’ genome project and genetic diversity in the Euro-Mediterranean basin. Invited conference. In Third European Bois Noir Workshop (pp. 11–13). E. Torres, A. Lavina, A. Batlle (Eds.). Barcelona.
Gatineau, F., Larrue, J., Clair, D., Lorton, F., Richard-Molard, M., & Boudon-Padieu, E. (2001). A new natural planthopper vector of stolbur phytoplasma in the genus Pentastiridius (Hemiptera: Cixiidae). European Journal of Plant Pathology, 107, 263–271.
Gatineau, F., Jacob, N., Vautrin, S., Larrue, J., Lherminier, J., Richard-Molard, M., & Boudon-Padieu, E. (2002). Association with the syndrome “Basses Richesses” of sugar beet of a phytoplasma and a bacterium-like organism transmitted by a Pentastiridius sp. Phytopathology, 92, 384–392.
Holzinger, W. E., Kammerlander, I., & Nickel, H. (2003). The Auchenorrhyncha of Central Europe. Fulgoromorpha, Cicadomorpha Excl. Cicadellidae (p. 673). Leiden: Brill Academic Publishers.
Huelsenbeck, J. P., & Ronquist, F. (2001). MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics, 17, 754–755.
Johannesen, J., & Riedle‐Bauer, M. (2014). Origin of a sudden mass occurrence of the stolbur phytoplasma vector Hyalesthes obsoletus (Cixiidae) in Austria. Annals of Applied Biology, 165, 488–495.
Johannesen, J., Foissac, X., Kehrli, P., & Maixner, M. (2012). Impact of vector dispersal and host-plant fidelity on the dissemination of an emerging plant pathogen. PLoS ONE, 7, e51809.
Jović, J., Cvrković, T., Mitrović, M., Krnjanjić, S., Petrović, A., Redinbaugh, M. G., Pratt, R. C., Hogenhout, S. A., & Toševski, I. (2009). Stolbur phytoplasma transmission to maize by Reptalus panzeri and the disease cycle of maize redness in Serbia. Phytopathology, 99, 1053–1061.
Jović, J., Ember, I., Mitrović, M., Cvrković, T., Krstić, O., Krnjajić, S., Acs, Z., Kolber, M., & Toševski, I. (2011). Molecular detection of potato stolbur phytoplasma in Serbia. Bulletin of Insectology, 64, 83–84.
Kostadinovska, E., Quaglino, F., Mitrev, S., Casati, P., Bulgari, D., & Bianco, P. A. (2014). Multiple gene analyses identify distinct “bois noir” phytoplasma genotypes in the Republic of Macedonia. Phytopathologia Mediterranea, 53, 300–310.
Langer, M., & Maixner, M. (2004). Molecular characterisation of grapevine yellows associated phytoplasmas of the stolbur-group based on RFLP-analysis of non-ribosomal DNA. Vitis, 43, 191–199.
Lessio, F., Tedeschi, R., & Alma, A. (2007). Population dynamics, host plants and infection rate with Stolbur phytoplasma of Hyalesthes obsoletus Signoret in north-western Italy. Journal of Plant Pathology, 89, 97–102.
Maixner, M. (1994). Transmission of German grapevine yellows (Vergilbungskrankheit) by the planthopper Hyalesthes obsoletus (Auchenorrhyncha: Cixiidae). Vitis, 33, 103–104.
Murolo, S., Marcone, C., Prota, V., Garau, R., Foissac, X., & Romanazzi, G. (2010). Genetic variability of the stolbur phytoplasma vmp1 gene in grapevines, bindweeds and vegetables. Journal of Applied Microbiology, 109, 2049–2059.
Murolo, S., Marcone, C., Prota, V., Garau, R., Foissac, X., & Romanazzi, G. (2013). Genetic variability of the stolbur phytoplasma vmp1 gene in grapevines, bindweeds and vegetables. Corrigendum. Journal of Applied Microbiology, 115, 631–633.
Orenstein, S., Zahavi, T., Nestel, D., Sharon, R., Barkalifa, M., & Weintraub, P. (2003). Spatial dispersion patterns of potential leafhopper and planthopper (Homoptera) vectors of phytoplasma in wine vineyards. Annals of Applied Biology, 142, 341–348.
Pacifico, D., Alma, A., Bagnoli, B., Foissac, X., Pasquini, G., Tessitori, M., & Marzachi, C. (2009). Characterization of Bois noir Isolates by restriction fragment length polymorphism of a Stolbur-specific putative membrane protein gene. Phytopathology, 99, 711–715.
Pinzauti, F., Trivellone, V., & Bagnoli, B. (2008). Ability of Reptalus quinquecostatus (Hemiptera: Cixiidae) to inoculate stolbur phytoplasma to artificial feeding medium. Annals of Applied Biology, 153, 299–305.
Power, A. G. (1992). Host plant dispersion, leafhopper movement and disease transmission. Ecological Entomology, 17, 63–68.
Quaglino, F., Zhao, Y., Casati, P., Bulgari, D., Bianco, P. A., Wei, W., & Davis, R. E. (2013). ‘Candidatus Phytoplasma solani’, a novel taxon associated with stolbur- and bois noir-related diseases of plants. International Journal of Systematic and Evolutionary Microbiology, 63, 2879–2894.
Radonjić, S., Hrnčić, S., Jović, J., Cvrković, T., Krstić, O., Krnjajić, S., & Toševski, I. (2009). Occurrence and distribution of grapevine yellows caused by stolbur phytoplasma in Montenegro. Journal of Phytopathology, 157, 682–685.
Rambaut, A. (2012). FigTree. URL http://www.tree.bio.ed.ac.uk/software/figtree.
Rambaut, A., & Drummond, A. J. (2009). Tracer v1.5, URL http://beast.bio.ed.ac.uk/Tracer.
Riedle-Bauer, M., Sára, A., & Regner, F. (2008). Transmission of a stolbur phytoplasma by the agalliinae leafhopper Anaceratagallia ribauti (Hemiptera, Auchenorrhyncha, Cicadellidae). Journal of Phytopathology, 156, 687–690.
Šeruga, M., Škorić, D., Kozina, B., Mitrev, S., Krajačić, M., & Ćurković Perica, M. (2003). Molecular identification of a phytoplasma infecting grapevine in the Republic of Macedonia. Vitis, 42, 181–184.
Swofford, D. L. (2002). Paup*. Phylogenetic Analysis Using Parsimony (*and Other Methods). Sunderland: Sinauer Associates.
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., & Kumar, S. (2011). MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance and maximum parsimony methods. Molecular Biology and Evolution, 28, 2731–2739.
Trivellone, V., Pinzauti, F., & Bagnoli, B. (2005). Reptalus quinquecostatus (Dufour) (Auchenorrhyncha Cixiidae) as a possible vector of Stolbur-phytoplasma in a vineyard in Tuscany. Redia, 88, 103–108.
Weintraub, P. G., & Beanland, L. (2006). Insect vectors of phytoplasmas. Annual Review of Entomology, 51, 91–111.
Acknowledgments
The authors are grateful to Xavier Foissac (INRA, Bordeaux-France) and Michael Maixner (JKI, Siebeldingen-Germany) for providing ‘Ca. P. solani’ reference strains. This study was funded by Ministry of Education, Science and Technological Development of the Republic of Serbia (grant no. III43001) and partly by the SCOPES program of the Swiss National Science Foundation (IZ73Z0_152414).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Atanasova, B., Jakovljević, M., Spasov, D. et al. The molecular epidemiology of bois noir grapevine yellows caused by ‘Candidatus Phytoplasma solani’ in the Republic of Macedonia. Eur J Plant Pathol 142, 759–770 (2015). https://doi.org/10.1007/s10658-015-0649-0
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10658-015-0649-0