Abstract
The Generation R Study is a population-based prospective cohort study from fetal life until adulthood. The study is designed to identify early environmental and genetic causes and causal pathways leading to normal and abnormal growth, development and health from fetal life, childhood and young adulthood. In total, 9,778 mothers were enrolled in the study. Data collection in children and their parents include questionnaires, interviews, detailed physical and ultrasound examinations, behavioural observations, Magnetic Resonance Imaging and biological samples. Efforts have been conducted for collecting biological samples including blood, hair, faeces, nasal swabs, saliva and urine samples and generating genomics data on DNA, RNA and microbiome. In this paper, we give an update of the collection, processing and storage of these biological samples and available measures. Together with detailed phenotype measurements, these biological samples provide a unique resource for epidemiological studies focused on environmental exposures, genetic and genomic determinants and their interactions in relation to growth, health and development from fetal life onwards.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The Generation R Study is a population-based prospective cohort study from fetal life until young adulthood. The background and specific research projects of the study have been described in detail [1–6]. Briefly, the Generation R Study is designed to identify early environmental and genetic causes of normal and abnormal growth, development and health from fetal life until young adulthood. The study focuses on six areas of research: (1) maternal health; (2) growth and physical development; (3) behavioural and cognitive development; (4) respiratory health and allergies; (5) diseases in childhood; and (6) health and healthcare for children and their parents.
Main exposures and outcomes studied in the Generation R Study are new or well-known risk factors for common diseases in childhood or adulthood. As published in the European Journal of Epidemiology, many of these risk factors have been related to common outcomes throughout the life course such as pregnancy and early childhood outcomes [7–17], cardiovascular disease [18–50], diabetes [51–60], obesity [61–69], metabolic diseases [70–75], respiratory diseases [76–85], neurological diseases [86–90] and other common outcomes [91–94]. The main outcomes and exposures in the Generation R Study are presented in Tables 1 and 2. A detailed and extensive data collection has been conducted over the years, starting in the prenatal phase and currently in childhood [4]. This data collection also comprises biological samples including blood, hair, faeces, nasal swabs, saliva and urine samples. These biological samples form the Generation R Study Biobank, which enables epidemiological studies focused on environmental exposures, genetic determinants and their interactions in relation to growth, health and development during fetal life, childhood and adulthood. We have previously described the design of this Biobank [5]. In this paper, we give an update of the collection, processing and storage of the biological samples collected during fetal life and childhood. In addition we give an overview of currently available measures in these samples.
Study cohort
In total, 9,778 mothers were enrolled in the study. Of these mothers, 91 % (n = 8,879) was enrolled in pregnancy. Fathers from mothers enrolled during pregnancy were invited to participate. In total, 71 % (n = 6,347) of all fathers was enrolled. Of all participating mothers, data are available in early pregnancy in 72 % (n = 7,069), in mid-pregnancy in 16 % (n = 1,594), late pregnancy in 2 % (n = 216) and from birth of their child in 9 % (n = 899). A total of 1,232 pregnant women and their children were enrolled in a subgroup of Dutch children for additional detailed studies until the age of 4 years. Of all eligible children in the study area, 61 % participated in the study at birth [4]. The largest ethnic groups are the Dutch, Surinamese, Turkish and Moroccan mothers.
Biological samples collection
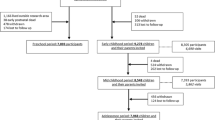
A detailed overview of the complete data collection in mothers, fathers and children until the age of 9 years is given elsewhere [4]. Figure 1 gives an overview of the collection of biological samples. During pregnancy, biological materials have been collected in early, mid- and late pregnancy and at birth. All samples were collected during a visit to one of our dedicated research centers. The planned amounts of blood taken by antecubital venipuncture were 35 ml in early pregnancy and 20 ml in mid-pregnancy from the mother and 10 ml from her partner. When mothers were enrolled in mid- and late pregnancy, 35 ml of blood was taken at the first visit. Urine samples (65 ml) were added to the data collection between February 2003 and November 2005. Directly after delivery, midwives or obstetricians collected maximum 30 ml cord blood from the umbilical vein [5].
Design and response biological sample collection until the age of 9 years. *Number of eligible subjects at enrolment reflects those participating in the study during pregnancy and who visited our research center in early, mid- or late pregnancy. **The number of urine samples at enrolment is lower since this data-collection was added after starting the study. ***The number of eligible children at enrolment reflects the number of live born children of mothers who were enrolled in the prenatal phase of the study. ****The number of hair samples at the age of 5 years is lower since collection was added after starting the study (2 years later)
During the preschool period, children participating in the Dutch subgroup have been invited six times to a dedicated research center were we collected biological samples. At the age of 6, 14 and 24 months a maximum amount of 20 ml blood was taken by antecubital venipuncture. At the age of 1.5, 6, 14, 24 and 36 months nasal samples were collected. At the age 6, 14, 24 and 36 months saliva was collected in 1.5 ml Eppendorf tubes using Saliva Collection device (ORACOL, Malvern Medical UK) and frozen at −20 °C and stored at −80 °C. In addition, at the age of 14 months, parents were asked to collect five saliva samples at home using Salivette sampling devices (Sarstedt, Rommelsdorf, Germany).
From the age of 6 years, all participating mothers and children are invited to a well-equipped and dedicated research center in the Erasmus MC-Sophia Children’s Hospital every 3 years (age 6 years visit completed, age 9 years visit ongoing, and age 12 visit planned) [4]. At the child’s age of 6 years, a maximum amount of 22.5 ml blood and a sample of hair (1–2 cm) was collected in both mother and child and nasal swabs, saliva (1.5 ml) and urine (80 ml) samples were collected in children only. At the age of 9 years, we are currently collecting blood (maximum 22.5 ml), hair (1–2 cm) and urine (54 ml) in both mother and child and nasal swabs only from the child. In a random group of 1,000 children we collect 2.5 ml of blood in a 2.5 ml PAXgene™ blood RNA tube (PAXgene Tubes-Becton–Dickinson). RNA will be isolated using a PAXgene Blood RNAkit-Qiagen (Qiagen, Hilden, Germany). In addition, parents are asked to collect faeces for intestinal microbiome analysis from the children at home.
Logistics
Blood and urine
All biological samples are bar coded with a unique and anonymous laboratory number. Blood or urine samples collected at one visit have the same bar code with an additional unique tube number. All following steps in processing, storing and data management of the samples are linked to this unique tube number.
During the prenatal phase, all samples from the mother and father were taken by research nurses and temporally stored at our research center or one of the obstetric departments at room temperature for a maximum of 3 h. The urine samples were stored at 4 °C and transported to the STAR-MDC laboratory for further processing within 24 h of receipt. All samples were transported to a dedicated laboratory facility of the regional laboratory in Rotterdam, the Netherlands (STAR-MDC) for further processing and storage. Cord blood samples collected at home or at one of the hospitals in Rotterdam were collected by a midwife or obstetrician. Subsequently, courier services with a 7/24 availability were responsible for transportation of cord blood samples to our laboratory within 2 h.
During childhood, all blood and urine samples collected at our research center in the Erasmus MC-Sophia’s Children’s Hospital are stored for a maximum of 4 h at 4 °C with the exception of blood collected for further immunologic analysis (room temperature) or RNA isolation (2–4 h at room temperature before storage at −20 °C). Twice a day blood and urine samples are transported to the STAR-MDC for further processing and storage.
After collection and transportation, blood and urine samples are centrally processed and stored at the STAR-MDC laboratory. Samples for DNA extraction are stored as EDTA whole blood samples at −20 °C. All collected EDTA plasma and serum samples are processed within 4 h after venous puncture. Total processing time takes 15 min. The samples are spun and the plasma and serum volume is distributed into 250 µl aliquots and transferred to 0.65 ml polypropylene tubes (Micronic) by a Tecan automatic liquid handler. Aliquots tubes from one plasma and serum sample are divided over four different microliter trays and immediately stored at −80 °C. The four trays are divided over four different freezers with different power supplies at one location (STAR-MDC laboratory). Each Micronic tube is uniquely coded on the bottom with the Traxis® 2D code.
The urine samples were distributed manually in one 5 ml (only during pregnancy) and three 20 ml tubes. The 5 ml urine tube was sent to the Department of Medical Microbiology, Erasmus MC. The remaining urine tubes (20 ml) were stored at −20 °C at the STAR-MDC laboratory. At the age of 9 years onwards, the urine is filled out in 4.5 ml tubes and stored at −20 °C.
DNA
DNA from mothers, fathers and children has been isolated from whole blood EDTA tubes. DNA extraction from all children has been conducted manually using the Qiagen FlexiGene Kit (Qiagen Hilden, Germany) [95]. DNA extraction, plating and normalization from 5 ml whole blood samples from the mothers and fathers was performed at the Human Genotyping and Sequencing Facility of the Genetic Laboratory at the Department of Internal Medicine, Erasmus MC, by a Hamilton STAR multi-channel robot using AGOWA magnetic bead technology.
Extracted DNA has been automatically collected in stock tubes (2D Matrix, Micronic) organized in 96-wells format, which can be individually addressed. These stock samples are split into two tubes that are stored at different locations. One of the stocks is used for normalizing the DNA concentrations in 96 deep well (DW) plates. This protocol, as well as other manipulations of the DNA samples, are performed on a Caliper ALH3000 pipetting robot (8/96/384 channels) with a Twister module and a 96/384 wells Tecan GENios plus UV reader. A random selection of 5 % of the total number of samples is put into separate 96 DW plates for control purposes. For normalization, sample and diluent volumes are automatically calculated and produced to obtain equal DNA concentrations for all samples (50 ng/µl). From these stock 96-DW plates, 384 DW plates are created, with 1 ng/µl per sample. Subsequently, replica PCR plates (384 wells) with a concentration of 2 ng/µl are created for genotype studies.
Stock 96-well plates (DW and 2D Matrix, Micronic) are stored at −20 °C at the Genetic Epidemiology Laboratory, Erasmus MC. All genotyping and sequencing studies with Generation R DNA material are performed in-house at the Human Genotyping and Sequencing Facility of the Genetic Laboratory of the Department of Internal Medicine, Erasmus MC.
Other biological samples
Nasal swabs for bacterial cultures are transported in Amies transport medium to the Medical Microbiology laboratory, Erasmus MC within 6 h of sampling for further processing and storage.
Parents were asked to collect five saliva samples at home using Salivette sampling devices (Sarstedt, Rommelsdorf, Germany) at the age of 14 months in the Dutch subgroup and at the age of 6 years in all participating children. Parents received detailed written instructions with pictures concerning the saliva sampling. They collected five saliva samples during one single weekday: immediately after awakening, 30 min later, around noon, between 1500 hours and 1600 hours, and at bedtime. The salivettes were mailed to the Genetic Epidemiology Laboratory, Erasmus MC. Here, the samples were centrifuged and frozen at −80 °C. After completion of the data collection at the age of 14 months, all frozen samples were sent on dry ice in one batch by courier to the laboratory of the Department of Biological Psychology laboratory at the Technical University of Dresden for analysis. Saliva samples were transported to the Clinical Chemistry laboratory, Erasmus MC for storage.
Hair samples are stored at room temperature and transported to the laboratory of Internal Medicine, Erasmus MC for further processing and storage.
Microbiome analyses focused on the gut microbiome are planned but other eligible samples are collected as well such as nasal swabs, saliva, and urine. Faecal samples collected at home are mailed by post to the laboratory of the Department of Gastroenterlogy and Hepatology, Erasmus MC and DNA is isolated from the faecal samples and stored at −80 °C and used for subsequent 16S microbiome genetic analysis.
Response rates
Overall response rates for the collection of biological samples are presented in Fig. 1. To get these high response rates, we collected blood samples from veni punctures that were already planned in routine care whenever possible. Blood for DNA extraction was available from 89, 82 and 61 % of the eligible mothers, fathers and children. The larger number of missings in fathers and children were mainly due to non-participation of fathers and logistical constraints during delivery, respectively. Absolute numbers of urine samples are lower than the absolute number of blood samples because the urine sample collection was performed during a limited period in the prenatal phase of the study. Efforts have been carried out for completing DNA collection in the children samples with the collection of whole blood samples at the age of 6 and now in 9 years. DNA is available for analysis in 78 % of all mothers, 76 % of all participating fathers and 58 % of all live born children.
At the age of 6 years the overall response rate for blood collection in mothers was 89 %. In children the response was 69 % due to lack of consent by the parents or non-successful venous punctures. The overall response was 97 % for urine samples, 96 % for nasal swabs and 97 % for saliva samples. The collection of hair started at a later stage. Collecting biological samples by the parents at home led to lower response rates (43 % for saliva swabs).
Available measures
Blood for phenotypes
Results of analyses performed in blood samples (EDTA plasma) for routine care for pregnant women (Hb, Ht, HIV, HBsAg, Lues, Rhesus factor and irregular antibodies) were obtained from midwife and obstetric registries [96–99]. Additional measurements performed in serum and plasma samples collected in mothers and children are shown in Table 3 [100–145].
Genomics data
All Genomics Data are generated at the Human Genotyping and Sequencing Facility of the Genetic Laboratory of the Department of Internal Medicine (www.glimdna.org), where the GWAS datasets of the Rotterdam Study were also created, a prospective cohort study among over 10,000 adults [146–148]. The Generation R Genomics data currently include GWAS data, DNA Exome Array data, DNA methylation data, and single polymorphisms data (e.g., individual SNPs, VNTRs).
Genome wide association study (GWAS) database
Genetic data have been generated by a genome wide association scan (GWAS) using Illumina HumanHap 610 or 660 Quad chips (Illumina Inc., San Diego, USA), depending on time of collection. The GWAS dataset underwent a stringent QC process, which has been described in detail previously [3, 149]. Most GWAS analyses are strongly embedded in the Early Growth Genetics (EGG) Consortium and Early Genetics and Longitudinal Epidemiology (EAGLE) Consortium, in which several birth cohort studies combine their GWAS efforts focused on multiple outcomes in fetal life, childhood and adolescence. These efforts have already led to successful identification of various common genetic variants related to birth weight, birth length, infant head circumference, childhood adiposity, body mass index, bone mineral density, atopic dermatitis and other outcomes [149–167].
Single-nucleotide polymorphisms (SNPs)
As GWAS is not yet available in parents, genotyping is mainly performed using Taqman allelic discrimination assay (Applied Biosystems, Foster City, CA) and Abgene QPCR ROX mix (Abgene, Hamburg, Germany) in mother and father. To confirm the accuracy of the genotyping results, 276 randomly selected samples from the Generation R Study are genotyped for a second time with the same method. The error rate was less than 1 %. In Table 4 all available SNPs and VNTRs in mothers and fathers are shown. In children, individual genotype data are extracted from the genomewide Illumina 610 or 660 Quad chips. If SNPs were not directly genotyped, we used MACH (version 1.0.15) software to impute genotypes using the HapMap II CEU (release 22) as reference set or SNPs were genotyped using the same method as the parents. SNPs were used for various candidate gene, replication and Mendelian randomization studies [120, 143, 168–201].
Exome array variant database
Variants in exomes were measured using an Illumina exome chip v1.1 array in a DNA samples of a subgroup of ~ 1,000 Dutch children. The array contains 270.000 genetic variants in exonic sequences and can be used to study common and rare coding variants next to the GWAS data.
Epigenome wide association study (EWAS) database
DNA methylation was measured on a genome wide level in cord blood samples in a subgroup of ~1,000 Dutch children using the Illumina 450 K Infinium BeadChip (Illumina Inc., San Diego, USA), which contains 485,553 methylation sites at a single nucleotide resolution. Quality control of analyzed samples was performed using standardized criteria. Samples were excluded in case of low sample call rate (<99 %), colour balance >3, low staining efficiency, poor extension efficiency, poor hybridization performance, low stripping efficiency after extension and poor bisulfite conversion. Current analyses are based on cord blood samples. We also plan to measure DNA methylation at different ages.
Candidate gene methylation
DNA methylation was assessed at different loci in genomic DNA isolated from cord blood samples and has been used to study the association with ADHD and fetal and infant growth [202, 203]. Candidate genes for methylation were selected on the basis of their potential involvement in neurotransmitter systems and neurodevelopment. Additionally regions were selected of the IGF2DMR and H19 gene that are implicated in fetal growth. Isolated genomic DNA (500 ng) from cord blood samples was treated with sodium bisulphite for 16 h using the EZ-96 DNA methylation kit (Shallow) (Zymo Research, Irvine, CA, USA). This was followed by PCR amplification, fragmentation after reverse transcription and analysis on a mass spectrometer (Sequenom, Inc, San Diego, USA). This generated mass spectra that were translated into quantitative DNA methylation levels of different CpG sites by MassARRAY EpiTYPER Analyzer software (v1.0, build 1.0.6.88 Sequenom, Inc, San Diego, USA).
Samples were randomly divided over bisulphite conversion and PCR amplification batches. For each individual, the assays were amplified from the same bisulphite-treated DNA. All methylation measurements were done in triplicate from the same bisulphite-treated DNA.
Saliva
Salivary cortisol concentrations were measured using a commercial immunoassay with chemiluminescence detection (CLIA; IBL Hamburg, Germany) with which various studies have been performed to determine risk factors in relation to infant cortisol rhythms and stress reactivity [204–208]. Samples collected at the age of 6 years will be processed at a later stage.
Nasal swabs
Nasal swabs were taken with rayon tipped dacron pernasal swabs (Copan Italia, Brescia, Italy), transported in Amies transport medium and plated within 6 h of sampling on a blood agar plate with 5 % sheep blood, a chocolate agar plate and a Heamophilus selective agar plate. The plates are kept at 35 °C in a CO2 rich environment for 2 days. Bacterial growth was determined daily. All bacteria were determined by standard methods. Various studies have been performed studying colonization of the nasopharynx by potential pathogens [122, 123, 125, 126, 209–216]. At the age of 9 years all swabs are stored at −80 °C after collection and will be processed at a later stage.
Urine
In urine samples collected during pregnancy, levels for various environmental exposures are measured like organophosphorous pesticide, bisphenol A, and phthalate levels to study their effect on fetal development and is planned to be done in samples obtained at the age of 6 years [217–220]. To validate the use of cannabis, urine was tested on the presence of 11-nor-Δ 9-THC-9-COOH using the DRI® Cannabinoid Assay (Microgenics) [221]. To determine C. trachomatis infection during pregnancy, urine samples were tested by PCR [222]. In children at the age of 6 years, kidney function was determined by creatinine and albumin levels using the Beckman Coulter AU analyzer, creatinine levels were measured according to the Jaffe method [135–137]. Dietary intake of iodine during pregnancy was assessed by measuring iodine in urine [111, 223, 224].
Data management and privacy protection
All blood and urine samples stored at the STAR-MDC laboratory are registered in Labosys (Philips) [225]. Both the anonymous person unique study number and all sample numbers are registered which enables matching of each sample to a study subject. A backup of this registration is available at the Erasmus MC. All other laboratories use different systems for registration in which laboratory number, tube number and date of processing is registered. Our data management receives a backup of these registrations every 3 months and is responsible for creating all selection lists and registers the picking-out of the samples, the number of samples and remaining total volume in storage and the number of freeze counts. The sample numbers are removed from the dataset before the results become available for the researchers. The dataset for researchers include a subject unique identification number that enables feedback about a sample of the subject to the data manager but do not enable identification of that particular subject.
Collaboration
The Generation R Study is not the only birth-cohort study with an extensive collection of biological samples. Generation R collaborates with several other cohort studies for replication or meta-analyses.
The Generation R study has an open policy with regard to collaboration with other research groups. Request for collaboration and use of biological samples should primarily be addressed to Vincent Jaddoe (v.jaddoe@erasmusmc.nl), Principal Investigator of the Generation R study. These requests are discussed in the Generation R Study Management Team regarding their scientific merits, study aims, overlap with ongoing studies, logistic consequences and financial contributions. General policy is that collaborating researchers and groups are responsible themselves for the financial requirements. Laboratory tests are preferably conducted in the Erasmus Medical Center, but this is not required. After approval of the project by the Generation R Study Management Team and the Medical Ethical Committee of the Erasmus Medical Center, the collaborative research project is embedded in one of the research areas supervised by the specific principal investigator.
References
Hofman A, Jaddoe VW, Mackenbach JP, Moll HA, Snijders RF, Steegers EA, et al. Growth, development and health from early fetal life until young adulthood: the Generation R Study. Paediatr Perinat Epidemiol. 2004;18(1):61–72.
Jaddoe VW, van Duijn CM, van der Heijden AJ, Mackenbach JP, Moll HA, Steegers EA, et al. The Generation R Study: design and cohort update until the age of 4 years. Eur J Epidemiol. 2008;23(12):801–11.
Jaddoe VW, van Duijn CM, van der Heijden AJ, Mackenbach JP, Moll HA, Steegers EA, et al. The Generation R Study: design and cohort update 2010. Eur J Epidemiol. 2010;25(11):823–41.
Jaddoe VW, van Duijn CM, Franco OH, van der Heijden AJ, van Iizendoorn MH, de Jongste JC, et al. The Generation R Study: design and cohort update 2012. Eur J Epidemiol. 2012;27(9):739–56.
Jaddoe VW, Bakker R, van Duijn CM, van der Heijden AJ, Lindemans J, Mackenbach JP, et al. The Generation R Study Biobank: a resource for epidemiological studies in children and their parents. Eur J Epidemiol. 2007;22(12):917–23.
White T, El Marroun H, Nijs I, Schmidt M, van der Lugt A, Wielopolki PA, et al. Pediatric population-based neuroimaging and the Generation R Study: the intersection of developmental neuroscience and epidemiology. Eur J Epidemiol. 2013;28(1):99–111.
Firoozi F, Lemiere C, Beauchesne MF, Perreault S, Forget A, Blais L. Impact of maternal asthma on perinatal outcomes: a two-stage sampling cohort study. Eur J Epidemiol. 2012;27(3):205–14.
Baba S, Wikstrom AK, Stephansson O, Cnattingius S. Influence of smoking and snuff cessation on risk of preterm birth. Eur J Epidemiol. 2012;27(4):297–304.
Bluming AZ. Breast cancer following diethylstilbestrol exposure in utero: a tragedy? Eur J Epidemiol. 2012;27(4):317–8 (author reply 8).
Niederkrotenthaler T, Rasmussen F, Mittendorfer-Rutz E. Perinatal conditions and parental age at birth as risk markers for subsequent suicide attempt and suicide: a population based case–control study. Eur J Epidemiol. 2012;27(9):729–38.
Jacobsen BK, Knutsen SF, Oda K, Fraser GE. Obesity at age 20 and the risk of miscarriages, irregular periods and reported problems of becoming pregnant: the Adventist Health Study-2. Eur J Epidemiol. 2012;27(12):923–31.
Naver KV, Secher NJ, Ovesen PG, Gorst-Rasmussen A, Lundbye-Christensen S, Nilas L. Offspring preterm birth and birth size are related to long-term risk of maternal diabetes. Eur J Epidemiol. 2013;28(5):427–32.
Ludvigsson JF, Zugna D, Cnattingius S, Richiardi L, Ekbom A, Ortqvist A, et al. Influenza H1N1 vaccination and adverse pregnancy outcome. Eur J Epidemiol. 2013;28(7):579–88.
Nezvalova-Henriksen K, Spigset O, Nordeng H. Triptan safety during pregnancy: a Norwegian population registry study. Eur J Epidemiol. 2013;28(9):759–69.
Kelly-Irving M, Lepage B, Dedieu D, Bartley M, Blane D, Grosclaude P, et al. Adverse childhood experiences and premature all-cause mortality. Eur J Epidemiol. 2013;28(9):721–34.
Rozendaal AM, van Essen AJ, te Meerman GJ, Bakker MK, van der Biezen JJ, Goorhuis-Brouwer SM, et al. Periconceptional folic acid associated with an increased risk of oral clefts relative to non-folate related malformations in the Northern Netherlands: a population based case–control study. Eur J Epidemiol. 2013;28(11):875–87.
Luque-Fernandez MA, Franco M, Gelaye B, Schomaker M, Garitano IG, D’Este C, et al. Unemployment and stillbirth risk among foreign-born and Spanish pregnant women in Spain, 2007-2010: a multilevel analysis study. Eur J Epidemiol. 2013;28(12):991–9.
Juutilainen A, Kastarinen H, Antikainen R, Peltonen M, Salomaa V, Tuomilehto J, et al. Trends in estimated kidney function: the FINRISK surveys. Eur J Epidemiol. 2012;27(4):305–13.
Petersen CB, Gronbaek M, Helge JW, Thygesen LC, Schnohr P, Tolstrup JS. Changes in physical activity in leisure time and the risk of myocardial infarction, ischemic heart disease, and all-cause mortality. Eur J Epidemiol. 2012;27(2):91–9.
Costanza MC, Beer-Borst S, James RW, Gaspoz JM, Morabia A. Consistency between cross-sectional and longitudinal SNP: blood lipid associations. Eur J Epidemiol. 2012;27(2):131–8.
van Oeffelen AA, Agyemang C, Bots ML, Stronks K, Koopman C, van Rossem L, et al. The relation between socioeconomic status and short-term mortality after acute myocardial infarction persists in the elderly: results from a nationwide study. Eur J Epidemiol. 2012;27(8):605–13.
Cook NR, Cole SR, Buring JE. Aspirin in the primary prevention of cardiovascular disease in the Women’s Health Study: effect of noncompliance. Eur J Epidemiol. 2012;27(6):431–8.
Nuesch E, Dale CE, Amuzu A, Kuper H, Bowling A, Ploubidis GB, et al. Inflammation, coagulation and risk of locomotor disability in elderly women: findings from the British Women’s Heart and Health Study. Eur J Epidemiol. 2012;27(8):633–45.
Hansson J, Galanti MR, Hergens MP, Fredlund P, Ahlbom A, Alfredsson L, et al. Use of snus and acute myocardial infarction: pooled analysis of eight prospective observational studies. Eur J Epidemiol. 2012;27(10):771–9.
Grau M, Sala C, Sala J, Masia R, Vila J, Subirana I, et al. Sex-related differences in prognosis after myocardial infarction: changes from 1978 to 2007. Eur J Epidemiol. 2012;27(11):847–55.
Nordahl H, Rod NH, Frederiksen BL, Andersen I, Lange T, Diderichsen F, et al. Education and risk of coronary heart disease: assessment of mediation by behavioral risk factors using the additive hazards model. Eur J Epidemiol. 2013;28(2):149–57.
Larsson SC, Orsini N, Wolk A. Long-chain omega-3 polyunsaturated fatty acids and risk of stroke: a meta-analysis. Eur J Epidemiol. 2012;27(12):895–901.
Karppi J, Kurl S, Makikallio TH, Ronkainen K, Laukkanen JA. Low levels of plasma carotenoids are associated with an increased risk of atrial fibrillation. Eur J Epidemiol. 2013;28(1):45–53.
Kiefte-de Jong JC, Chowdhury R, Franco OH. Fish intake or omega-3 fatty acids: greater than the sum of all parts? Eur J Epidemiol. 2012;27(12):891–4.
Clays E, De Bacquer D, Janssens H, De Clercq B, Casini A, Braeckman L, et al. The association between leisure time physical activity and coronary heart disease among men with different physical work demands: a prospective cohort study. Eur J Epidemiol. 2013;28(3):241–7.
Pequignot R, Tzourio C, Peres K, Ancellin ML, Perier MC, Ducimetiere P, et al. Depressive symptoms, antidepressants and disability and future coronary heart disease and stroke events in older adults: the Three City Study. Eur J Epidemiol. 2013;28(3):249–56.
Brown MC, Best KE, Pearce MS, Waugh J, Robson SC, Bell R. Cardiovascular disease risk in women with pre-eclampsia: systematic review and meta-analysis. Eur J Epidemiol. 2013;28(1):1–19.
Mishra GD, Chiesa F, Goodman A, De Stavola B, Koupil I. Socio-economic position over the life course and all-cause, and circulatory diseases mortality at age 50–87 years: results from a Swedish birth cohort. Eur J Epidemiol. 2013;28(2):139–47.
Kunutsor SK, Apekey TA, Steur M. Vitamin D and risk of future hypertension: meta-analysis of 283,537 participants. Eur J Epidemiol. 2013;28(3):205–21.
Virta JJ, Heikkila K, Perola M, Koskenvuo M, Raiha I, Rinne JO, et al. Midlife cardiovascular risk factors and late cognitive impairment. Eur J Epidemiol. 2013;28(5):405–16.
Behrens G, Fischer B, Kohler S, Park Y, Hollenbeck AR, Leitzmann MF. Healthy lifestyle behaviors and decreased risk of mortality in a large prospective study of U.S. women and men. Eur J Epidemiol. 2013;28(5):361–72.
Threapleton DE, Greenwood DC, Burley VJ, Aldwairji M, Cade JE. Dietary fibre and cardiovascular disease mortality in the UK Women’s Cohort Study. Eur J Epidemiol. 2013;28(4):335–46.
Westerlund A, Bellocco R, Sundstrom J, Adami HO, Akerstedt T, Trolle Lagerros Y. Sleep characteristics and cardiovascular events in a large Swedish cohort. Eur J Epidemiol. 2013;28(6):463–73.
Wu Y, Li S, Zhang D. Circulating 25-hydroxyvitamin D levels and hypertension risk. Eur J Epidemiol. 2013;28(7):611–6.
Ahmadi-Abhari S, Luben RN, Wareham NJ, Khaw KT. Seventeen year risk of all-cause and cause-specific mortality associated with C-reactive protein, fibrinogen and leukocyte count in men and women: the EPIC-Norfolk study. Eur J Epidemiol. 2013;28(7):541–50.
Rahman I, Humphreys K, Bennet AM, Ingelsson E, Pedersen NL, Magnusson PK. Clinical depression, antidepressant use and risk of future cardiovascular disease. Eur J Epidemiol. 2013;28(7):589–95.
Jorde R, Schirmer H, Njolstad I, Lochen ML, Bogeberg Mathiesen E, Kamycheva E, et al. Serum calcium and the calcium-sensing receptor polymorphism rs17251221 in relation to coronary heart disease, type 2 diabetes, cancer and mortality: the Tromso Study. Eur J Epidemiol. 2013;28(7):569-78.
Faeh D, Rohrmann S, Braun J. Better risk assessment with glycated hemoglobin instead of cholesterol in CVD risk prediction charts. Eur J Epidemiol. 2013;28(7):551–5.
Simone B, De Stefano V, Leoncini E, Zacho J, Martinelli I, Emmerich J, et al. Risk of venous thromboembolism associated with single and combined effects of factor V Leiden, prothrombin 20210A and methylenetethraydrofolate reductase C677T: a meta-analysis involving over 11,000 cases and 21,000 controls. Eur J Epidemiol. 2013;28(8):621–47.
Malerba S, Turati F, Galeone C, Pelucchi C, Verga F, La Vecchia C, et al. A meta-analysis of prospective studies of coffee consumption and mortality for all causes, cancers and cardiovascular diseases. Eur J Epidemiol. 2013;28(7):527–39.
Julin B, Wolk A, Thomas LD, Akesson A. Exposure to cadmium from food and risk of cardiovascular disease in men: a population-based prospective cohort study. Eur J Epidemiol. 2013;28(10):837–40.
Hou L, Lloyd-Jones DM, Ning H, Huffman MD, Fornage M, He K, et al. White blood cell count in young adulthood and coronary artery calcification in early middle age: coronary artery risk development in young adults (CARDIA) study. Eur J Epidemiol. 2013;28(9):735–42.
Kershaw KN, Droomers M, Robinson WR, Carnethon MR, Daviglus ML, Monique Verschuren WM. Quantifying the contributions of behavioral and biological risk factors to socioeconomic disparities in coronary heart disease incidence: the MORGEN study. Eur J Epidemiol. 2013;28(10):807–14.
Schmidt M, Botker HE, Pedersen L, Sorensen HT. Adult height and risk of ischemic heart disease, atrial fibrillation, stroke, venous thromboembolism, and premature death: a population based 36-year follow-up study. Eur J Epidemiol. 2014;29(2):111–8.
Kunutsor SK, Burgess S, Munroe PB, Khan H. Vitamin D and high blood pressure: causal association or epiphenomenon? Eur J Epidemiol. 2014;29(1):1–14.
Ma L, Oei L, Jiang L, Estrada K, Chen H, Wang Z, et al. Association between bone mineral density and type 2 diabetes mellitus: a meta-analysis of observational studies. Eur J Epidemiol. 2012;27(5):319–32.
Kharazmi E, Lukanova A, Teucher B, Gross ML, Kaaks R. Does pregnancy or pregnancy loss increase later maternal risk of diabetes? Eur J Epidemiol. 2012;27(5):357–66.
Demakakos P, Marmot M, Steptoe A. Socioeconomic position and the incidence of type 2 diabetes: the ELSA study. Eur J Epidemiol. 2012;27(5):367–78.
Tamayo T, Jacobs DR Jr, Strassburger K, Giani G, Seeman TE, Matthews K, et al. Race- and sex-specific associations of parental education with insulin resistance in middle-aged participants: the CARDIA study. Eur J Epidemiol. 2012;27(5):349–55.
Galli L, Salpietro S, Pellicciotta G, Galliani A, Piatti P, Hasson H, et al. Risk of type 2 diabetes among HIV-infected and healthy subjects in Italy. Eur J Epidemiol. 2012;27(8):657–65.
Lagerros YT, Cnattingius S, Granath F, Hanson U, Wikstrom AK. From infancy to pregnancy: birth weight, body mass index, and the risk of gestational diabetes. Eur J Epidemiol. 2012;27(10):799–805.
Schottker B, Herder C, Rothenbacher D, Perna L, Muller H, Brenner H. Serum 25-hydroxyvitamin D levels and incident diabetes mellitus type 2: a competing risk analysis in a large population-based cohort of older adults. Eur J Epidemiol. 2013;28(3):267–75.
Satman I, Omer B, Tutuncu Y, Kalaca S, Gedik S, Dinccag N, et al. Twelve-year trends in the prevalence and risk factors of diabetes and prediabetes in Turkish adults. Eur J Epidemiol. 2013;28(2):169–80.
Min JY, Min KB. Serum C-peptide levels as an independent predictor of diabetes mellitus mortality in non-diabetic individuals. Eur J Epidemiol. 2013;28(9):771–4.
Buijsse B, Boeing H, Hirche F, Weikert C, Schulze MB, Gottschald M, et al. Plasma 25-hydroxyvitamin D and its genetic determinants in relation to incident type 2 diabetes: a prospective case-cohort study. Eur J Epidemiol. 2013;28(9):743–52.
Johnson PC, Logue J, McConnachie A, Abu-Rmeileh NM, Hart C, Upton MN, et al. Intergenerational change and familial aggregation of body mass index. Eur J Epidemiol. 2012;27(1):53–61.
Iversen LB, Strandberg-Larsen K, Prescott E, Schnohr P, Rod NH. Psychosocial risk factors, weight changes and risk of obesity: the Copenhagen City Heart Study. Eur J Epidemiol. 2012;27(2):119–30.
Rossi IA, Bochud M, Bovet P, Paccaud F, Waeber G, Vollenweider P, et al. Sex difference and the role of leptin in the association between high-sensitivity C-reactive protein and adiposity in two different populations. Eur J Epidemiol. 2012;27(5):379–84.
Nan C, Guo B, Warner C, Fowler T, Barrett T, Boomsma D, et al. Heritability of body mass index in pre-adolescence, young adulthood and late adulthood. Eur J Epidemiol. 2012;27(4):247–53.
Park SY, Wilkens LR, Murphy SP, Monroe KR, Henderson BE, Kolonel LN. Body mass index and mortality in an ethnically diverse population: the Multiethnic Cohort Study. Eur J Epidemiol. 2012;27(7):489–97.
Rosenblad A, Nilsson G, Leppert J. Intelligence level in late adolescence is inversely associated with BMI change during 22 years of follow-up: results from the WICTORY study. Eur J Epidemiol. 2012;27(8):647–55.
Lissner L, Lanfer A, Gwozdz W, Olafsdottir S, Eiben G, Moreno LA, et al. Television habits in relation to overweight, diet and taste preferences in European children: the IDEFICS study. Eur J Epidemiol. 2012;27(9):705–15.
Behrens G, Matthews CE, Moore SC, Freedman ND, McGlynn KA, Everhart JE, et al. The association between frequency of vigorous physical activity and hepatobiliary cancers in the NIH-AARP Diet and Health Study. Eur J Epidemiol. 2013;28(1):55–66.
Sabanayagam C, Wong TY, Xiao J, Shankar A. Serum cystatin C and prediabetes in non-obese US adults. Eur J Epidemiol. 2013;28(4):311–6.
Abbasi A, Corpeleijn E, Peelen LM, Gansevoort RT, de Jong PE, Gans RO, et al. External validation of the KORA S4/F4 prediction models for the risk of developing type 2 diabetes in older adults: the PREVEND study. Eur J Epidemiol. 2012;27(1):47–52.
Claessen H, Brenner H, Drath C, Arndt V. Repeated measures of body mass index and risk of health related outcomes. Eur J Epidemiol. 2012;27(3):215–24.
Brinks R, Tamayo T, Kowall B, Rathmann W. Prevalence of type 2 diabetes in Germany in 2040: estimates from an epidemiological model. Eur J Epidemiol. 2012;27(10):791–7.
Miettola S, Hartikainen AL, Vaarasmaki M, Bloigu A, Ruokonen A, Jarvelin MR, et al. Offspring’s blood pressure and metabolic phenotype after exposure to gestational hypertension in utero. Eur J Epidemiol. 2013;28(1):87–98.
Dai X, Yuan J, Yao P, Yang B, Gui L, Zhang X, et al. Association between serum uric acid and the metabolic syndrome among a middle- and old-age Chinese population. Eur J Epidemiol. 2013;28(8):669–76.
Aune D, Norat T, Romundstad P, Vatten LJ. Whole grain and refined grain consumption and the risk of type 2 diabetes: a systematic review and dose-response meta-analysis of cohort studies. Eur J Epidemiol. 2013;28(11):845–58.
Pekkanen J, Lampi J, Genuneit J, Hartikainen AL, Jarvelin MR. Analyzing atopic and non-atopic asthma. Eur J Epidemiol. 2012;27(4):281–6.
Duijts L. Fetal and infant origins of asthma. Eur J Epidemiol. 2012;27(1):5–14.
Gabriele C, Silva LM, Arends LR, Raat H, Moll HA, Hofman A, et al. Early respiratory morbidity in a multicultural birth cohort: the Generation R Study. Eur J Epidemiol. 2012;27(6):453–62.
Heikkinen SA, Quansah R, Jaakkola JJ, Jaakkola MS. Effects of regular exercise on adult asthma. Eur J Epidemiol. 2012;27(6):397–407.
Patel S, Henderson J, Jeffreys M, Davey Smith G, Galobardes B. Associations between socioeconomic position and asthma: findings from a historical cohort. Eur J Epidemiol. 2012;27(8):623–31.
Broms K, Norback D, Sundelin C, Eriksson M, Svardsudd K. A nationwide study of asthma incidence rate and its determinants in Swedish pre-school children. Eur J Epidemiol. 2012;27(9):695–703.
Figarska SM, Boezen HM, Vonk JM. Dyspnea severity, changes in dyspnea status and mortality in the general population: the Vlagtwedde/Vlaardingen study. Eur J Epidemiol. 2012;27(11):867–76.
Baughman P, Marott JL, Lange P, Martin CJ, Shankar A, Petsonk EL, et al. Combined effect of lung function level and decline increases morbidity and mortality risks. Eur J Epidemiol. 2012;27(12):933–43.
Brostrom EB, Akre O, Katz-Salamon M, Jaraj D, Kaijser M. Obstructive pulmonary disease in old age among individuals born preterm. Eur J Epidemiol. 2013;28(1):79–85.
MacIntyre EA, Carlsten C, MacNutt M, Fuertes E, Melen E, Tiesler CM, et al. Traffic, asthma and genetics: combining international birth cohort data to examine genetics as a mediator of traffic-related air pollution’s impact on childhood asthma. Eur J Epidemiol. 2013;28(7):597–606.
Azevedo Da Silva M, Singh-Manoux A, Brunner EJ, Kaffashian S, Shipley MJ, Kivimaki M, et al. Bidirectional association between physical activity and symptoms of anxiety and depression: the Whitehall II study. Eur J Epidemiol. 2012;27(7):537–46.
Smithers LG, Golley RK, Mittinty MN, Brazionis L, Northstone K, Emmett P, et al. Dietary patterns at 6, 15 and 24 months of age are associated with IQ at 8 years of age. Eur J Epidemiol. 2012;27(7):525–35.
Chowdhury R, Stevens S, Ward H, Chowdhury S, Sajjad A, Franco OH. Circulating vitamin D, calcium and risk of cerebrovascular disease: a systematic review and meta-analysis. Eur J Epidemiol. 2012;27(8):581–91.
Novak M, Toren K, Lappas G, Kok WG, Jern C, Wilhelmsen L, et al. Occupational status and incidences of ischemic and hemorrhagic stroke in Swedish men: a population-based 35-year prospective follow-up study. Eur J Epidemiol. 2013;28(8):697–704.
Mons U, Schottker B, Muller H, Kliegel M, Brenner H. History of lifetime smoking, smoking cessation and cognitive function in the elderly population. Eur J Epidemiol. 2013;28(10):823–31.
Adami HO, Lagiou P, Trichopoulos D. Breast cancer following diethylstilbestrol exposure in utero: insights from a tragedy. Eur J Epidemiol. 2012;27(1):1–3.
Tennant PW, Parker L, Thomas JE, Craft SA, Pearce MS. Childhood infectious disease and premature death from cancer: a prospective cohort study. Eur J Epidemiol. 2013;28(3):257–65.
Anic GM, Madden MH, Sincich K, Thompson RC, Nabors LB, Olson JJ, et al. Early life exposures and the risk of adult glioma. Eur J Epidemiol. 2013;28(9):753–8.
Raoult D, Foti B, Aboudharam G. Historical and geographical parallelism between the incidence of dental caries, Streptococcus mutans and sugar intake. Eur J Epidemiol. 2013;28(8):709–10.
Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16(3):1215.
Mook-Kanamori DO, Steegers EA, Eilers PH, Raat H, Hofman A, Jaddoe VW. Risk factors and outcomes associated with first-trimester fetal growth restriction. JAMA. 2010;303(6):527–34.
Tromp II, Gaillard R, Kiefte-de Jong JC, Steegers EA, Jaddoe VW, Duijts L, et al. Maternal hemoglobin levels during pregnancy and asthma in childhood: the Generation R Study. Ann Allergy Asthma Immunol. 2014;112(3):263–5.
Gaillard R, Eilers PH, Yassine S, Hofman A, Steegers EA, Jaddoe VW. Risk factors and consequences of maternal anaemia and elevated haemoglobin levels during pregnancy: a population-based prospective cohort study. Paediatr Perinat Epidemiol. 2014;28(3):213–26.
Welten M, Gaillard R, Hofman A, de Jonge L, Jaddoe V. Maternal haemoglobin levels and cardio-metabolic risk factors in childhood: the Generation R Study. BJOG. 2014.
Coolman M, Timmermans S, de Groot CJ, Russcher H, Lindemans J, Hofman A, et al. Angiogenic and fibrinolytic factors in blood during the first half of pregnancy and adverse pregnancy outcomes. Obstet Gynecol. 2012;119(6):1190–200.
Bouwland-Both MI, Steegers EA, Lindemans J, Russcher H, Hofman A, Geurts-Moespot AJ, et al. Maternal soluble fms-like tyrosine kinase-1, placental growth factor, plasminogen activator inhibitor-2, and folate concentrations and early fetal size: the Generation R study. Am J Obstet Gynecol. 2013;209(2):121 e1–11.
Brown ZA, Schalekamp-Timmermans S, Tiemeier HW, Hofman A, Jaddoe VW, Steegers EA. Fetal sex specific differences in human placentation: a prospective cohort study. Placenta. 2014;35(6):359–64.
Henrichs J, Bongers-Schokking JJ, Schenk JJ, Ghassabian A, Schmidt HG, Visser TJ, et al. Maternal thyroid function during early pregnancy and cognitive functioning in early childhood: the generation R study. J Clin Endocrinol Metab. 2010;95(9):4227–34.
Ghassabian A, Bongers-Schokking JJ, Henrichs J, Jaddoe VW, Visser TJ, Visser W, et al. Maternal thyroid function during pregnancy and behavioral problems in the offspring: the generation R study. Pediatr Res. 2011;69(5 Pt 1):454–9.
Medici M, de Rijke YB, Peeters RP, Visser W, de Muinck Keizer-Schrama SM, Jaddoe VV, et al. Maternal early pregnancy and newborn thyroid hormone parameters: the Generation R study. J Clin Endocrinol Metab. 2012;97(2):646–52.
Ghassabian A, Bongers-Schokking JJ, de Rijke YB, van Mil N, Jaddoe VW, de Muinck Keizer-Schrama SM, et al. Maternal thyroid autoimmunity during pregnancy and the risk of attention deficit/hyperactivity problems in children: the Generation R Study. Thyroid. 2012;22(2):178–86.
van Mil NH, Steegers-Theunissen RP, Bongers-Schokking JJ, El Marroun H, Ghassabian A, Hofman A, et al. Maternal hypothyroxinemia during pregnancy and growth of the fetal and infant head. Reprod Sci. 2012;19(12):1315–22.
Medici M, Timmermans S, Visser W, de Muinck Keizer-Schrama SM, Jaddoe VW, Hofman A, et al. Maternal thyroid hormone parameters during early pregnancy and birth weight: the Generation R Study. J Clin Endocrinol Metab. 2013;98(1):59–66.
Korevaar TI, Medici M, de Rijke YB, Visser W, de Muinck Keizer-Schrama SM, Jaddoe VW, et al. Ethnic differences in maternal thyroid parameters during pregnancy: the Generation R study. J Clin Endocrinol Metab. 2013;98(9):3678–86.
Roman GC, Ghassabian A, Bongers-Schokking JJ, Jaddoe VW, Hofman A, de Rijke YB, et al. Association of gestational maternal hypothyroxinemia and increased autism risk. Ann Neurol. 2013;74(5):733–42.
Medici M, Ghassabian A, Visser W, de Muinck Keizer-Schrama SM, Jaddoe VW, Visser WE, et al. Women with high early pregnancy urinary iodine levels have an increased risk of hyperthyroid newborns: the population-based Generation R Study. Clin Endocrinol (Oxf). 2014;80(4):598–606.
Korevaar TI, Schalekamp-Timmermans S, de Rijke YB, Visser WE, Visser W, de Muinck Keizer-Schrama SM, et al. Hypothyroxinemia and TPO-antibody positivity are risk factors for premature delivery:the generation R study. J Clin Endocrinol Metab. 2013;98(11):4382–90.
Godoy GA, Korevaar TI, Peeters RP, Hofman A, de Rijke YB, Bongers-Schokking JJ, et al. Maternal thyroid hormones during pregnancy, childhood adiposity and cardiovascular risk factors: the Generation R Study. Clin Endocrinol (Oxf). 2014;81(1):117–25.
Ghassabian A, Henrichs J, Tiemeier H. Impact of mild thyroid hormone deficiency in pregnancy on cognitive function in children: lessons from the Generation R Study. Best Pract Res Clin Endocrinol Metab. 2014;28(2):221–32.
Ghassabian A, El Marroun H, Peeters RP, Jaddoe VW, Hofman A, Verhulst FC, et al. Downstream effects of maternal hypothyroxinemia in early pregnancy: nonverbal IQ and brain morphology in school-age children. J Clin Endocrinol Metab. 2014;99(7):2383–90.
Korevaar TI, Steegers EA, Schalekamp-Timmermans S, Ligthart S, de Rijke YB, Visser WE, et al. Soluble Flt1 and placental growth factor are novel determinants of newborn thyroid (dys)function: the generation R study. J Clin Endocrinol Metab. 2014;99(9):E1627–34.
Medici M, Korevaar TI, Schalekamp-Timmermans S, Gaillard R, de Rijke YB, Visser WE, et al. Maternal early-pregnancy thyroid function is associated with subsequent hypertensive disorders of pregnancy: the Generation R Study. J Clin Endocrinol Metab. 2014;jc20141505.
Gishti O, Gaillard R, Durmus B, Hofman A, Duijts L, Franco OH, et al. Infant diet and metabolic outcomes in school-age children. The Generation R Study. Eur J Clin Nutr. 2014;68(9):1008–15.
Duijts L, Bakker-Jonges LE, Labout JA, Jaddoe VW, Hofman A, Steegers EA, et al. Perinatal stress influences lymphocyte subset counts in neonates. The Generation R Study. Pediatr Res. 2008;63(3):292–8.
Duijts L, Bakker-Jonges LE, Mook-Kanamori DO, Labout JA, Hofman A, van Duijn CM, et al. Variation in the IGF-1 gene is associated with lymphocyte subset counts in neonates: the Generation R Study. Clin Endocrinol (Oxf). 2009;70(1):53–9.
Duijts L, Bakker-Jonges LE, Labout JA, Jaddoe VW, Hofman A, Steegers EA, et al. Fetal growth influences lymphocyte subset counts at birth: the Generation R Study. Neonatology. 2009;95(2):149–56.
Lebon A, Verkaik NJ, de Vogel CP, Hooijkaas H, Verbrugh HA, van Wamel WJ, et al. The inverse correlation between Staphylococcus aureus and Streptococcus pneumoniae colonization in infants is not explained by differences in serum antibody levels in the Generation R Study. Clin Vaccine Immunol. 2011;18(1):180–3.
Lebon A, Verkaik NJ, Labout JA, de Vogel CP, Hooijkaas H, Verbrugh HA, et al. Natural antibodies against several pneumococcal virulence proteins in children during the pre-pneumococcal-vaccine era: the generation R study. Infect Immun. 2011;79(4):1680–7.
Ernst GD, de Jonge LL, Hofman A, Lindemans J, Russcher H, Steegers EA, et al. C-reactive protein levels in early pregnancy, fetal growth patterns, and the risk for neonatal complications: the Generation R Study. Am J Obstet Gynecol. 2011;205(2):132 e1–12.
Verhaegh SJ, de Vogel CP, Riesbeck K, Lafontaine ER, Murphy TF, Verbrugh HA, et al. Temporal development of the humoral immune response to surface antigens of Moraxella catarrhalis in young infants. Vaccine. 2011;29(34):5603–10.
Verkaik NJ, Nguyen DT, de Vogel CP, Moll HA, Verbrugh HA, Jaddoe VW, et al. Streptococcus pneumoniae exposure is associated with human metapneumovirus seroconversion and increased susceptibility to in vitro HMPV infection. Clin Microbiol Infect. 2011;17(12):1840–4.
de Jonge LL, Steegers EA, Ernst GD, Lindemans J, Russcher H, Hofman A, et al. C-reactive protein levels, blood pressure and the risks of gestational hypertensive complications: the Generation R Study. J Hypertens. 2011;29(12):2413–21.
van den Hooven EH, de Kluizenaar Y, Pierik FH, Hofman A, van Ratingen SW, Zandveld PY, et al. Chronic air pollution exposure during pregnancy and maternal and fetal C-reactive protein levels: the Generation R Study. Environ Health Perspect. 2012;120(5):746–51.
Kiefte-de Jong JC, Jaddoe VW, Uitterlinden AG, Steegers EA, Willemsen SP, Hofman A, et al. Levels of antibodies against tissue transglutaminase during pregnancy are associated with reduced fetal weight and birth weight. Gastroenterology. 2013;144(4):726-35 e2.
Sonnenschein-van der Voort AM, Jaddoe VW, Moll HA, Hofman A, van der Valk RJ, de Jongste JC, et al. Influence of maternal and cord blood C-reactive protein on childhood respiratory symptoms and eczema. Pediatr Allergy Immunol. 2013;24(5):469–75.
den Hollander WJ, Holster IL, den Hoed CM, van Deurzen F, van Vuuren AJ, Jaddoe VW, et al. Ethnicity is a strong predictor for Helicobacter pylori infection in young women in a multi-ethnic European city. J Gastroenterol Hepatol. 2013;28(11):1705–11.
den Hollander WJ, Holster IL, van Gilst B, van Vuuren AJ, Jaddoe VW, Hofman A, et al. Intergenerational reduction in Helicobacter pylori prevalence is similar between different ethnic groups living in a Western city. Gut. 2014.
Jansen MA, Tromp II, Kiefte-de Jong JC, Jaddoe VW, Hofman A, Escher JC, et al. Infant feeding and anti-tissue transglutaminase antibody concentrations in the Generation R Study. Am J Clin Nutr. 2014;100(4):1095–101.
Jansen MA, Kiefte-de Jong JC, Gaillard R, Escher JC, Hofman A, Jaddoe VW, et al. Growth trajectories and bone mineral density in anti-tissue transglutaminase antibody-positive children: the Generation R study. Clin Gastroenterol Hepatol. 2014.
Bakker H, Kooijman MN, van der Heijden AJ, Hofman A, Franco OH, Taal HR, et al. Kidney size and function in a multi-ethnic population-based cohort of school-age children. Pediatr Nephrol. 2014;29(9):1589–98.
Bakker H, Gaillard R, Franco OH, Hofman A, van der Heijden AJ, Steegers EA, et al. Fetal and infant growth patterns and kidney function at school age. J Am Soc Nephrol. 2014;25(11):2607–15.
Kooijman MN, Bakker H, van der Heijden AJ, Hofman A, Franco OH, Steegers EA, et al. Childhood kidney outcomes in relation to fetal blood flow and kidney size. J Am Soc Nephrol. 2014;25(11):2616–24.
Gaillard R, Steegers EA, Duijts L, Felix JF, Hofman A, Franco OH, et al. Childhood cardiometabolic outcomes of maternal obesity during pregnancy: the Generation R Study. Hypertension. 2014;63(4):683–91.
Jaddoe VW, de Jonge LL, Hofman A, Franco OH, Steegers EA, Gaillard R. First trimester fetal growth restriction and cardiovascular risk factors in school age children: population based cohort study. BMJ. 2014;348:g14.
Gaillard R, Rurangirwa AA, Williams MA, Hofman A, Mackenbach JP, Franco OH, et al. Maternal parity, fetal and childhood growth, and cardiometabolic risk factors. Hypertension. 2014;64(2):266–74.
Kiefte-de Jong JC, Timmermans S, Jaddoe VW, Hofman A, Tiemeier H, Steegers EA, et al. High circulating folate and vitamin B-12 concentrations in women during pregnancy are associated with increased prevalence of atopic dermatitis in their offspring. J Nutr. 2012;142(4):731–8.
Bergen NE, Jaddoe VW, Timmermans S, Hofman A, Lindemans J, Russcher H, et al. Homocysteine and folate concentrations in early pregnancy and the risk of adverse pregnancy outcomes: the Generation R Study. BJOG. 2012;119(6):739–51.
Steenweg-de Graaff J, Roza SJ, Steegers EA, Hofman A, Verhulst FC, Jaddoe VW, et al. Maternal folate status in early pregnancy and child emotional and behavioral problems: the Generation R Study. Am J Clin Nutr. 2012;95(6):1413–21.
van der Valk RJ, Kiefte-de Jong JC, Sonnenschein-van der Voort AM, Duijts L, Hafkamp-de Groen E, Moll HA, et al. Neonatal folate, homocysteine, vitamin B12 levels and methylenetetrahydrofolate reductase variants in childhood asthma and eczema. Allergy. 2013;68(6):788–95.
Steenweg-de Graaff J, Ghassabian A, Jaddoe VW, Tiemeier H, Roza SJ. Folate concentrations during pregnancy and autistic traits in the offspring. The Generation R Study. Eur J Public Health. 2014.
Hofman A, van Duijn CM, Franco OH, Ikram MA, Janssen HL, Klaver CC, et al. The Rotterdam Study: 2012 objectives and design update. Eur J Epidemiol. 2011;26(8):657–86.
Ikram MA, van der Lugt A, Niessen WJ, Krestin GP, Koudstaal PJ, Hofman A, et al. The Rotterdam Scan Study: design and update up to 2012. Eur J Epidemiol. 2011;26(10):811–24.
Hofman A, Darwish Murad S, van Duijn CM, Franco OH, Goedegebure A, Ikram MA, et al. The Rotterdam Study: 2014 objectives and design update. Eur J Epidemiol. 2013;28(11):889–926.
Medina-Gomez C, Kemp JP, Estrada K, Eriksson J, Liu J, Reppe S, et al. Meta-analysis of genome-wide scans for total body BMD in children and adults reveals allelic heterogeneity and age-specific effects at the WNT16 locus. PLoS Genet. 2012;8(7):e1002718.
Freathy RM, Mook-Kanamori DO, Sovio U, Prokopenko I, Timpson NJ, Berry DJ, et al. Variants in ADCY5 and near CCNL1 are associated with fetal growth and birth weight. Nat Genet. 2010;42(5):430–5.
Paternoster L, Standl M, Chen CM, Ramasamy A, Bonnelykke K, Duijts L, et al. Meta-analysis of genome-wide association studies identifies three new risk loci for atopic dermatitis. Nat Genet. 2012;44(2):187–92.
Taal HR, Verwoert GC, Demirkan A, Janssens AC, Rice K, Ehret G, et al. Genome-wide profiling of blood pressure in adults and children. Hypertension. 2012;59(2):241–7.
Bradfield JP, Taal HR, Timpson NJ, Scherag A, Lecoeur C, Warrington NM, et al. A genome-wide association meta-analysis identifies new childhood obesity loci. Nat Genet. 2012;44(5):526–31.
Ikram MA, Fornage M, Smith AV, Seshadri S, Schmidt R, Debette S, et al. Common variants at 6q22 and 17q21 are associated with intracranial volume. Nat Genet. 2012;44(5):539–44.
Taal HR, St Pourcain B, Thiering E, Das S, Mook-Kanamori DO, Warrington NM, et al. Common variants at 12q15 and 12q24 are associated with infant head circumference. Nat Genet. 2012;44(5):532–8.
Horikoshi M, Yaghootkar H, Mook-Kanamori DO, Sovio U, Taal HR, Hennig BJ, et al. New loci associated with birth weight identify genetic links between intrauterine growth and adult height and metabolism. Nat Genet. 2013;45(1):76–82.
Benyamin B, Pourcain B, Davis OS, Davies G, Hansell NK, Brion MJ, et al. Childhood intelligence is heritable, highly polygenic and associated with FNBP1L. Mol Psychiatry. 2014;19(2):253–8.
Cousminer DL, Berry DJ, Timpson NJ, Ang W, Thiering E, Byrne EM, et al. Genome-wide association and longitudinal analyses reveal genetic loci linking pubertal height growth, pubertal timing and childhood adiposity. Hum Mol Genet. 2013;22(13):2735–47.
Bonnelykke K, Sleiman P, Nielsen K, Kreiner-Moller E, Mercader JM, Belgrave D, et al. A genome-wide association study identifies CDHR3 as a susceptibility locus for early childhood asthma with severe exacerbations. Nat Genet. 2014;46(1):51–5.
van der Valk RJ, Duijts L, Timpson NJ, Salam MT, Standl M, Curtin JA, et al. Fraction of exhaled nitric oxide values in childhood are associated with 17q11.2-q12 and 17q12-q21 variants. J Allergy Clin Immunol. 2014;134(1):46–55.
Benke KS, Nivard MG, Velders FP, Walters RK, Pappa I, Scheet PA, et al. A genome-wide association meta-analysis of preschool internalizing problems. J Am Acad Child Adolesc Psychiatry. 2014;53(6):667–76.
Kemp JP, Medina-Gomez C, Estrada K, St Pourcain B, Heppe DH, Warrington NM, et al. Phenotypic dissection of bone mineral density reveals skeletal site specificity and facilitates the identification of novel loci in the genetic regulation of bone mass attainment. PLoS Genet. 2014;10(6):e1004423.
Stergiakouli E, Gaillard R, Tavare JM, Balthasar N, Loos RJ, Taal HR, et al. Genome-wide association study of height-adjusted BMI in childhood identifies functional variant in ADCY3. Obesity (Silver Spring). 2014;22(10):2252–9.
Parmar PG, Marsh JA, Rob Taal H, Kowgier M, Uitterlinden AG, Rivadeneira F, et al. Polymorphisms in genes within the IGF-axis influence antenatal and postnatal growth. J Dev Orig Health Dis. 2013;4(2):157–69.
Rietveld CA, Esko T, Davies G, Pers TH, Turley P, Benyamin B, et al. Common genetic variants associated with cognitive performance identified using the proxy-phenotype method. Proc Natl Acad Sci USA. 2014;111(38):13790–4.
St Pourcain B, Cents RA, Whitehouse AJ, Haworth CM, Davis OS, O’Reilly PF, et al. Common variation near ROBO2 is associated with expressive vocabulary in infancy. Nat Commun. 2014;5:4831.
van der Valk RJ, Kreiner-Moller E, Kooijman MN, Guxens M, Stergiakouli E, Saaf A, et al. A novel common variant in DCST2 is associated with length in early life and height in adulthood. Hum Mol Genet. 2014.
Mook-Kanamori DO, Miranda Geelhoed JJ, Steegers EA, Witteman JC, Hofman A, Moll HA, et al. Insulin gene variable number of tandem repeats is not associated with weight from fetal life until infancy: the Generation R Study. Eur J Endocrinol. 2007;157(6):741–8.
Geelhoed JJ, Mook-Kanamori DO, Witteman JC, Hofman A, van Duijn CM, Moll HA, et al. Variation in the IGF1 gene and growth in foetal life and infancy. The Generation R Study. Clin Endocrinol (Oxf). 2008;68(3):382–9.
van Houten VA, Mook-Kanamori DO, van Osch-Gevers L, Steegers EA, Hofman A, Moll HA, et al. A variant of the IGF-I gene is associated with blood pressure but not with left heart dimensions at the age of 2 years: the Generation R Study. Eur J Endocrinol. 2008;159(3):209–16.
Mook-Kanamori DO, de Kort SW, van Duijn CM, Uitterlinden AG, Hofman A, Moll HA, et al. Type 2 diabetes gene TCF7L2 polymorphism is not associated with fetal and postnatal growth in two birth cohort studies. BMC Med Genet. 2009;10:67.
Mook-Kanamori DO, Steegers EA, Uitterlinden AG, Moll HA, van Duijn CM, Hofman A, et al. Breast-feeding modifies the association of PPARgamma2 polymorphism Pro12Ala with growth in early life: the Generation R Study. Diabetes. 2009;58(4):992–8.
de Kort SW, Mook-Kanamori DO, Jaddoe VW, Hokken-Koelega AC. Interactions between TCF7L2 genotype and growth hormone-induced changes in glucose homeostasis in small for gestational age children. Clin Endocrinol (Oxf). 2010;72(1):47–52.
Geelhoed MJ, Steegers EA, Koper JW, van Rossum EF, Moll HA, Raat H, et al. Glucocorticoid receptor gene polymorphisms do not affect growth in fetal and early postnatal life. The Generation R Study. BMC Med Genet. 2010;11:39.
Maas JA, Mook-Kanamori DO, Ay L, Steegers EA, van Duijn CM, Hofman A, et al. Insulin VNTR and IGF-1 promoter region polymorphisms are not associated with body composition in early childhood: the generation R study. Horm Res Paediatr. 2010;73(2):120–7.
Geelhoed JJ, van Duijn C, van Osch-Gevers L, Steegers EA, Hofman A, Helbing WA, et al. Glucocorticoid receptor-9beta polymorphism is associated with systolic blood pressure and heart growth during early childhood. The Generation R Study. Early Hum Dev. 2011;87(2):97–102.
Mook-Kanamori DO, Ay L, Hofman A, van Duijn CM, Moll HA, Raat H, et al. No association of obesity gene FTO with body composition at the age of 6 months. The Generation R Study. J Endocrinol Invest. 2011;34(1):16–20.
Mook-Kanamori DO, Marsh JA, Warrington NM, Taal HR, Newnham JP, Beilin LJ, et al. Variants near CCNL1/LEKR1 and in ADCY5 and fetal growth characteristics in different trimesters. J Clin Endocrinol Metab. 2011;96(5):E810–5.
Sovio U, Mook-Kanamori DO, Warrington NM, Lawrence R, Briollais L, Palmer CN, et al. Association between common variation at the FTO locus and changes in body mass index from infancy to late childhood: the complex nature of genetic association through growth and development. PLoS Genet. 2011;7(2):e1001307.
Leermakers ET, Taal HR, Bakker R, Steegers EA, Hofman A, Jaddoe VW. A common genetic variant at 15q25 modifies the associations of maternal smoking during pregnancy with fetal growth: the generation R study. PLoS ONE. 2012;7(4):e34584.
Taal HR, van den Hil LC, Hofman A, van der Heijden AJ, Jaddoe VW. Genetic variants associated with adult blood pressure and kidney function do not affect fetal kidney volume. The Generation R Study. Early Hum Dev. 2012;88(9):711–6.
Tyrrell J, Huikari V, Christie JT, Cavadino A, Bakker R, Brion MJ, et al. Genetic variation in the 15q25 nicotinic acetylcholine receptor gene cluster (CHRNA5-CHRNA3-CHRNB4) interacts with maternal self-reported smoking status during pregnancy to influence birth weight. Hum Mol Genet. 2012;21(24):5344–58.
Gaillard R, Durmus B, Hofman A, Mackenbach JP, Steegers EA, Jaddoe VW. Risk factors and outcomes of maternal obesity and excessive weight gain during pregnancy. Obesity (Silver Spring). 2013;21(5):1046–55.
Luijk MP, Velders FP, Tharner A, van Ijzendoorn MH, Bakermans-Kranenburg MJ, Jaddoe VW, et al. FKBP5 and resistant attachment predict cortisol reactivity in infants: gene–environment interaction. Psychoneuroendocrinology. 2010;35(10):1454–61.
Luijk MP, Roisman GI, Haltigan JD, Tiemeier H, Booth-Laforce C, van Ijzendoorn MH, et al. Dopaminergic, serotonergic, and oxytonergic candidate genes associated with infant attachment security and disorganization? In search of main and interaction effects. J Child Psychol Psychiatry. 2011;52(12):1295–307.
Luijk MP, Tharner A, Bakermans-Kranenburg MJ, van Ijzendoorn MH, Jaddoe VW, Hofman A, et al. The association between parenting and attachment security is moderated by a polymorphism in the mineralocorticoid receptor gene: evidence for differential susceptibility. Biol Psychol. 2011;88(1):37–40.
Pluess M, Velders FP, Belsky J, van Ijzendoorn MH, Bakermans-Kranenburg MJ, Jaddoe VW, et al. Serotonin transporter polymorphism moderates effects of prenatal maternal anxiety on infant negative emotionality. Biol Psychiatry. 2011;69(6):520–5.
Szekely E, Herba CM, Arp PP, Uitterlinden AG, Jaddoe VW, Hofman A, et al. Recognition of scared faces and the serotonin transporter gene in young children: the Generation R Study. J Child Psychol Psychiatry. 2011;52(12):1279–86.
Cents RA, Tiemeier H, Velders FP, Jaddoe VW, Hofman A, Verhulst FC, et al. Maternal smoking during pregnancy and child emotional problems: the relevance of maternal and child 5-HTTLPR genotype. Am J Med Genet B Neuropsychiatr Genet. 2012;159B(3):289–97.
Tiemeier H, Velders FP, Szekely E, Roza SJ, Dieleman G, Jaddoe VW, et al. The Generation R Study: a review of design, findings to date, and a study of the 5-HTTLPR by environmental interaction from fetal life onward. J Am Acad Child Adolesc Psychiatry. 2012;51(11):1119-35 e7.
Velders FP, De Wit JE, Jansen PW, Jaddoe VW, Hofman A, Verhulst FC, et al. FTO at rs9939609, food responsiveness, emotional control and symptoms of ADHD in preschool children. PLoS ONE. 2012;7(11):e49131.
Velders FP, Dieleman G, Cents RA, Bakermans-Kranenburg MJ, Jaddoe VW, Hofman A, et al. Variation in the glucocorticoid receptor gene at rs41423247 moderates the effect of prenatal maternal psychological symptoms on child cortisol reactivity and behavior. Neuropsychopharmacology. 2012;37(11):2541–9.
Kok R, Bakermans-Kranenburg MJ, van Ijzendoorn MH, Velders FP, Linting M, Jaddoe VW, et al. The role of maternal stress during pregnancy, maternal discipline, and child COMT Val158Met genotype in the development of compliance. Dev Psychobiol. 2013;55(5):451–64.
Cents RA, Kok R, Tiemeier H, Lucassen N, Szekely E, Bakermans-Kranenburg MJ, et al. Variations in maternal 5-HTTLPR affect observed sensitive parenting. J Child Psychol Psychiatry. 2014;55(9):1025–32.
Pappa I, Mileva-Seitz VR, Szekely E, Verhulst FC, Bakermans-Kranenburg MJ, Jaddoe VW, et al. DRD4 VNTRs, observed stranger fear in preschoolers and later ADHD symptoms. Psychiatry Res. 2014.
van der Knaap NJ, El Marroun H, Klumpers F, Mous SE, Jaddoe VW, Hofman A, et al. Beyond classical inheritance: the influence of maternal genotype upon child’s brain morphology and behavior. J Neurosci. 2014;34(29):9516–21.
Windhorst DA, Mileva-Seitz VR, Linting M, Hofman A, Jaddoe VW, Verhulst FC, et al. Differential susceptibility in a developmental perspective: DRD4 and maternal sensitivity predicting externalizing behavior. Dev Psychobiol. 2014.
van der Valk RJ, Duijts L, Kerkhof M, Willemsen SP, Hofman A, Moll HA, et al. Interaction of a 17q12 variant with both fetal and infant smoke exposure in the development of childhood asthma-like symptoms. Allergy. 2012;67(6):767–74.
Raat H, van Rossem L, Jaddoe VW, Landgraf JM, Feeny D, Moll HA, et al. The Generation R study: a candidate gene study and genome-wide association study (GWAS) on health-related quality of life (HRQOL) of mothers and young children. Qual Life Res. 2010;19(10):1439–46.
Liebrechts-Akkerman G, Liu F, Lao O, Ooms AH, van Duijn K, Vermeulen M, et al. PHOX2B polyalanine repeat length is associated with sudden infant death syndrome and unclassified sudden infant death in the Dutch population. Int J Legal Med. 2014;128(4):621–9.
Bouthoorn SH, van Lenthe FJ, Kiefte-de Jong JC, Taal HR, Wijtzes AI, Hofman A, et al. Genetic taste blindness to bitter and body composition in childhood: a Mendelian randomization design. Int J Obes (Lond). 2014;38(7):1005–10.
Bouwland-Both MI, van Mil NH, Stolk L, Eilers PH, Verbiest MM, Heijmans BT, et al. DNA methylation of IGF2DMR and H19 is associated with fetal and infant growth: the generation R study. PLoS ONE. 2013;8(12):e81731.
van Mil NH, Steegers-Theunissen RP, Bouwland-Both MI, Verbiest MM, Rijlaarsdam J, Hofman A, et al. DNA methylation profiles at birth and child ADHD symptoms. J Psychiatr Res. 2014;49:51–9.
Saridjan NS, Huizink AC, Koetsier JA, Jaddoe VW, Mackenbach JP, Hofman A, et al. Do social disadvantage and early family adversity affect the diurnal cortisol rhythm in infants? The Generation R Study. Horm Behav. 2010;57(2):247–54.
Luijk MP, Saridjan N, Tharner A, van Ijzendoorn MH, Bakermans-Kranenburg MJ, Jaddoe VW, et al. Attachment, depression, and cortisol: deviant patterns in insecure-resistant and disorganized infants. Dev Psychobiol. 2010;52(5):441–52.
Kiefte-de Jong JC, Saridjan NS, Escher JC, Jaddoe VW, Hofman A, Tiemeier H, et al. Cortisol diurnal rhythm and stress reactivity in constipation and abdominal pain: the Generation R Study. J Pediatr Gastroenterol Nutr. 2011;53(4):394–400.
Saridjan NS, Henrichs J, Schenk JJ, Jaddoe VW, Hofman A, Kirschbaum C, et al. Diurnal cortisol rhythm and cognitive functioning in toddlers: the Generation R Study. Child Neuropsychol. 2014;20(2):210–29.
Saridjan NS, Velders FP, Jaddoe VW, Hofman A, Verhulst FC, Tiemeier H. The longitudinal association of the diurnal cortisol rhythm with internalizing and externalizing problems in pre-schoolers. The Generation R Study. Psychoneuroendocrinology. 2014;50C:118–29.
Labout JA, Duijts L, Arends LR, Jaddoe VW, Hofman A, de Groot R, et al. Factors associated with pneumococcal carriage in healthy Dutch infants: the generation R study. J Pediatr. 2008;153(6):771–6.
Lebon A, Labout JA, Verbrugh HA, Jaddoe VW, Hofman A, van Wamel W, et al. Dynamics and determinants of Staphylococcus aureus carriage in infancy: the Generation R Study. J Clin Microbiol. 2008;46(10):3517–21.
Lebon A, Labout JA, Verbrugh HA, Jaddoe VW, Hofman A, van Wamel WJ, et al. Role of Staphylococcus aureus nasal colonization in atopic dermatitis in infants: the Generation R Study. Arch Pediatr Adolesc Med. 2009;163(8):745–9.
Verhaegh SJ, Lebon A, Saarloos JA, Verbrugh HA, Jaddoe VW, Hofman A, et al. Determinants of Moraxella catarrhalis colonization in healthy Dutch children during the first 14 months of life. Clin Microbiol Infect. 2010;16(7):992–7.
Verkaik NJ, Lebon A, de Vogel CP, Hooijkaas H, Verbrugh HA, Jaddoe VW, et al. Induction of antibodies by Staphylococcus aureus nasal colonization in young children. Clin Microbiol Infect. 2010;16(8):1312–7.
Lebon A, Moll HA, Tavakol M, van Wamel WJ, Jaddoe VW, Hofman A, et al. Correlation of bacterial colonization status between mother and child: the Generation R Study. J Clin Microbiol. 2010;48(3):960–2.
Labout JA, Duijts L, Lebon A, de Groot R, Hofman A, Jaddoe VV, et al. Risk factors for otitis media in children with special emphasis on the role of colonization with bacterial airway pathogens: the Generation R study. Eur J Epidemiol. 2011;26(1):61–6.
Verhaegh SJ, Snippe ML, Levy F, Verbrugh HA, Jaddoe VW, Hofman A, et al. Colonization of healthy children by Moraxella catarrhalis is characterized by genotype heterogeneity, virulence gene diversity and co-colonization with Haemophilus influenzae. Microbiology. 2011;157(1):169–78.
Ye X, Pierik FH, Hauser R, Duty S, Angerer J, Park MM, et al. Urinary metabolite concentrations of organophosphorous pesticides, bisphenol A, and phthalates among pregnant women in Rotterdam, the Netherlands: the Generation R study. Environ Res. 2008;108(2):260–7.
Ye X, Pierik FH, Angerer J, Meltzer HM, Jaddoe VW, Tiemeier H, et al. Levels of metabolites of organophosphate pesticides, phthalates, and bisphenol A in pooled urine specimens from pregnant women participating in the Norwegian Mother and Child Cohort Study (MoBa). Int J Hyg Environ Health. 2009;212(5):481–91.
Snijder CA, Heederik D, Pierik FH, Hofman A, Jaddoe VW, Koch HM, et al. Fetal growth and prenatal exposure to bisphenol A: the generation R study. Environ Health Perspect. 2013;121(3):393–8.
Jusko TA, Shaw PA, Snijder CA, Pierik FH, Koch HM, Hauser R, et al. Reproducibility of urinary bisphenol A concentrations measured during pregnancy in the Generation R Study. J Expo Sci Environ Epidemiol. 2014;24(5):532–6.
El Marroun H, Tiemeier H, Jaddoe VW, Hofman A, Verhulst FC, van den Brink W, et al. Agreement between maternal cannabis use during pregnancy according to self-report and urinalysis in a population-based cohort: the Generation R Study. Eur Addict Res. 2011;17(1):37–43.
Rours GI, Duijts L, Moll HA, Arends LR, de Groot R, Jaddoe VW, et al. Chlamydia trachomatis infection during pregnancy associated with preterm delivery: a population-based prospective cohort study. Eur J Epidemiol. 2011;26(6):493–502.
van Mil NH, Tiemeier H, Bongers-Schokking JJ, Ghassabian A, Hofman A, Hooijkaas H, et al. Low urinary iodine excretion during early pregnancy is associated with alterations in executive functioning in children. J Nutr. 2012;142(12):2167–74.
Ghassabian A, Steenweg-de Graaff J, Peeters RP, Ross HA, Jaddoe VW, Hofman A, et al. Maternal urinary iodine concentration in pregnancy and children’s cognition: results from a population-based birth cohort in an iodine-sufficient area. BMJ Open. 2014;4(6):e005520.
Available from: Website: http://www.healthcare.philips.com/nl_nl/products/healthcare_informatics/products/LaboratoriumIT/.
Acknowledgments
The Generation R Study is conducted by the Erasmus Medical Center in close collaboration with the School of Law and Faculty of Social Sciences of the Erasmus University Rotterdam, the Municipal Health Service Rotterdam area, Rotterdam, the Rotterdam Homecare Foundation, Rotterdam and the Stichting Trombosedienst & Artsenlaboratorium Rijnmond (STAR-MDC), Rotterdam. We gratefully acknowledge the contribution of children and parents, general practitioners, hospitals, midwives and pharmacies in Rotterdam. The authors thank Ms. Letty Jacobs, Mr. Ronald van den Nieuwenhof, Ms. Patricia van Sichem-Maeijer, and Ms. Karien Toebes for their assistance in logistics and data management. We thank Mr. Andy A.L.J. van Oosterhout, Ms. Jeannette Vergeer-Drop, Ms. Andrea J.M. Vermeij-Verdoold from the Genetic Epidemiology Laboratory, Erasmus MC for their assistance and advice in logistics and laboratory procedures. From the Human Genotyping Facility of the Genetic Laboratory of the Department of Internal Medicine, Erasmus MC, the Netherlands, we thank the following people: A: Ms. Mila Jhamai, Mr. Pascal Arp, and Mr. Marijn Verkerk for their help in creating the Generation R GWAS database; B: Mr. Michael Verbiest, Ms. Mila Jhamai, Ms. Sarah Higgins, Mr. Marijn Verkerk, Dr. Lisette Stolk and Dr. Janine F. Felix for their help in creating the Generation R EWAS database, and C: Ms. Mila Jhamai, Ms. Sarah Higgins, and Mr. Marijn Verkerk for their help in creating the Generation R Exome Chip database, and Carolina Medina-Gomez, BSc, Lennart Karsten, BSc, and Dr. Linda Broer for QC and variant calling. We thank Dr. Robert Kraaij from the Genetic Laboratory of the Department of Internal Medicine, Erasmus MC, the Netherlands in coordinating the faeces collection and 16S microbiome analysis. The general design of Generation R Study is made possible by financial support from the Erasmus MC, Rotterdam, the Erasmus University Rotterdam, the Netherlands Organization for Health Research and Development (ZonMw), the Netherlands Organisation for Scientific Research (NWO), the Ministry of Health, Welfare and Sport. O.H. Franco works in ErasmusAGE, a center for aging research across the life course funded by Nestlé Nutrition (Nestec Ltd.); Metagenics Inc.; and AXA. Nestlé Nutrition (Nestec Ltd.); Metagenics Inc.; and AXA had no role in design and conduct of the study; collection, management, analysis, and interpretation of the data; and preparation, review or approval of the manuscript. Vincent Jaddoe received an additional grant from the Netherlands Organization for Health Research and Development (NWO, ZonMw-VIDI 016.136.361).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kruithof, C.J., Kooijman, M.N., van Duijn, C.M. et al. The Generation R Study: Biobank update 2015. Eur J Epidemiol 29, 911–927 (2014). https://doi.org/10.1007/s10654-014-9980-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10654-014-9980-6