Abstract
The genetic structure of ten grape phylloxera populations, sampled in summer of 2006 and 2007 from four distinct viticultural areas in Austria, Daktulosphaira vitifoliae (Fitch) (Homoptera: Phylloxeridae) was analyzed using six SSR markers (Dvit1-Dvit6). Leaf-feeding populations were chosen from similar ecological habitats, where susceptible rootstock hosts have overtaken scions in abandoned vineyards and produce grape phylloxera populations. To study population structures and test for dominating genotypes, population genetic measures were performed. The genetic diversity detected within the entire set of 315 genotypes was high, with 223 distinct multilocus genotypes (MLGs). Excess of heterozygotes and significant deviations from Hardy-Weinberg equilibrium in some populations indicated that the major reproduction mode in these populations is asexual but, sexual reproduction also confirmed by the sign. P sex values. The genetic diversity within populations was higher than between populations, although only three overlapping genotypes could be found. MLGs were rare, indicating that no candidate for superclones were detected in the leaf-feeding populations studied.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Grape phylloxera Daktulosphaira vitifoliae (Fitch) is an important, highly destructive insect pest on susceptible or partially resistant grape varieties where it may feed on roots and leaves (Forneck and Huber 2009). The aphid is native to North America, was introduced to Europe about 1855 and into Austria in 1867 and led to a widespread destruction of vineyards of all European grapevine varieties (Vitis vinifera L.). Galls (nodosities) are induced in the meristematic zone of the root tip and deformed root tissue (tuberosity) is formed on lignified root parts. In association with soil borne saprophytes and other abiotic stresses, nodosities and tuberosities lead to severe yield losses and eventual death of the plant (Powell et al. 2013). Modern viticulture relies on grafting V. vinifera varieties on partially resistant rootstocks mainly bred from V. berlandieri Planch. V. riparia Michx. V. rupestris Scheele which allow limited reproduction of phylloxera on roots. However reports indicate the appearance of aggressive biotypes that overcome the partial resistance if environmental stresses affect the grapevine host (Powell et al. 2013). Phylloxera feed on commercial vineyards on partially resistant rootstocks (mainly root-feeding). Populations can also build in abandoned vineyards in which rootstocks overtake the V. vinifera scions (root- and leaf feeding) supporting leaf-feeding populations (e.g., Kocsis and Bari 2013). These perennial habitats foster large “long-living” populations to develop on roots and leaves. From there phylloxera lineages and populations have been observed to infest nearby commercial vineyards (Müller 2010).
European gallicole phylloxera populations show high genotypic variability (Forneck et al. 2000, 2001; Vorwerk and Forneck 2007) however parthenogenesis has been proposed to be the major reproductive mode of these phylloxera (Vorwerk and Forneck 2006). Recent studies of radicole (root-feeding) phylloxera populations in California indicate that both asexual and sexual reproduction occurs (Lund 2013). A population can be temporarily and spatially studied with microsatellite markers (Corrie et al. 2002; Vorwerk and Forneck 2006; Islam et al. 2013; Riaz et al. 2014). These markers also allow the assessment of the genetic structure of populations and interpretation of biological processes such as reproduction mode, migration of clones and abundance of common clonal lineages (Sunnucks et al. 1997; Downie et al. 2000; Forneck et al. 2001). Such studies are needed for a better understanding of the genetic diversity and fine scale population structure of phylloxera, so that the evolutionary potential to adapt to resistant V. vinifera (leaves) and resistant rootstock (roots) can be elucidated.
High abundance aphid clones, so called superclones, have been shown to occur spatially and temporarily and selection must force a population to limit its genetic potential and count on single genotypes. Such genotypes are thought to be “general-purpose genotypes” (Lynch 1984) which show “broad ecological tolerance resulting from interclonal selection in temporally variable environments” (Vorburger et al. 2003). The reasons for their population success are still unknown, neither host adaptation (Fenton et al. 1998), nor insecticide resistance or agronomical practices (Vorburger et al. 2003) could yet explain their dominance. Two radicole Australian phylloxera genotypes G1 and G4 have been termed superclones (Powell et al. 2013) exhibiting broader geographical distribution and higher virulence levels (Herbert et al. 2006).
In the presented study of the fine scale genetic population structure was studied searching for the occurrence of phylloxera dominant genotypes in leaf galling habitats to provide data to further the understanding of the population structure of clonal insect pests.
The objectives of this study were: (1) to analyze the reproduction mode of ten leaf feeding phylloxera populations, (2) to understand the genetic structure within and among populations in different Austrian wine regions, and (3) to search for dominating distinct multilocus genotypes (MGLs) within the population.
Material and methods
Phylloxera populations
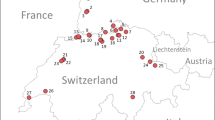
Leaves with third-generation leaf galling D. vitifoliae were collected in 2007 and 2008 from abandoned vineyards and wild growing rootstocks climbing on native hedges at ten locations throughout major Austrian viticulture regions (AU1, AU2: Eisenstadt, AU3: Krems, AU4: Wien-Hackenberg, AU5: Gols, AU6: Bernhardtal, AU7: Watzelsdorf, AU8: Großriedental, AU9: Klöch, AU10: Rechnitz) (Fig. 1). Sampling was conducted in a hierarchical manner: three leaf-galled shoots (1–2 m long) were randomly chosen - one from the center and two from the periphery of each location. Four to five leaves were randomly chosen from each shoot and two to three single leaf-feeding adults were obtained by dissecting the galls. Single individual adults were used for genotyping.
Sampling sites of Phylloxera leaf populations for performance and genotyping studies. Sampling was conducted in spring of 2007 and 2008 in Weinviertel (Watzelsdorf. Bernhardthal). Wachau (Krems). Wagram (Großriedenthal). Wien (Wien Hackenberg). Neusiedlersee (Gols). Neusiedlersee Hügelland (Eisenstadt. Großhöflein). Südburgenland (Rechnitz). Südoststeiermark (Klöch). Performance studies were conducted with populations from Bernhardthal. Gols. Krems. Wien and adapted single founder lineages established from Eisenstadt and Eisenstadt Großhöflein. Underlined locations represent field populations used in this study
Genotyping of phylloxera individuals obtained in populations
Single adults obtained from leaf galls were used for DNA extraction and genotyping with six SSR markers. Sampling of leaf-galled shoots was performed hierarchically as previously described (Vorwerk and Forneck 2006). Total genomic DNA was extracted from a single adult using a column-based DNA extraction kit (QIAGEN, Hilden, Germany) as described earlier (Vorwerk and Forneck 2006). Subsequent PCR reactions were performed in 10 μl reaction volumes containing: 800 nM of each primer, 200 μM dNTPs each (Fermentas, St. Leon Rot, Germany), 1× PCR Buffer 1,5 mM MgCl2, 0,25U Taq DNA Polymerase (Invitrogen, Karlsruhe, Germany) and approximately 5 ng of template DNA. PCR primers for amplification of the loci Dvit1, Dvit2, Dvit3, Dvit4 and Dvit5 (Corrie et al. 2002), and Dvit6 (Vorwerk and Forneck 2006) were applied. Amplified alleles were separated in a 6 % polyacrylamid gel, and visualized with an automated LI-COR 4300 DNA analyzer (Licor Biosciences GmbH, Bad Homburg, Germany).
Data analysis
Population genetic analysis
Genetic data for all six loci were combined and genotypes assigned to each sample. Genotypes bearing missing data were eliminated from calculations. To prevent distortions of estimates for heterozygosity and F-statistics a single copy of each multilocus genotype was applied in the data analysis for F-statistics, estimates for heterozygosity, Hardy-Weinberg (HW) exact tests and linkage disequilibrium according to Sunnucks et al. (1997). Allele frequencies, mean number of alleles per locus, observed heterozygosity (Hobs) and unbiased estimates of heterozygosity expected under Hardy-Weinberg assumptions (HE) (Nei 1978) were processed using Genepop v. 3.4 implemented on the web (http://wbiomed.curtin.edu.au/genepop/; Raymond and Rousset 1995). This program was also employed to test data sets for Hardy-Weinberg equilibrium (HWE) and linkage disequilibrium (LD). Exact P-values for these tests were calculated using the Markov chain method (Guo and Thompson 1992). Fstat 2.9.3 (Goudet 1995) was employed, carrying out F-statistics for all population pairs and confidence intervals based on bootstrap re-sampling were estimated according to Weir and Cockerham (1984). Clonal diversity within populations was calculated for each population as k = G/N, where G is the number of different multilocus genotypes present in the sample and N is the sample size. P sex values were calculated with the program MLGsim2.0 (http://www.rug.nl/research/theoretical-biology/downloads) for every multicopy genotype in each population as suggested by Halkett et al. (2005). Thresholds for P sex values were estimated for each population from Monte Carlo simulations. Significant P sex values indicate that multicopy genotypes are statistically overrepresented in a population and therefore a product of clonal amplification. Population pairwise F ST values were calculated using Fstat 2.9.3 (Goudet 1995). F ST measures were transformed into F ST / (1- F ST) and a permuted correlation was computed against the natural logarithm (Ln) of distances between the sampling sites in kilometers, using the program Isolde (Genepop v. 3.4, http://wbiomed.curtin.edu.au/genepop/; Raymond and Rousset, 1995). The observed Spearman rank - correlation coefficient was compared under the null hypotheses that the two variables are independent. The distribution of alleles and genotypes across all populations was tested using Genepop v. 3.4 unbiased tests of population pairwise differentiation (Fishers exact test).
Results
Genotypic diversity and population assignment
Three hundred and fifteen grape phylloxera samples from 10 fine scale populations obtained from leaf galling habitats were analyzed from four viticultural regions. In total 226 distinct genotypes were identified within all populations tested based on the combination of allelic data from six SSR markers. The average number of alleles per locus in the populations ranged from 2 to 3. No significant null alleles (>0.005 frequency) were found in the SSR data (data not shown). Across all loci and populations the expected heterozygosity (He) was lower than the observed heterozygosity (Hobs) which ranged from 0.4484 (AU6) to 0.7012 (AU8) within the populations. Test of HW Equilibrium showed the presence of excess of heterozygosity (F IS < 0) in several loci in each population (Table 1).
Multilocus repeated genotypes were observed in each population (Table 1) indicating reproduction diversity (G:N ratio) ranging from 0.333 to 0.9667. HW exact probability tests showed significant deviations in four out of ten populations (Table 1) and mostly negative F IS values (in nine out of ten populations) indicating higher portions of heterozygotes and suggesting primarily asexual reproduction. There were frequent deviations from HWE in the direction of heterozygote excess, a common property of microsatellite data sets from obligate parthenogens proposed to diversify by mutation (Simon et al. 1999). Locus-wise comparisons showed high variation within the populations (data not shown) where the distribution of both positive and negative F IS were observed at various loci. The probability of independently produced repeated genotype by sexual reproduction was determined by numbers of significances of P sex values in MLG simulations (Table 2). The numbers of clonal and sexually produced MLGs suggest both sexual and asexually produced genotypes in all populations studied.
Three overlapping genotypes could be found throughout the entire set of genotypic classes in three of the ten population (AU7, AU8 and AU10).
F ST values showed differentiation in differing levels among the phylloxera habitats and regions studied. The indices ranged from 0.0150 to 0.3817 signifying different levels of shared genetic diversity (a values of 0 implies panmixis, 1 implies two populations do not share any genetic diversity). The population pairwise F ST comparision showed that some do and some do not differ significantly in allele frequencies (Table 2).
Upon closer inspection of allele frequencies no association among geographic location or other potential environmenal parameters responsible for divergent selection in populations were found (data not shown), furthermore there was more genetic variation within the populations than between populations.
Discussion
As an introduced pest phylloxera has invaded most viticultural regions worldwide from its native range on wild Vitis species in North America. In its native habitat phylloxera mostly feeds on leaves and reproduces through cyclical parthenogenesis (Lund 2013). Invasion of phylloxera occurred through human mediated spread of infested plant materials and occurred from multiple origins (Downie 2002) into European vineyards. Within the last decades the insect has invaded a new niche to feed on susceptible leaves of rootstocks (from abandoned vineyards where rootstock shoots overtake scions). Here phylloxera finds similar conditions as in the native habitat, being able to feed and reproduce both on leaves and roots. Previous studies of leaf-feeding populations in Europe showed high genotypic diversity (Forneck et al. 2000, 2001; Vorwerk and Forneck 2006, 2007) although reproduction was primarily asexual and clonal. Invasive insects tend to transition to obligate or functional parthenogenesis (Caron et al. 2014). An invasion that produces clones may be characterized by a number of fixed clones or a complex reproduction scheme of cyclical parthenogenesis and recurrent generation of new clones (Caron et al. 2014). Natural selection will act quickly on large clonal populations in which pre-adapted genotypes will be favored and new phenotypes may evolve through mutation and develop to superclones. Vineyards provide an excellent habitat for the invasion and evolution of root-feeding superclones, especially in newly planted vineyards with identical rootstocks and optimal root growing conditions. Two radicole genotypes, G1 and G4, have been singled out as ‘superclones’ (Umina et al. 2007) in Australia with broader geographic distribution and high virulence levels. The potential of leaf-feeding niches to host phylloxera superclone candidate was studied because of the important implications for genotypes and phenotype diversity in phylloxera and their ability to adapt to environments and consequently for control strategies.
Limited gene flow among populations
This present study showed very limited, extremely low gene flow among populations within a 200 km range and confirmed an earlier study of leaf-feeding populations over a larger geographical distribution (Vorwerk and Forneck 2006). Continuous gene flow would prevent populations from diverging. As a result of isolation and the subsequent limitation of gene exchange, populations will diverge by genetic drift or by selection or adaptation to its local niche. Gene flow supports introduction of new genes into new niches (vineyards) distant form the original site of mutation to introduce new genes, which displace old less adapted ones. In clonal populations gene flow may occur in which each clone has several mutations that differentiate from pre-existing clones. Taking into account that multiple genes may flow together in clones, a genotypic flow (of clonal lineages) may occur between populations. Leaf-feeding habitats studied are isolated by various factors and thus promote genetic diversification and evolution of new clonal lineages as shown in the previous studies. The migration of phylloxera depends on the phylloxera phenotype, host plants available and environmental conditions (Powell et al. 2013). Phylloxera migrates through wind dispersal as first instars or winged sexuapare over several kms and requires host plants (Vitis spp.) to bridge migration of active insects. Dispersal of phylloxera may occur through human transportation over broader geographic ranges.
Our results show, that genotypic flow of dominant MLGs do rarely occur isolating the habitat and evolving high level diversity clonal populations. If superclones could evolve in this habitats it may be more likely for them to invade adjacent vineyards where they may feed on partially resistant leaves and initiate new populations. These incidents have been observed in recent years (Müller 2010; Koennecke et al. 2011; Vidart et al. 2013) however genotypic studies are still underway.
Reproduction mode of leaf-feeding populations
Our results confirm earlier results showing that the predominat reproduction mode of introduced phylloxera populations is predominately asexual (Vorwerk and Forneck 2006, 2007; Islam et al. 2013; Sun et al. 2009; Lund 2013) according to genetic parameters suggested by Halkett et al. (2005). Because the genetic diversity within populations is remarkably high for asexually reproducing phylloxera previous work assumed that either rare sexual events occur(ed) before or after the introduction of phylloxera in the 1850s. Recent work (Lund 2013) provided the first evidence that phylloxera leaf feeding populations from American native habitats are mainly sexually reproducing. It may be likely that a high proportion of allelic diversity was introduced by invasive single founders from multiple origins over a large time period since 1850s and have evolved clonally since then. However, sexual reproduction cannot be excluded. Sexual morphs exist and can be frequently observed migrating in root-feeding populations. All leaf-feeding populations studied also had minor feeding on the feeder roots where first instars overwinter as hibernating morphs.
Why not superclones?
We provided evidence that within 10 Austrian and six European leaf-feeding populations no dominating MLGs were found to be suitable candidate for superclones. Given the genetic structure and ecological habitats of these populations the chances of finding them are in retrospect rather unlikely. Superclones of phylloxera in Europe would have first evolved originally shortly after invading their new habitat of the 19th Century habitat, own-rooted cultivars of V. vinifera ssp. The successful phenotype would have been adapted to root-feeding, a new Vitis species and parthenogenesis. Assuming that such an adaptation had occurred and superclones arose, they would have spread and eventually destroyed their habitat. By the gradual introduction of partially resistant rootstocks, new genes were required promoting further evolution of clonal lineages, leading to more aggressive strains capable of out-competing the first invaders.
Vinifera adapted clonal lineages (e.g., G1, G4; biotype A, B) with vinifera parentage are rarely found in Europe where the majority of viticulture uses resistant rootstocks. Such cases could be found in both root- and leaf-feeding habitats, which have undergone massive selection pressure for new invading phylloxera. Vineyards where phylloxera have adapted on partially resistant rootstocks, as preported previously, where they have invaded un-infested vineyards, or when new rootstocks have been planted in phylloxerated soils. Invasion of leaf-feeding commercial vineyards may select for superclone candidates which would have to adapt to the new host species. High variability in host adaptation between and within populations has been demonstrated in earlier studies (Corrie et al. 2002; Granett and Kocsis 2000; Granett et al. 2001; Forneck et al. 2000; Ritter et al. 2007) in American, European and Australian regions. Studies are underway to elucidate where habitats promote phylloxera superclones.
Population genetic parameters in phylloxera populations
We studied the population genetic parameters suggested by Halkett et al. (2005) and Arnaud‐Haond et al. (2007) and followed the suggestions made by the authors. We are aware of potential errors arising by interpreting the data by neglecting the fact that phylloxera populations are non host alternating, the aphid is sessile and the natural migration distance is very low; parameters indicating clonality may be underestimated.
Isolation of populations will be the rule due to the biology of the insect and the host habitat structure in Europe. Sampling of phylloxera leaf-feeding populations may lead to additional errors, since insects will hatch and form clusters in closest meristematic leaves, which growth is affected by environmental conditions and canopy management (in the case of commercial vineyards).
Conclusion
The ten leaf-feeding phylloxera populations studied in Austrian viticultural regions did not have superclone candidates. Introduced phylloxera do have the potential to form such superclones since they reproduce asexually, have extremely reduced genotypic flow, consist of many diverge clonal lineages and have very high population sizes. The populations are bridged between seasons by few overwintering asexual morphs that represent the successful host adapted clonal lineages that found or sustain the existing fittest clonal lineage. Such superclones will challenge viticulture and vineyard management strategies that promote the evolution of phylloxera superclones need to be carefully observed.
References
Arnaud‐Haond, et al. (2007). Standardizing methods to address clonality in population studies. Molecular Ecology, 16, 5115–5139.
Caron, V., Ede, F.J., Sunnucks, P. (2014) Unravelling the paradox of loss of genetic variation during invasion: Superclones may explain the success of a clonal invader. PLoS ONE, 9(6), art. no. e97744.
Corrie, A. M., Crozier, R. H., van Heswijck, R., & Hoffman, A. A. (2002). Clonal reproduction and population genetic structure of grape phylloxera Daktulospaira vitifoliae. in Australia. Heredity, 88, 203–211.
Downie, D. A. (2002). Locating the sources of an invasive pest, grape phylloxera, using a mitochondrial DNA gene genealogy. Molecular Ecology, 11(10), 2013–2026.
Downie, D. A., Grannett, J., & Fisher, J. R. (2000). Distribution and abundance of leaf galling and foliar sexual morphs of grape phylloxera and Vitis species in the central and eastern United States. Environmental Entomology, 29, 979–986.
Fenton, B., Woodford, J. A. T., & Malloch, G. (1998). Analysis of clonal diversity of the peach-potato Myzus persicae (Sulzer) in Scotland. UK and evidence for the existence of a predominant clone. Molecular Ecology, 7, 1475–1487.
Forneck, A., & Huber, L. (2009). (A)sexual reproduction – a review of life cycles of grape phylloxera. Daktulosphaira vitifoliae. Entomologia Experimentalis et Applicata, 131, 1–10.
Forneck, A., Walker, M. A., & Blaich, R. (2000). Genetic structure of an introducted pest. Grape phylloxera (Daktulopshaira vitifoliae Fitch) in Europe. Genome, 41, 669–678.
Forneck, A., Walker, M. A., & Blaich, R. (2001). Ecological and genetics aspects of Grape Phylloxera’s (Daktulosphaira vitifoliae Fitch) performance on rootstock hosts. Bulletin of Entomological Research, 91, 445–451.
Goudet, J. (1995). FSTAT Version 1.2: a computer program to calculate F-statistics. The Journal of Heredity, 86, 485–486.
Granett, J., & Kocsis, L. (2000). Populations of grape phylloxera gallicoles on rootstock foliage in Hungary. Vitis, 39, 37–41.
Granett, J., Walker, A. M., Kocsis, L., & Omer, A. D. (2001). Biology and management of grape phylloxera. Annual Review of Entomology, 46, 387–412.
Guo, S. W., & Thompson, E. A. (1992). Performing the exact test of Hardy-Weinberg proportion for multiple alleles. Biometrics, 48, 361–372.
Halkett, F., Plantegenest, M., Prunier-Leterme, N., Mieuzet, L., Delmotte, F., & Simon, J. C. (2005). Admixed sexual and facultatively asexual aphid lineages at mating sites. Molecular Ecology, 14(1), 325–336.
Herbert, K. S., Hoffmann, A. A., & Powell, K. S. (2006). Changes in grape phylloxera abundance in ungrafted vineyards. Journal of Economic Entomology, 99(5), 1774–1783.
Islam, M.S., Roush, T.L., Walker, M.A., Granett, J., Lin, H. (2013) Reproductive mode and fine-scale population genetic structure of grape phylloxera (Daktulosphaira vitifoliae) in a viticultural area in California. BMC Genetics, 14, art. no. 123.
Kocsis, L. & Bari, I. (2013) Preference and performance of leaf-galling grape phylloxera on Vitis sp. Cultivars. 6th International Symposium on Phylloxera. Bordeaux. France.
Koennecke, T., Aigner, C., Specht, S., Lawo, N. C., & Forneck, A. (2011). A stepwise assessment of Daktulosphaira vitifoliae infested grapevines in a Viennese vineyard site. Acta Horticulturae, 904, 59–62.
Lund, K. (2013) Phylloxera Biodiversity. PhD Thesis University of California, Davis. 120pp.
Lynch, C. M. (1984). Destabilizing hybridization. general-purpose genotypes and geographic parthenogenesis. Quarterly Review of Biology, 59, 257–290.
Müller, N. (2010) Verwilderte Reben an den Böschungen des Kaiserstuhls. Diplomarbeit. Albert-Ludwigs-Universtität Freiburg i.Br. Fakultät für Biologie. 77 pp
Nei, M. (1978). Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics, 89(3), 583–590.
Powell, K. S., Cooper, P. D., & Forneck, A. (2013). The biology, physiology and host-plant interactions of grape phylloxera daktulosphaira vitifoliae. Advances in Insect Physiology, 45, 159–218.
Raymond, M., & Rousset, F. (1995). GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism. Journal of Heredity, 86:248–249
Riaz, S., Lund, K., Lin, H., & Walker, M. A. (2014). Development and characterization of a large set of microsatellite markers for grape phylloxera (Daktulosphaira vitifoliae Fitch). Vitis, 53(2), 95–101.
Ritter, A., Vorwerk, S., Blaich, R., & Forneck, A. (2007). Adaptational potential of grape phylloxera (Daktulosphaira vitifoliae) clonal lineages. Mitteilungen Klosterneuburg, 57, 116–122.
Simon, J. C., Baumann, S., Sunnucks, P., Hebert, P. D., Pierre, J. S., Le Gallic, J. F., & Dedryver, C. A. (1999). Reproductive mode and population genetic structure of the cereal aphid Sitobion avenae studied using phenotypic and microsatellite markers. Molecular Ecology, 8(4), 531–545.
Sun, Q.-H., Chen, Y.-C., Wang, H. B., Downie, D. A., & Zhai, H. (2009). Origin and genetic diversity of grape phylloxera in China. Acta Entomologica Sinica, 52(8), 885–894.
Sunnucks, P., De Barro, P. J., Lushai, G., Maclean, N., & Hales, D. F. (1997). Genetic structure of an aphid studied using microsatellites: cyclic parthenogenesis differentiated lineages and host specialisation. Molecular Ecology, 6, 1059–1073.
Umina, P. A., Corrie, A. M., Herbert, K. S., White, V. L., Powell, K. S., & Hoffmann, A. A. (2007). The use of DNA markers for pest management - Clonal lineages and population biology of grape phylloxera. Acta Horticulturae, 733, 183–195.
Vidart, M. V., Mujica, M. V., Bao, L., Duarte, F., Bentancourt, C. M., Franco, J., & Scatoni, I. B. (2013). Life history and assessment of grapevine phylloxera leaf galling incidence on Vitis species in Uruguay. SpringerPlus, 2(1), 1–9.
Vorburger, C., Lancaster, M., & Sunnecks, P. (2003). Environmentally related patterns of reproductive modes in the aphid Myzus persicae and the predominance of two “superclones” in Victoria. Australia. Molecular Ecology, 12, 3493–3504.
Vorwerk, S., & Forneck, A. (2006). Reproductive mode of grape phylloxera (Daktulosphaira vitifoliae. Homoptera: Phylloxeridae) in Europe: molecular evidence for predominantly asexual populations and a lack of gene flow between them. Genome, 49(6), 678–687.
Vorwerk, S., & Forneck, A. (2007). Analysis of genetic variation within clonal lineages of grape phylloxera (Daktulosphaira vitifoliae Fitch) using AFLP fingerprinting and DNA sequencing. Genome, 50(7), 660–667.
Weir, B. S., & Cockerham, C. C. (1984). Estimating F-statistics for the analysis of population structure. Evolution, 38(6), 1358–1370.
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Forneck, A., Anhalt, U.C.M., Mammerler, R. et al. No evidence of superclones in leaf-feeding forms of austrian grape phylloxera (Daktulosphaira vitifoliae). Eur J Plant Pathol 142, 441–448 (2015). https://doi.org/10.1007/s10658-015-0624-9
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10658-015-0624-9