Abstract
Inland (Menidia beryllina) and tidewater silversides (M. peninsulae) are the most common of the Menidia fishes along the Texas coast, USA. Though genetically distinct, these species are morphologically very similar and often segregate by salinity with M. beryllina occupying fresh to brackish salinities and M. peninsulae occupying brackish to oceanic salinities. Their ranges are known to overlap, suggesting the possibility for interspecific competition, and significant hybridization has been documented. This study utilizes an integrative approach of morphological and molecular techniques to examine if divergent environmental constraints (inland freshwater impoundments versus coastal saline bays) could potentially result in parallel changes in morphology among populations of Menidia spp. occupying similar habitat types or if Menidia spp. occupying the same areas exhibit divergent changes in morphology (i.e., character displacement) suggesting historic interspecific competition. While both coastal (sympatric) and inland (allopatric) forms of M. beryllina showed significant morphological separation from M. peninsulae, coastal populations were slightly more similar to M. peninsulae than inland populations. Intraspecific comparisons revealed significant morphological and molecular variation among M. beryllina populations and limited variation among M. peninsulae populations. These findings coupled with a review of past Menidia work suggest that salinity regimes may drive the presence of M. beryllina in coastal bays, with increased salinity restricting movement of this species among river drainages resulting in both interspecific and intraspecific isolation. Thus, morphological and molecular variation in Menidia seems to be driven primarily by environmental conditions with interspecific competitive interactions of little significance. This study is the most comprehensive comparison of these species to date and conclusions may be applied broadly to the process and sequence of speciation of Menidia fishes in the Gulf of Mexico.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The genus Menidia is comprised of New World silverside fishes (Family: Atherinopsidae), with eight marine, fresh, and brackish water species ranging from the North Atlantic through the Gulf of Mexico to the South Atlantic (Froese and Pauly 2013). Four species are known from coastal Texas, USA: Menidia beryllina, M. peninsulae, M. clarkhubbsi, and M. audens. In Texas, M. audens has only been identified along the lower Sabine River (north of river mile 50) and is not known to venture into the saltwater environment (Suttkus et al. 2005). Menidia beryllina generally occupy inland freshwater and coastal brackish water habitats and M. peninsulae occupy more saline, oceanic habitats in the coastal region (Echelle and Echelle 1997; Hoese and Moore 1998). Menidia clarkhubbsi is much less common and was first described on the Texas coast (Echelle and Mosier 1982) as an all-female unisexual species complex, which presumably resulted from the hybridization of ancestral forms of M. peninsulae and M. beryllina (Echelle et al. 1983).
Taxonomic differentiation of Menidia fishes has often been unclear. Methods of meristic and morphometric distinction of M. beryllina and M. peninsulae have been addressed by a number of researchers (Gosline 1948; Edwards et al. 1978; Chernoff et al. 1981; Echelle and Mosier 1982) though the universality and utility of these methods is debated. Traditional meristic counts have been shown to largely overlap between the two species (Hoese and Moore 1998). Johnson (1975) first suggested that M. beryllina and M. peninsulae could be distinguished via the posterior extension of the gas bladder though Edwards et al. (1978) further assessed the utility of this trait and found that its posterior extension could be altered with salinity, compromising its taxonomic usefulness. In contrast, Echelle and Mosier (1982) demonstrated the taxonomic utility of the gas bladder in differentiating M. beryllina from M. peninsulae even in the presence of M. clarkhubbsi. In spite of the disagreement in meristic divergence of these species, it is generally agreed they are genetically distinct (Johnson 1975; Echelle and Mosier 1982) and tend to separate based on the salinity of surrounding waters (Echelle and Echelle 1997; Hoese and Moore 1998; Suttkus and Mettee 1998). Echelle and Echelle (1997) assessed patterns of sympatric Menidia spp. distributions in Copano Bay, Texas, USA and found that M. beryllina dominated the fresher waters within the bay and tidal creeks, while M. peninsulae was abundant within the more saline waters of the bay, though the two species were observed in the same sampling areas on multiple occasions. In Perdido Bay, Alabama, USA, Suttkus and Mettee (1998) observed similar distributional patterns of Menidia spp. abundance relative to salinity, though in this case, the two species were completely segregated among samples.
Fluker et al. (2011) examined morphological and molecular variation among allopatric and sympatric populations of M. audens and M. beryllina along an estuarine-freshwater gradient. Using geometric morphometrics they observed clinal variation in body shape of M. audens with distance upstream, but such variation was not observed for M. beryllina. Nonetheless, they noted significant differences in body shape between the two species throughout the range of the study. The patterns of morphological divergence observed by Fluker et al. (2011) outline the potential for similar morphological divergence among populations of M. beryllina and M. peninsulae. Speciation in atherinopsids is thought to have been largely driven by geographic separation and adaptation to differing environments (Gosline 1948; Barbour 1973; Bamber and Henderson 1988; Fluker et al. 2011), though it’s clear that there remains a high level of intraspecific phenotypic plasticity among many populations.
Given that populations of M. peninsulae and M. beryllina are thought to occur in sympatry at the estuarine-freshwater interface, character displacement (Brown and Wilson 1956; Grant 1972) may occur in response to resource competition. Thus, Menidia spp. occupying the same areas (i.e., occurring in sympatry) may exhibit divergent changes in morphology, which would suggest historic interspecific competition. In contrast, divergent environmental constraints (inland freshwater impoundments versus coastal saline bays and river mouths) could potentially result in parallel changes in morphology among populations of Menidia spp. occupying similar habitat types via phenotypic plasticity or parallel evolution via ecological selection. More specifically, it would be interesting to know whether there are distinct morphological characters associated with M. beryllina that persist across distinct freshwater drainage systems, and if these characters are absent in the coastal M. peninsulae. Numerous factors have been shown to affect external shape development among species (e.g. diet, habitat, interaction, climate, other stressors), however between species morphological differences may be due to functional trait differences (Loy et al. 2000; Borcherding and Magnhagen 2008; Cakmak and Alp 2010).
In this study, meristic and truss-based morphological body measurements were used to characterize body size and shape in samples of Menidia spp. from inland and coastal waters of Texas, USA. These data were used to determine whether there are quantifiable divergent phenotypes between interspecific and intraspecific populations of M. beryllina and M. peninsulae, or even among geographically isolated populations of M. beryllina. The morphology data were accompanied by a phylogenetic analysis of mitochondrial DNA (mtDNA) haplotype data to test the hypothesis that observed phenotypic differences can be significantly related to genetic lineage. The paradigm of historical isolation as the likely mechanism driving speciation in this system was thus evaluated using the genetic data; such a mechanism would necessarily dictate that contemporary hybrids are the result of secondary contact. This study helps to elucidate the nature of adaptation and interaction and is the most comprehensive comparison of these species to date. Additionally, these data further elucidate the process and sequence of speciation and may apply broadly to Menidia fishes in the Gulf of Mexico.
Methods
Sample collection
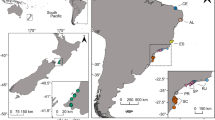
Coastal samples of Menidia spp. were collected via bag seine in coordination with the Texas Parks and Wildlife Department (TPWD) routine Marine Resource Monitoring Program (Martinez-Andrade and Fisher 2012). Fish were collected from a range of salinities in the Upper Laguna Madre (ULM), Aransas Bay (AB), Galveston Bay (GB), Sabine Lake (SL), and Cedar Lakes (CL; Fig. 1). Additional collections were made from inland populations of M. beryllina in Lake Texana (TEX), Lake Choke Canyon (CHO), Lake Mathis (MAT), Lake Houston (HOU), and Lake Somerville (SOM). Sampling occurred continuously from February 2014 to August 2015. Fish were immediately placed in sealed bags and on ice after collection and transported back to the field station where they were frozen for genetic and morphological analysis.
Morphological analysis
A sub-sample of fish to be genetically identified (n = 161) were thawed and processed for meristic counts and truss-based morphological body measurements (no hybrids were used for the morphological portion of the analysis). Fish used for morphology tests were examined for freshness and firmness prior to photographs, any misshapen fish were excluded from these analyses. Fish from HOU and GB were not included in this portion of the analysis due to poor condition. Selected meristic counts were based on past studies, which attempted to differentiate these two species of Menidia (Hoese and Moore 1998) including counts of predorsal scales, first dorsal fin spines, second dorsal fin soft rays, and anal fin soft rays. Condition (appearance and extension) of the swim bladder was also examined but was excluded from the formal analysis due to difficulties in quantifying this trait (Edwards et al. 1978; Echelle and Echelle 1997). A second reader was periodically utilized to check for accuracy of counts. Morphological body measurements were employed in a truss-based system to examine whether these body measurements could be used to significantly distinguish species. Digital photographs of the left sides of fish were taken using a 10.2 megapixel digital camera with a 18-55 mm Nikkor zoom lens (D40x, Nikon Corp., Tokyo, Japan), following procedures outlined in Muir et al. (2012). Photographs were processed and nine landmarks were identified that yielded 20 truss-style measurements (Fig. 2). Measurements were taken between identified landmarks using SigmaScan Pro 5.0 software (Systat Inc., Richmond, California).
Truss style analysis differs from the more recent geometric morphometric approach in that it is founded in distance-based measures of morphological character while the geometric morphometric approach is founded on shape-based measures of morphological character (Strauss and Bookstein 1982; Webster and Sheets 2010). While some authors suggest that the geometric morphometric approach is more robust and allows for a better visualization of shape differences (Parsons et al. 2003; Maderbacher et al. 2008), the truss-based approach is more useful in assessing the degree of differentiation as it relates to functional morphology (Bhagat et al. 2011). Additionally, comparisons of the two methods have found only minor deviation in final results (Bhagat et al. 2011).
Both meristic count variables and truss style morphological body measurement variables were combined for all statistical analyses. All morphological body measurements were standardized by standard length. Principle component analysis (PCA) was used to reduce multivariate dimensionality to uncorrelated principle components (PCs). Prior to PCA, variables were standardized to a mean of zero and variance of one to account for the differing scales used for meristics (count) and morphological measurements (length in mm). We thus used the first ten PCs for all downstream permutational multivariate analysis of variance (perMANOVA) and discriminant analyses (DA; Geiger et al. 2016). Hierarchical perMANOVA (Anderson 2001; 1000 permutations) was used to assess differences in body meristics and measurements between species (with collection site nested within species). For the purposes of this study, M. beryllina and M. peninsulae were assumed to occur in sympatry in the coastal bays (ULM, AB, CL, GB, and SL) and M. beryllina population from inland lakes (TEX, CHO, MAT, HOU, and SOM) considered allopatric to coastal populations of M. peninsulae. In other words the following comparisons are inland M. beryllina versus M. peninsulae (which only occur on the coastal bays) and coastal M. beryllina versus M. peninsulae. Thus two analyses were conducted. The first analysis was between allopatric M. peninsulae (ULM, AB, and CL) and M. beryllina (TEX, CHO, MAT, and SOM) and the second analysis was conducted between sympatric M. peninsulae (ULM, AB, and CL) and M. beryllina (CL and SL). To avoid inflation of the type-I error rate, we utilized a Bonferroni correction of the alpha values (α = 0.05/2 = 0.025). As PCA is simply a dimensionality reduction technique and does not attempt to categorize data based on a priori group designations, post-hoc discriminant analysis with jackknifed cross-validation procedures among allopatric M. peninsulae and M. beryllina and sympatric M. peninsulae and M. beryllina was used to further examine and validate these relationships. Additionally, these DA procedures were applied among M. beryllina collection sites and M. peninsulae collection sites on the first ten PCs of these data sets to examine intraspecific variation in body meristics and measurements. Principle component analysis and DA were conducted in JMP Version 9.0.0 (SAS Institute Inc., Cary, NC 1989–2007) while perMANOVA and all plotting were conducted in R Version 3.1.1 (R Core Team 2014).
Genetic species identification
All specimens were thawed prior to excision of tissue for DNA isolation. Each specimen was sub-sampled (approximately 20 mg tissue) for DNA isolation. The DNA was extracted from tissue sample using Gentra Puregene Tissue Kits (Qiagen Inc., Valencia, CA), following the manufacturer’s instructions. Final elution volume was 200 uL for all DNA samples.
The cytochrome B locus of the mitochondrial genome was used to determine species in Menidia specimens. This locus was first amplified from genomic DNA using polymerase chain reaction (PCR). The primers used were novel primers devised by alignment of Menidia spp. using online sequences (forward 5′-CGGCTGTACCTTACGTGGGCAACGC, reverse 5′-CTCTGAGGCTCTGAGCTACCAGGGC). Aliquots of each PCR product were purified with Exo-Sapit® PCR purification reagent (Affymetrix, Inc., Santa Clara, CA), and directly sequenced with the forward primer from PCR. Sequencing reactions were carried out in 20 μL volumes with DTCS Quick Start Master Mix (BeckmanCoulter Inc., Brea, CA), followed by sequencing with a CEQ 8000 capillary sequencer (Beckman Coulter, Inc.). Sequence traces were aligned with the software package Sequencher (vers. 5.2.4; Gene Codes Corp., Ann Arbor, MI). Species determination was made after comparison with published online sequences of both M. peninsulae (NCBI accession #KC736345.1 and #JQ282038.1) and M. beryllina (NCBI accession #KC736408.1 and #JQ282033.1).
Because hybridization is known to occur between Menidia spp. (Echelle and Echelle 1997), it is expected that mtDNA haplotype data may occasionally be misleading for species identification, particularly in the presence of F1 hybrids or backcrosses. For this reason, we explored the microsatellite data set initially developed for M. menidia by Sbrocco and Barber (2011) for potential diagnostic markers using a small subset of specimens. Of the ten described markers from Sbrocco and Barber (2011), two were identified in initial sampling that amplified both species and appeared to have diagnostic range differences. We used three non-mixed samples of M. peninsulae (two from upper Laguna Madre and one from Aransas Bay; overall n = 96) and two non-mixed samples of M. beryllina (one from Lake Somerville, and one from Lake Texana; overall n = 115) to characterize allele frequencies in these two markers. Non-mixed samples were designated as those sampled from the same locale, on the same day, that had only one species mtDNA lineage represented. At one locus (Mm108), the species were nearly fixed for different alleles (M. peninsulae allele 235 frequency = 0.964; M. beryllina allele 239 frequency = 0.963), with some overlap between the two, as well as the presence of rare alleles in both species. At the second locus (Mm204), there was a range difference between species, with M. peninsulae alleles occurring generally between 170 and 216 bp, and M. beryllina alleles occurring between 216 and 300 bp, although there was occasional overlap of these ranges between the species at the 216 allele. For hybrid identification, we assumed that only specimens that had diagnostic alleles from both species at both loci were first-generation hybrids. Individuals which had diagnostic hybrid genotypes at one locus but not the other, or which had microsatellite genotypes of one species and mtDNA haplotypes of the other species, were considered to be later-generation hybrids.
Intra- and interspecific molecular variation
Molecular variation within and among Menidia spp. was examined by statistical analysis of mtDNA haplotype distributions. For these analyses, 400 consensus base pairs were examined in a subsample of the individuals observed in this study (n = 775). The number of haplotypes and haplotype diversity were assessed using the software DnaSP version 4.10 (Rozas et al. 2003). A phylogenetic network of haplotypes was constructed for each of two divergent lineages observed in the initial analysis. Networks were constructed using the programs Network 4.6 and Network Publisher 2.0 (Fluxus Technology Ltd., UK). Networks were used to qualitatively examine the geographic relationships between sampled haplotypes within each lineage.
The analysis of molecular variance (AMOVA, Excoffier et al. 1992) was used to quantify statistical association of haplotypes among sample areas. Because of the deep divergence observed initially between species lineages, AMOVA was used to assess genetic divergence independently for each lineage. The mtDNA haplotypes of hybrids were sorted by lineage and were included in the AMOVA regardless of species status. Conducting the analysis in this way assumes that cross-lineage hybrid haplotypes arose from recent hybridization, rather than historical incomplete lineage sorting. We believe this to be a valid assumption given the high rate of hybrids found in previous studies of Menidia spp. in Texas coastal waters (Echelle et al. 1988; Echelle and Echelle 1997), and the otherwise deep genetic divergence between lineages indicated here.
The distance method chosen for the minimum spanning network in AMOVA was simple pairwise difference. Individuals were grouped by sample (geographic area), and AMOVA was carried out using the software Arlequin 3.5 (Excoffier and Lischer 2010). The AMOVA model was permuted across samples, such that the variation in distribution of haplotypes was partitioned into among-sample and within-sample components. The variance components were used to generate an estimate of θst, roughly corresponding to the percentage of variation accounted for by among-sample comparisons. The statistical significance of θst was tested using 1000 random data permutations in Arlequin, under the null hypothesis of no differentiation among samples (Ho: θst = 0; Ha: θst > 0). Significant values of θst were further interpreted using post-hoc examination of pairwise sample estimates of θst. Pairwise θst were estimated in Arlequin using the pairwise difference method, and 1000 permutations of the data among samples were used to assess statistical significance, again under the null hypothesis of θst = 0. The cutoff value for statistical significance (α = 0.05) was adjusted to account for multiple tests performed simultaneously using a sequential Bonferroni procedure (Holm 1979).
Results
A total of 904 Menidia spp. were collected among the ten sampling locations (Table 1). This included 518 pure M. peninsulae, all of which were sampled at coastal locations (GB, CL, AB, and ULM). There were also 266 pure M. beryllina. These were found primarily in inland lakes (HOU, SOM, TEX, MAT, and CHO), although two were found in coastal CL, and 18 were found in coastal SL. Finally, 120 F1 hybrids and back-crosses were identified in two coastal sites, CL (n = 105), and ULM (n = 15). Coastal CL was the only location where pure forms of both species were found along with hybrids. Of 205 fish collected at CL, 48%were genetically identified as M. peninsulae, 27 % were identified as back-crossed hybrids, 24 % as F1 hybrids, and only two M. beryllina were identified.
Morphological analysis
The first 10 PCs on allopatric populations of M. beryllina and M. peninsulae accounted for 93.3 % of variation. Results from the perMANOVA found allopatric populations of these species to be morphologically distinct (p = 0.0009) with the nested site factor also significant (p = 0.0009; Table 2). The DA between allopatric populations yielded a cross-validation error rate of 2.8 % with PC1 and PC3 largely driving the group separation (Fig. 3). PC1 and PC3 were primarily loaded by mid-body measurements and dorsal scale counts (Fig. 4). Similarly, the first ten PCs on sympatric populations of M. beryllina and M. peninsulae accounted from 93.5 % of variation. Results from the perMANOVA suggested that sympatric populations of these species are also morphologically distinct (p = 0.009, Table 3), though here, the nested site factor was not found to be significant (p = 0.996, Table 3). The DA between sympatric populations yielded a cross-validation error rate of 5.6 % with group separation again largely driven by PC1 and PC3 (Fig. 5). PC1 and PC3 were, again, primarily loaded by mid-body measurements and dorsal scale counts (Fig. 4).
Heaviest loadings (red) on principle component (PC) 1 and PC3 for principle component analysis on meristic counts and morphological body measurements of allopatric (left) and sympatric (right) populations of Menidia beryllina and M. peninsulae. PC1 and PC3 were found to contribute most to group (species) separation in the discriminant analysis
The first 10 PCs from the PCA on M. beryllina data accounted for 91.3 % of variation. The DA for M. beryllina among sites yielded a cross-validation error rate of 21.8 %. Menidia beryllina from SOM were most dissimilar from all other sites while fish from CHO and MAT were almost indistinguishable. Menidia beryllina from TEX were most similar to coastal M. beryllina from SL (Fig. 6). Group separation was largely driven by PC1 and PC6. PC1 and PC6 were primarily loaded by mid-body measurements and measurements surrounding the head and jaw area (Fig. 7). The first ten PCs from the PCA on M. peninsulae data accounted for 93.9 % of variation. Less group separation was apparent among sites for M. peninsulae with the DA yielding a cross-validation error rate of 32.1 % (Fig. 8). Group separation was largely driven by PC1 and PC6. PC1 and PC6 were, again, primarily loaded by mid-body measurements and measurements surrounding the head and jaw area (Fig. 7). Full outputs from all PCAs are given in Appendix A and full outputs from all DAs are given in Appendix B.
Scores (and 95 % confident interval ellipses) from a discriminant analysis on the first ten principle components of meristic counts and morphological body measurements among Menidia beryllina collection locations: Lake Somerville (SOM; x, dotted black line), Lake Choke Canyon (CHO;  , solid red line), Mathis Lake (MAT; ο,
dashed black line), Sabine Lake (SL; ο, dashed red line), and Lake Texana (TEX; •,
solid black line)
, solid red line), Mathis Lake (MAT; ο,
dashed black line), Sabine Lake (SL; ο, dashed red line), and Lake Texana (TEX; •,
solid black line)
Heaviest loadings (red) on principle component (PC) 1 and PC6 for principle component analysis on meristic counts and morphological body measurements of Menidia beryllina (left) and M. peninsulae (right). PC1 and PC6 were found to contribute most to group (site) separation in the discriminant analysis
Scores (and 95 % confidence interval ellipses) from a discriminant analysis on the first ten principle components of meristic counts and morphological body measurements among Menidia peninsulae collection locations Aransas Bay (AB; •, solid black line), Cedar Lakes (CL;  , solid red line), and Upper Laguna Madre (ULM; ο, dashed black line)
, solid red line), and Upper Laguna Madre (ULM; ο, dashed black line)
Though not quantified and included in the formal analysis, condition of the swim bladder was found to be species specific among all comparisons. Menidia peninsulae was qualitatively characterized by opaque coloration and little extension beyond the anal fin spine while M. beryllina was characterized by transparent coloration and extension well beyond the anal fin spine.
Molecular analysis
There were two main lineages in the mtDNA data set that were separated from one another by 21 nucleotide substitutions. One of these lineages (lineage “MB”) was observed in all of the M. beryllina sampled in inland lakes, and is assumed here to be characteristic of the historical mtDNA profile of this species (Fig. 9). There were 30 haplotypes observed in lineage MB, in 293 sampled individuals (Table 4). In addition to inland lake samples, lineage MB was present in a single pure M. beryllina collected in CL, in 18 pure M. beryllina individuals collected in coastal SL, and was also present in some presumptive hybrid and back-crossed individuals from CL and ULM. When individuals containing lineage MB were pooled in these samples, there were highly significant differences in the distribution of mtDNA haplotypes among samples based on AMOVA (θst =0.591, P < 0.0001).
The mtDNA phylogenetic network for lineage MB, or Menidia beryllina sampled in inland and coastal areas in Texas. Haplotypes are represented as pie charts, the size of the chart roughly correlates to the respective frequency of each haplotype, and links between charts represent phylogenetic linkage. Reticulations on the network represent unresolved linkage among multiple haplotypes and/or unsampled (inferred) haplotypes. Haplotype charts are color-coded based on relative presence of each of five geographic samples containing lineage MB. The figure legend titles are arranged in descending order based on geographic distance from the coast
The post-hoc examination of θst calculated among pairwise samples of lineage MB suggested that the significant AMOVA result was driven primarily by divergent samples observed in the coastal ULM, and in inland lakes SOM, CHO, and MAT (Table 5). These samples were significantly diverged from other inland and coastal samples, and from one another, and the magnitude of divergence associated with these samples was an order of magnitude higher than among other such comparisons. Pairwise estimates of divergence among inland sites TEX and HOU and coastal sites CL and SL were markedly lower than comparisons among other sites. Qualitative examination of the haplotype network for lineage MB suggests two additional findings with regard to ULM, SOM, CHO and MAT: (1) these samples were comprised primarily of individuals that carry mtDNA haplotypes that are rare in other samples, (2) the overall number of haplotypes (haplotype diversity) sampled in these areas was low in comparison to other samples. The main haplotypes from SOM and ULM appeared to be phylogenetically related to each other. Similarly the primary haplotypes observed in CHO and MAT appeared to be phylogenetically linked.
The second observed lineage “MP” was present in most coastal individuals sampled in CL, ULM, GB and AB, excluding those individuals mentioned above. There were 104 haplotypes observed among 482 individuals. Haplotype diversity was high in all four sites where MP was found (Table 4). There was no evidence for genetic divergence among sampling locales in lineage MP (Fig. 10). The AMOVA of this lineage included four coastal sample sites (AB, CL, GB and ULM), and indicated no significant divergence among samples (θst = 0.003, P = 0.143), nor were there significant pairwise values of θst between any two samples after adjustment for multiple simultaneous tests.
The mtDNA phylogenetic network for lineage MP, or Menidia peninsulae sampled in coastal areas in Texas. Because of initial complexity contained in the MP network, star-contraction was employed, in which star-like groups on the edges of the network were collapsed on themselves to simplify the network. Haplotypes are represented as pie charts, the size of the chart roughly correlates to the frequency of each haplotype, and links between charts represent phylogenetic linkage. Haplotype charts are color-coded based on relative presence of each of three geographic samples containing lineage MP. Reticulations on the network represent unresolved linkage among haplotypes
Discussion
While the validity of morphological and meristic distinction of M. beryllina and M. peninsulae has often been debated (Gosline 1948; Edwards et al. 1978; Chernoff et al. 1981; Echelle and Mosier 1982), the suite of morphological and meristic traits examined here were able to consistently differentiate allopatric and sympatric forms of these species, though sympatric populations tend to be slightly more similar. Specific traits driving morphological differentiation between allopatric forms of these species were similar to those observed for sympatric forms in both the heaviest loaded PCs in the DA and specific variables loaded on those PCs. No single variable could be said to drive group differentiation between species for either allopatric or sympatric comparisons. This suggests that it is the general anatomical form that best differentiates these species, making identification outside of a multivariate framework very difficult. Additionally, this precludes drawing direct comparisons to differential functionality of these traits between species. Nonetheless, the heaviest loaded body measurements on the PCs thought to impact species separation were primarily located in the mid-section of the body surrounding the dorsal and anal fins (Fig. 4). Such measurements may be largely influenced by expansion and contraction of the gas bladder with salinity, which has often been used as a differentiating characteristic (Johnson 1975). Edwards et al. (1978) deemed this characteristic to be inappropriate given its physiological plasticity with salinity. Salinity among the coastal sites and between coastal and inland sites varied substantially, thus the variation observed among allopatric forms of M. peninsulae may be due at least partially to expansion and contraction of the gas bladder with salinity. The qualitative nature of the gas bladder data collected here precludes meaningful comparison of this trait among conspecifics, though it does support the observed variation between allopatric and sympatric forms of M. peninsulae and M. beryllina.
Regardless, the comparisons of group separation by meristic and morphological variables in allopatric and sympatric forms of these species suggest that sympatric (coastal) populations are not competing to any significant extent that would result in character displacement (Brown and Wilson 1956; Grant 1972). In short, character displacement is said to occur when two or more species that exist in sympatry display divergent characteristics from one another as compared to allopatric forms of the same species (Brown and Wilson 1956). Such divergence is an adaptation to avoid competition in sympatry that is not experienced in allopatry. Grant (1972) expanded on this phenomena and suggested that interactive segregation (behavior or ecological) often precedes morphological character displacement. McEachran and Martin (1977) observed this phenomenon among species of Rajid skate in the Gulf of St. Lawrence and observed that, while a number of closely related species coexisted in the Gulf of St. Lawrence, only two existed in sympatry and exhibited morphological character displacement. They speculated that the remaining species were able to establish contiguous distributions so as to avoid competition for the same resources (i.e., competitive exclusion), though limited niche space resulted in distributional overlap and morphological character displacement for the two species in question (McEachran and Martin 1977). It is possible that, while M. beryllina and M. peninsulae have been observed in the same habitats (e.g., Echelle and Echelle 1997), coarse segregation based on salinity may be sufficient to preclude significant competition between the two species. Nonetheless, given the spatially course nature of our sample regimented, addition study is certainly warranted to further test these observations.
Additionally, our findings indicate a significant amount of meristic and morphological variation among populations of M. beryllina. The DA suggests that group differentiation among M. beryllina collection sites may be roughly related to distance, with populations that are geographically closer to one another being anatomically more similar (e.g., CHO and MAT) than more geographically isolated populations (e.g., SOM). While, again, no single trait could be said to drive this group separation (among sites), measurements in the mid-body region and head/jaw region were heaviest loaded on the PCs most influencing the DA (Fig. 7). Group separation by head/jaw measurements may be more ecologically meaningful than the mid-body measurements (as discussed above) and could suggest differential foraging adaptations among sites. However, given the scope of this work, such statements are highly speculative.
The molecular data largely support these anatomical observations and point out some additional relationships not examined in the morphological portion of this study. Specifically, extensive mtDNA divergence among M. beryllina populations was observed, which was largely driven by pure M. beryllina specimens from the inland lakes (CHO, MAT, and SOM), and by divergent back-crossed specimens from the coastal site ULM. Among these are four isolated populations, the genetic similarity (based on a low pairwise estimate of genetic divergence) between CHO and MAT conforms to expectations based on geography. These two sampling areas are reservoirs located approximately 50 km from one another along the same river drainage (Nueces River). The similarity of mtDNA haplotypes from SOM and ULM is more difficult to explain due to the geographic separation between these areas. Multiple references suggest anthropogenic introductions of silversides into inland Texas impoundments in the mid-1900’s (Bonn and Kemp 1952; Riggs and Bonn 1959; Hubbs 1982) and it is reasonable to suggest that such introductions could have come from limited coastal sources. A recent introduction of this species into SOM would be a probable explanation of our mtDNA haplotype data. Two lines of evidence support this interpretation. First, the genetic diversity in the SOM population of M. beryllina is extremely low compared to other areas. This is a common consequence of a “founder event,” such as the transplantation of a small number of individuals to a new habitat. Second, the linkage of the SOM mtDNA haplotypes and those sampled in the ULM would make sense if the ancestral stock released into SOM came from a lower coast drainage system.
In contrast to the relatively high levels of genetic divergence among most inland populations, there is relative similarity of haplotype distribution among individuals carrying MB lineage in coastal areas outside of ULM (CL and SL), as well as two inland lakes which are located <20 km from coastal areas (TEX and HOU). Among these four sampling areas, the only significant pairwise estimate of divergence was between the two inland lakes, although divergence was low relative to estimates between other inland lakes. This similarity between TEX and coastal populations of M. beryllina is also supported by the morphological portion of this study (Fig. 6). The general pattern of high divergence and low haplotype diversity among isolated inland sites, paired with relatively low genetic divergence and high haplotype diversity among coastal and near-coast sites, in addition to anatomical similarity among all of these sites, suggests that coastal areas may be an avenue of gene flow between otherwise isolated river drainages. This hypothesis is strengthened by the presence of M. beryllina haplotypes being carried in hybrids and back-crosses in both CL and ULM, as well as previous studies (Echelle et al. 1989), suggesting that mtDNA lineages may persist over several generations in coastal areas via introgression.
Much less anatomical variation was found among sampling locations of M. peninsulae. This was supported by the lack of evidence for genetic divergence. Such genetic mixing among geographically separated populations of fishes along the Texas coast is not surprising. Anderson and Karel (2009) suggest that the Gulf Intracoastal Waterway (GIWW), a deep shipping channel that connects the AB and ULM via Corpus Christi Bay, may serve as a corridor for eggs or larval stage fishes. This may additionally explain the slight morphological divergence observed in M. peninsulae from CL as compared to specimens from AB and ULM (Fig. 4) as the GIWW does not pass directly through this bay system. Alternatively, the morphological divergence may be explained by the relatively long geographic distance between CL and the other two sampled bay systems (average ~ 195 km), which themselves are in relatively close proximity to one another (~ 50 km). Although our genetic data indicate a lack of divergence among any of the sampled populations of M. peninsulae, it is nevertheless likely that AB and ULM share migrants more frequently based on distance and access alone, and therefore it is likely that these two areas are more demographically associated with one another.
The absence of M. beryllina from a majority of coastal collections is an interesting commentary on the transient nature of suitable habitat from year to year and the potential for salinity to effectively isolate populations of various river drainages. Further, this calls into question our distinction of coastal bay as areas of sympatry for these two species. Echelle and Echelle (1997) conducted extensive sampling throughout Aransas and Copano Bay (a secondary bay of Aransas Bay), Texas from June to August 1984. They covered a variety of habitat types (e.g., freshwater streams and bays, isolated pools, tidal creeks, shallow bays and tidal pools, and seaward bays) and captured 4144 specimens consisting of 37.3 % M. beryllina, 54.0 % M. peninsulae, 5.8 % M. beryllina x M. peninsulae hybrids, and 2.8 % unisexual M. clarkhubbsi. This work suggests that such bays do serve as areas of sympatry for these species. However, as discussed previously, work by Suttkus and Mettee (1998) found no overlap in collection of M. beryllina and M. peninsulae from the Perdido Bay region of Alabama and Florida. Although Suttkus and Mettee (1998) termed this an ‘apparent allopatric distribution’ of these two species, perhaps parapatry is a better designation. Mean salinity in the Aransas Bay system (including Copano Bay) in 1984 was 22.7 ppt contrasted with 30.8 ppt in 2014, on the tail end of a three-year drought (TPWD unpublished data). Multiple authors have noted segregation of M. beryllina and M. peninsulae based on salinity (Middaugh et al. 1986; Echelle and Echelle 1997; Suttkus and Mettee 1998), with M. peninsulae occupying higher oceanic salinities and M. beryllina occupying fresh to brackish salinities. While in the present study we did not sample as intensively as Echelle and Echelle (1997) and collections were not formally standardized with the intention of assessing abundance of various species, the study region did cover a similar spatial scale throughout Aransas and Copano Bay. The lack of M. beryllina collected here (and in other coastal sites) may have been due to drought conditions, which resulted in increased salinities and thus decreased habitat suitability for M. beryllina in coastal regions. It is likely that M. beryllina and M. peninsulae habitat in the coastal region expands and contracts with salinity regimes in coastal bays along with the potential for their interaction. Given the morphologically plastic nature of these species and importance of the coastal region for gene flow among river drainages, such increased salinity regimes may effectively isolate populations of M. beryllina. Such plasticity has been widely observed in atherinopsids and Bamber and Henderson (1988) hypothesized that this plastic nature primes this family of fishes for rapid speciation into freshwater environments containing vacant niches.
Aside from the suspected anthropogenic introduction of M. beryllina to SOM, it is plausible that historic speciation of Menidia fishes could have been driven by historic salinity regimes that restrict gene flow among river drainages through coastal bays. The deep divergence observed in mtDNA lineages between species observed here, coupled with commonly observed hybrids and back-crosses in this study and a previous study (Echelle and Echelle 1997) indeed suggests historical isolation of the species, facilitating divergence of mtDNA forms, followed by contemporary secondary contact. Recent periodic invasions of M. beryllina into coastal areas via changes in salinity may thus drive observed periodic hybridization. It should be noted also that haplotypes that were observed to be characteristic of M. peninsulae were never observed in inland sites, suggesting that interspecific gene flow occurs (geographically) from inland areas to coastal areas but not in reverse. These observations have implications for future interspecific interactions. Trends towards declining rainfall and anthropogenic changes to freshwater inflow could serve to further isolate populations of M. beryllina, resulting in (1) further genetic and morphological differentiation between M. peninsulae and M. beryllina, and (2) additional speciation among isolated inland populations of M. beryllina.
In summary, this study utilized an integrative approach of morphological and molecular techniques to examine interspecific variation of allopatric and sympatric populations of M. beryllina and M. peninsulae in addition to intraspecific variation of these two species. While both allopatric and sympatric morphological comparisons showed significant group separation between species, sympatric populations of these species were slightly more similar than allopatric populations. This suggests that competition among these species is not occurring to any extent that would result in divergent changes in morphology (i.e., character displacement). Additionally, both morphological and molecular data suggest extensive variation among populations of M. beryllina while variation among populations of M. peninsulae is much less definite. A review of past work on Menidia fishes in this region (Echelle and Echelle 1997) suggests that salinity regimes seem to drive the presence of M. beryllina in coastal bays with increased salinity restricting movement by this species among river drainages. Given the plastic nature of Menidia observed in this study and others (Bamber and Henderson 1988), such isolation may be responsible for speciation of Menidia fishes. Thus, morphological and molecular variation in Menidia spp. seems to be more so driven by isolation resulting from environmental conditions with interspecific competitive interactions being of little significance.
References
Anderson MJ (2001) A new method for non-parametric multivariate analysis of variance. Austral Ecol 26:32–46
Anderson JD, Karel WJ (2009) A genetic assessment of current management strategies for spotted seatrout in Texas. Marine and coastal fisheries: dynamics, management and ecosystem Science 1:121–132
Bamber RN, Henderson PA (1988) Pre-adaptive plasticity in atherinids and the estuarine seat of teleost evolution. J Fish Biol 33:17–23
Barbour CD (1973) A biogeographical history of Chirostoma (Pisces: Atherinidae): a species flock from the Mexican Plateau. Copeia 1973:533–556
Bhagat Y, Fox MJ, Ferreira MT (2011) Morphological diversification in introduced pumpkinseed (Lepomis gibbosus): assessing truss-based and geometric morphometric approaches. Fundam Appl Limnol 178:341–351
Bonn EW, Kemp RJ (1952) Records of fresh-water fishes from Texas. Copeia 1952:204–205
Borcherding J, Magnhagen C (2008) Food abundance affects both morphology and behaviour of juvenile perch. Ecol Freshw Fish 17:207–218
Brown WL, Wilson EO (1956) Character displacement. Syst Zool 5:49–64
Cakmak E, Alp A (2010) Morphological differneces among the Mesopotamian spiny eel, Mastacembelus mastacembelus (Banks & Solander 1794), populations. Turk J Fish Aquat Sci 10:87–92
Chernoff B, Conner JV, Bryan CF (1981) Systematics of the Menidia beryllina complex (Pices:Atherinidae) from the Gulf of Mexico and its tributaries. Copeia 1981:319–336
Echelle AA, Echelle AF (1997) Patters of abundance and distribution among members of the unisexual-bisexual complex of fishes (Atherinidae: Menidia. Copeia 2:249–259
Echelle AA, Mosier DT (1982) Menidia clarkhubbsi, n. sp. (Pisces: Atherinidae), an all-female species. Copeia 3:533–540
Echelle AA, Echelle AF, Crozier CD (1983) Evolution of an all-female fish, Menidia clarkhubbsi (Atherinidae). Evolution 37:772–784
Echelle AA, Echelle AF, DeBault LE, Durham DW (1988) Ploidy levels in silverside fishes (Atherinidae, Menidia) on the Texas coast: flow-cytometric analysis of the occurrence of allotriploidy. J Fish Biol 32:835–844
Echelle AA, Dowling TE, Moritz CC, Brown WM (1989) Mitochondrial-DNA diversity and the origin of the Menidia clarkhubbsi complex of unisexual fishes (Atherinidae). Evolution 43:984–993
Edwards RJ, Marsh E, Stevens FB Jr (1978) The utility of the air bladder position in determining specific relationships in atherinid genus Menidia. Contrib Mar Sci 21:1–7
Excoffier L, Lischer HEL (2010) Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour 10:564–567
Excoffier L, Smouse PE, Quattro JM (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131:479–491
Fluker BL, Pezold F, Minton RL (2011) Molecular and morphological divergence in the inland silverside (Menidia beryllina) along a freshwater-estuarine interface. Environ Biol Fish 91:311–325
Froese R, Pauly D Editors (2013) FishBase. World wide web electronic publication. www.fishbase.org. version (12/2013)
Geiger MF, Schreiner C, Delmastro GB, Herder F (2016) Combining geometric morphometrics with molecular genetics to investigate a putative hybrid complex: a case study with barbels Barbus spp. (Teleostei: Cyprinidae). J Fish Biol. doi:10.1111/jfb.12871
Gosline WA (1948) Speciation in the fishes of the genus Menidia. Evolution 2:306–313
Grant PR (1972) Convergent and divergent character displacement. Biol J Linn Soc 4:39–68
Hoese HD, Moore RH (1998) Fishes of the gulf of Mexico – Texas, Louisiana, and adjacent waters, 2nd edn. Texas A&M University Press, College Station
Holm S (1979) A simple sequentially rejective multiple test procedure. Scand J Stat 6:65–70
Hubbs C (1982) Life history dynamics of Menidia beryllina from Lake Texoma. Am Midl Nat 107:1–12
Johnson MS (1975) Biochemical systematics of the atherinid genus Menidia. Copeia 4:662–691
Loy A, Busilacchi S, Costa C, Ferlin L, Cataudella S (2000) Comparing geometric morphometrics and outline fitting methods to monitor fish shape variability of D. puntazzo (Teleostea: Sparidae. Aquac Res 21:271–283
McEachran JD, Martin CO (1977) Possible occurrence of character displacement in the sympatric skates Raja erinacea and R. ocellata (Pisces: Rajidae). Environ Biol Fish 2:121–130
Maderbacher M, Bauer C, Herler L, Postls L, Makasa L, Strumbauer C (2008) Assessment of traditional versus geometric morphometrics for discriminating populations of Tropheus moorii species complex (Teleostei: Cichlidae), a Lake Tanganyika model for allopatric speciation. J Zool Syst Evol Res 46:153–161
Martinez-Andrade F, Fisher M (2012) Marine resource monitoring operations manual. Texas parks and wildlife department. Texas, Austin
Middaugh DP, Hemmer MJ, Rose YL (1986) Laboratory spawning cues in Menidia beryllina and Menidia peninsulae (Pisces: Atherinidae) with notes on survival and growth of larvae at different salinities. Environ Biol Fish 15:107–117
Muir AM, Vecsei P, Krueger CC (2012) A perspective on perspectives: methods to reduce variation in shape analysis of digital images. Trans Am Fish Soc 141:1161–1170
Parsons KJ, Robinson BW, Hrbek T (2003) Getting into shape: an empirical comparison of traditional truss-based morphometric methods with a newer geometric method applied to new world cichlids. Environ Biol Fish 67:417–431
R Core Team (2014) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria http://www.R-project.org
Riggs CD, Bonn EW (1959) An annotated list of fishes of Lake Texoma, Oklahoma and Texas. Southwest Nat 4:157–168
Rozas J, Sánchez-DelBarrio JC, Messeguer X, Rozas R (2003) DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19:2496–2497
Sbrocco EJ, Barber PH (2011) Ten polymorphic microsatellite loci for the Atlantic Silverside, Menidia menidia. Cons Gen Res 3:585–587
Strauss RE, Bookstein FL (1982) The truss: body form reconstruction in morphometrics. Syst Zool 31:113–135
Suttkus RD, Mettee MF (1998) Morphometric, meristic, and natural history notes on Menidia beryllina and M. peninsulae in a marginal sympatric area in Perdido Bay, Alabama and Florida. Southeast Fish Counc Proc 37:7–13
Suttkus RD, Thompson BA, Blackburn JK (2005) An analysis of the Menidia complex in the Mississippi River Valley in two nearby minor drainages. Southeast Fish Couc Proc 48:1–9
Webster M, Sheets HD (2010) A practical introduction to landmark-based geometric morphometrics. Paleontol Soc Pap 16:163–188
Acknowledgments
We would like to thank field personnel from Texas Parks and Wildlife Department (TPWD) Coastal Fisheries Division in addition to Alice Best from TPWD Inland Fisheries Divisions for their help and expertise with planning and execution of field collections. Additionally, we’d like to thank Amy Gebhard and Autumn Torres for their aid in the collection of meristic data. Finally, we’d like to thank Fred Stevens and a number of internal and external reviewers for their insightful comments and edits to early versions of this manuscript.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
ESM 1
(DOCX 300 kb)
Rights and permissions
About this article
Cite this article
Olsen, Z., Anderson, J. & McDonald, D. Morphological and molecular variation among populations of tidewater (Menidia peninsulae) and inland (M. beryllina) silversides: insight into drivers of adaptation and speciation of silverside fishes. Environ Biol Fish 99, 857–871 (2016). https://doi.org/10.1007/s10641-016-0528-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10641-016-0528-3