Abstract
The quality of an oocyte can be defined by its potential to produce a normal and viable embryo. In Oncorhynchus mykiss (Walbaum), oocyte quality is highly variable under both natural and aquaculture conditions. To ensure the competitiveness and sustainability of rainbow trout farming, new tools are needed to evaluate oocyte potential. Considering that the abundance of certain maternal mRNAs incorporated during oogenesis can determine egg quality, the aim of the present study was to assess if significant differences existed in the abundances of the maternal mRNAs pou2 and zorba in low and high quality O. mykiss eggs. Analyses determined that survival until the end of gastrulation varied significantly between high and low quality egg batches, and significant correlations were established with posterior early development events. The abundance of pou2 and zorba transcripts varied between high and low quality groups, with higher relative expression recorded in the low quality group. Additionally, the abundances of these transcripts were significantly correlated with survival until blastopore closure, the earliest ontogenetic event evaluated as a quality attribute for this salmonid species. This correlation is supported by participations reported for both proteins in other teleost fish during embryonic development. These results form a basis for complementary studies and permit proposing maternal pou2 and zorba mRNA as potential markers for O. mykiss egg quality.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
One of the limiting factors in the reproductive success of rainbow trout Oncorhynchus mykiss (Walbaum) is gamete quality, which is highly variable under natural and culture conditions (Bobe and Labbé 2010). In salmonids, embryonic survival and development can be maternally (Springate et al. 1984) and paternally (Aas et al. 1991; Babiak et al. 1998; Gile and Ferguson 1995) influenced. Even females from the same stock that are maintained in the same tank and kept under identical culture conditions can produce eggs with variable survival rates (Bromage et al. 1992), with post-fertilization mortalities at times reaching 100 % (Craik and Harvey 1984; Ridelman et al. 1984).
By tracking O. mykiss families originating from one female and one male, it was found that variations in descendent survival until seven weeks post-fertilization were principally influenced by the female gamete (Nagler et al. 2000). Based on these results, the embryonic survival of O. mykiss could be directly linked to egg quality. High quality eggs have the potential to produce a viable alevin (Kjørsvik et al. 1990), or, in other words, quality fertilized eggs survive to reach the eye pigmentation, hatching, and first-feeding alevin stages, which increases the likelihood of giving rise to healthy, fast-growing juveniles (Bromage et al. 1992). Determining and ensuring this attribute is a key factor with considerable impact on the sustainability of fish farming as breeding programs are dependent on fish completing the lifecycle to maintain a broodstock (Bobe 2015).
Variations in egg quality are primarily dependent on the biological condition of the female, which can vary based on diet composition and nutrient availability during oocyte formation. It is during oogenesis that the incorporation, synthesis, and processing of oocyte components occurs, such as of amino acids, lipids, carbohydrates, vitamins, minerals, and metals required for enzymatic and other metabolic activities (Tata 1986; Brooks et al. 1997). Specifically, there is a period of intense messenger RNA (mRNA) synthesis during initial oocyte growth, and mRNA are stored in a non-transduced state until posterior activation at specific times during early embryonic development (Hake and Richter 1997). These maternal mRNA are highly important since embryos are initially transcriptionally inactive and are entirely dependent on these maternal gene products (Schier 2007; Ramachandra et al. 2008). Indeed, various processes are directed by maternally-derived mRNA, including oocyte activation, fertilization (Dosch et al. 2004), the formation of germinal cells (Hashimoto et al. 2004), growth regulation (Yang et al. 1999), embryogenesis (Wagner et al. 2004), initiation of zygotic transcription (Andeol 1994), formation of the embryonic dorsal-ventral axis (Schier 2001), and tissue morphogenesis (Abrams and Mullins 2009). Considering developmental relevancy, many studies have aimed to assess both egg and offspring quality by establishing relationships between maternally-derived, molecular-marker mRNAs and oocyte developmental competence (Bobe 2015).
While fertilization triggers a series of complex events that finalize with the alevin reabsorbing its yolk sac and feeding exogenously, there is evidence that the success rate of this process is defined during the early stages of embryonic development (Finch et al. 2009). One key event during this developmental phase is gastrulation, a process where harmonic movements establish and juxtapose the three primary germ layers that give rise to the embryonic axis, the ectoderm, mesoderm, and endoderm (Kunz 2004). In fish, this process, which can occur through epiboly and/or involution, culminates in blastopore closure (Gilbert 2010). In rainbow trout, this process can take between 9 and 10 days post-fertilization and is highly complex as epiboly advancement occurs simultaneously with the start of somitogenesis (Finch et al. 2009). Due to this, rainbow trout production procedures indicate that eggs should not be perturbed or moved until completing the eyed egg stage (Shelton 1994). For fish in general, gastrulation is a period highly sensitive to environmental factors (Strahle and Jesuthasan 1993; Steeger et al. 2001; Kjorsvik et al. 2004).
Among the maternal RNAs that could play a role in the gastrulation phase are pou and orb, both of which are involved in the process of epiboly. Pou2 is part of a family of transcription factors homologous to Oct-3/4/Pou5fl in mammals; these factors have been implicated in the control of gene expression during early development (Takeda et al. 1994), and its presence linked to maternal origin in several models of teleost fishes (Marandel et al. 2013). The functions of these factors include regulating dorsoventral modeling and morphogenesis (Reim and Brand 2006), modulating the morphogenetic process of epiboly, mediating the thinning and spreading of the blastoderm over the yolk (Abrams and Mullins 2009), and establishing the dorsoventral pattern and the convergence and extension of blastula (Khan et al. 2012). In zebra fish (Danio rerio), Pou2 is maternally expressed, and its transcripts are present from the one-cell stage until reaching 100 % epiboly in the gastrulation period. Following this, mRNA levels are undetectable, suggesting that this expression pattern is required to maintain a high degree of undifferentiated cells (Takeda et al. 1994). In O. mykiss, Pou2 is strongly expressed in pre-vitellogenic ovary tissue, suggesting the conservation of this gene in gonadal development and function (Bellaiche et al. 2014). For its part, the Orb/CEP-binding protein (CPEB) is a gene coding for a maternally-derived protein that participates in multiple stages of invertebrate and amphibian oogenesis (Lantz et al. 1992) by regulating the processes of mRNA polyadenylation and translation. Danio rerio presents a homolog of Orb/CPEB denominated Zorba (Zebrafish orbA), which is present during the first oocyte stages and which accumulates throughout development (Bally-Cuif et al. 1998). Studies in Xenopus laevis, an amphibian, have demonstrated the relation and participation of Orb/CPEB in oocyte maturation through the polyadenylation and translation of c-mos mRNA (Stebbins-Boaz et al. 1996; Suzuki et al. 2009). In invertebrates, mutations in this transcript result in eggs without proteins, blocked development, and anomalies in the localization of other mRNA involved in the formation of the dorsal-ventral axis (Christerson and Mckearin 1994; McKearin and Christerson 1994). Despite the importance of these proteins in the early stages of development, no reports currently exist for expression levels in trout eggs or on possible correlations between expression levels and the success of early ontogeny.
Considering that the abundance of certain maternal mRNAs could be significantly related with the quality of Oncorhynchus mykiss eggs, the aim of the present study was to determine if variations exist between different quality eggs in regards to the relative abundances of maternal pou2 and zorba mRNA. Egg quality was defined based on the survival during early-stage development, including the end of gastrulation, during blastopore closure.
Materials and methods
Experimental design
Rainbow trout eggs were obtained from an artisan fish farm (Piscicultura Salmones Pangue) located in central Chile (36.8°S, 72.8°W). During the sampling periods, adult rainbow trout were maintained in a flow-through system under water temperature and photoperiod conditions characteristic to the fall-winter months (10° ± 2 °C; LD 12:12). Three experiments were performed during each annual reproductive cycle between 2010 and 2012. For each experiment, spawning females in their 4th to 6th day post-ovulation were used, according to recommendations by Springate et al. (1984).
Males and females were first anesthetized via bath emersion in benzocaine (BZ-20®, Veterquimica, Santiago, Chile) for approximately 10 min. Prior to obtaining gametes, the length and weight of each female were recorded to obtain Fulton’s condition factor. After female spawning, two aliquots of ten unfertilized eggs were taken. These were stored in sterile tubes and kept at −80 °C until use in molecular analyses.
Eggs from each female were inseminated with pool of sperm from least three males. Following fertilization, two aliquots were taken from each of 100 eggs, which were separately incubated at an approximate temperature of 10 ± 2 °C and under constant water flow. Each replicate with 100 eggs was monitored daily to evaluate embryonic development, and dead embryos were periodically removed. In the first experiment (2010), the stage specific survival rates were estimated by developmental stage, quantifying the number of embryos to survive each of the following events: blastopore closure (survival rate from fertilization to blastopore closure); eyed egg (survival rate from blastopore closure to eye pigmentation); hatching (survival rate from eye pigmentation to hatching); and sac reabsorption (survival rate from hatching to yolk sac reabsorption).
During the second (2011) and third (2012) experiments, embryo survival was quantified only until blastopore closure. At experimental temperature this event occurs close to 9th day post fertilization between 80 or 100 C Temperature Units (CTU, Roberts and White 1992).
Molecular analysis
RNA was extracted from unfertilized eggs using the TRIzol® Reagent (Thermo Fisher, Waltham, MA, USA) according to the manufacturer’s instructions. Briefly, ten eggs were homogenized with 6 ml of TRIzol® until obtaining a homogenous solution that was incubated for 15 min at room temperature. This solution was then centrifuged at 5000 rpm for 15 min at 4 °C; the upper phase was transferred to a new tube, and 150 μL of chloroform per 6 mL of TRIzol® were added. The sample was agitated in a vortex for 20 s, incubated for 3 min at room temperature, and centrifuged at 12,000 rpm for 10 min at room temperature. The upper aqueous phase was recovered, to which 375 μL of isopropyl alcohol were added, followed by incubation for 10 min at room temperature, and centrifugation at 12,000 rpm for 10 min at room temperature.
The obtained pellet was washed with 800 μL of 75 % ethanol and centrifuged at 7500 rpm for 5 min at room temperature. The supernatant was then discarded, and this process was repeated three times. Finally, the obtained RNA pellet was resuspended in nuclease-free water. Due to the high vitellogenic content of eggs, the total RNA was purified in the E.Z.N.A.® Total RNA Kit (Omega Bio-tek, Norcross, GA, USA) according to the manufacturer’s instructions. RNA concentration and purity were estimated using a Nanodrop™ 2000 Spectrophotometer at optical density (Thermo Scientific, Waltham, MA, USA). The quality of extracted RNA was evaluated by electrophoresis (100 V, 45 min) on 1 % agarose gel stained by 5 mg/mL ethidium bromide.
To obtain cDNA, 3 μg of extracted RNA were treated with DNase I (Thermo Scientific, Waltham, MA, USA) for 30 min at 37 °C. After digestion, the reaction was stopped by adding 1.5 μL of EDTA followed by 15 min of incubation at 65 °C. The purified and DNase I-treated (approximate concentration of 5 ng/μL) RNA was hybridized with 100 ng of Oligo dT until a final volume of 14 μL. The hybridization solution was incubated for 5 min at 70 °C and, immediately thereafter, for 1 min at 4 °C. The reverse transcription (RT) reaction mixture contained 5 μL of 5X MMLV buffer, 10 mm of each dNTPs, and 200 IU of the RevertAid™ H Minus M-MuLV Reverse Transcriptase (Promega, Madison, WI, USA) in a final volume of 11 μL. This reaction mixture was added to the RNA/Oligo dT solution until obtaining a final volume of 25 μL. Following this, the solution was incubated for 60 min at 42 °C, and the cDNA was obtained by heating the solution for 15 min at 65 °C. The resulting cDNA was stored at –20 °C until qPCR amplification.
Primer design and qPCR
Specific primers were designed to amplify the mRNA of pou2 and zorba. From the paralogous sequence for Pou2 in D. rerio (Pou5f1; GenBank Accession Number: NM_131112.1), six homologous EST sequences were found in O. mykiss (GenBank Accession Numbers: BX876054.3, CX030782.1, CX037963.1, BX911359.3, CX031087.1, and CX038135.1). From the paralogous sequence for Zorba in D. rerio (Zorba; GenBank Accession Number: AF076918), three homologous EST sequences were found in O. mykiss (GenBank Accession Numbers: CX030865, CX031846, and CX041710). All EST sequences used in this study, excepting the pou2 sequences BX876054.3 and BX911359.3, were obtained from the O. mykiss oocytes library. Single-unit transcriptional sequences obtained for pou2 (BX876054.3; CX030782.1; CX037963.1) and zorba (CX030865; CX031846; CX041710) were aligned, and specific primers were designed using the Vector NTI v.10.0 software (Invitrogen, Carlsbad, CA, USA) (Table 1).
The primers used for amplifying the RNA of the housekeeping genes EF1-α and ubiquitin were described by Aegerter et al. (2005). Primer specificity was evaluated using the BLAST tool (http://www.ncbi.nlm.nih.gov/blast/). The amplifications were performed using 20 μL in a Rotor-Gene™ 6000 real-time system (Corbett Life Sciences, Qiagen, Venlo, Netherlands) for 40 cycles, followed by two cycles at 94 °C for 30 s, and alignment/extension at 60 °C for 30 s. The amplification efficiency for each of the studied mRNA was determined from a serial standard curve of cDNA synthesized from a pool of total RNA isolated from eggs with previously quantified distinct survival qualities. Relative abundance estimates of maternal mRNA in each of the eggs were determined through the ΔΔCt method, using EF1-α first and second experiments and ubiquitin (third experiment) as housekeeping genes.
Statistical analysis
Prior to statistical analysis, the survival rates were arcsine transformed, and normality (chi-squared) and homogeneity of variance (Bartlett) were checked. To compare among survival by developmental stage (experiment I) an analysis of variance nonparametric (Kruskal-Wallis test) followed by an “a posteriori” test of multiple comparisons, were applied. To determine possible correlations between the biological stages and between mRNA expression levels and the survival rate, the Pearson correlation coefficient was calculated. For the three experiments, the criteria used to classify eggs as high or low quality corresponded to the values of the first and final quintile of survival rates calculated for blastopore closure. Therefore, rates less than 20 % were low quality and rates greater than 80 % were high quality, which are values within the ranges described by other authors when classifying rainbow trout eggs (Aegerter et al. 2003; Bonnet et al. 2007a). To evaluate differences in the abundance of mRNAs in regards to variations in the quality of oocyte (high/low), a Mann-Whitney U test or a t-test were used, depending on data normality and homogeneity of variance. All statistical analyses were performed with the Statistica 7.0 software.
Results
In the three experiments, the conditions of the females were adequate, with Fulton’s condition factor values greater than 1.3 (Table 2). In each of the experiments, there was high variability in the degree of offspring survival from each female (batch), with extreme values between 0 and 100 %.
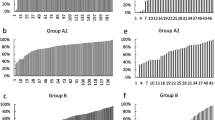
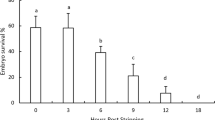
In Experiment I, in which early rainbow trout development was followed until yolk sac absorption, survival was evaluated at distinct ontogenetic stages, revealing important variations between the progeny of distinct females (and/or batches, Fig. 1). Extreme survival rates were from 0 to 100 %. However, the magnitude of survival between each ontogenetic stage varied significantly (K-W(3, N=98) = 16.9 p = 0.0007). Average values were higher between hatching and yolk sac absorption (> 80 %), a rate statistically different from early stages (blastopore closure 52.9 % and eyed egg 47.7 %; p < 0.05; Fig. 1). Coincidentally, there was a positive correlation between survival at blastopore closure and posterior events (eyed egg r = 0.820, p < 0.005; and hatching r = 0.532, p = 0.016, n = 28), whereas survival at the end of early development (yolk sac absorption) was only correlated with survival at hatching (r = 0.521, p = 0.018, n = 28).
When classifying batches as high or low quality as based on survival rate until blastopore closure (i.e. < 20 % low quality and >80 % high quality), the number of batches to analyze was reduced. Nevertheless, the significant differences between the high and low quality groups were maintained when evaluating the remaining early stages (Mann-Whitney U test, p < 0.05; Fig. 2). This result suggests that probably atend of blastopore closure, the trajectories for early development are already defined, therefore survival of this phase could be a valid criterion for initially classifying each batch.
Between experiments, the greatest survival rates until blastopore closure were recorded in Experiment III, in which the majority of eggs reached this stage (> 70 %; Table 2). Due to this, when classifying batches from each experiment as high or low quality, the number of batches (female) to consider in molecular analyses were reduced (N Exp I = 6, N Exp II = 8, and N Exp III = 6).
To relate the mRNA expression of pou2 and zorba with the quality of the previously selected eggs, specific primers were designed and expression levels were measured through qPCR. For analyses, batches from each experiment were grouped according to quality. For both pou2 and zorba transcripts, significant differences were found between expression levels in high and low quality eggs (Mann-Whitney U test, p < 0.005; Fig. 3). The observed tendency was that of greater expression of both messengers in low quality eggs, or, in other words, the expression levels were negatively and significantly correlated with survival until blastopore closure (Pou2 r = −0.50, p < 0.05; and Zorba r = −0.62, p = 0.001).
Discussion
Considering that the abundance of the certain maternal mRNA incorporated during oogenesis can significantly affect the quality of O. mykiss eggs, the aim of the present study was to evaluate if there were significant differences in the relative expression levels of the maternal mRNAs pou2 and zorba in eggs of different qualities. The early ontogenetic events used to determine egg quality included survival until blastopore closure, one of the key stages in vertebrate embryogenesis.
Early development monitoring in rainbow trout over three consecutive reproductive periods revealed high variability in the survival of this phase of the lifecycle, which is consistent with that described by various other authors in this species (Springate et al. 1984; Aegerter et al. 2004; Aegerter et al. 2005). Experiment I, which tracked eggs until yolk sac absorption, showed that survival until the eyed egg stage was less than that of later stages analyzed, reinforcing the importance of early ontogenetic events to posterior survival. Likewise, there was a significant positive correlation between survival until blastopore closure and survival of posterior events, which is similar to the positive relationship described for trout between fertilization and subsequent developmental stages, including eye pigmentation, hatching, and yolk sac absorption (Springate et al. 1984). While prior studies have not considered gastrulation success an indicator for egg quality, the present data do suggest gastrulation success indicative of egg quality. In other words, the present observations support initial batch classifications (i.e. high and low quality) according to survival until blastopore closure.
These data highlight the relevance of gastrulation in early ontogeny, especially for rainbow trout. In fish, the complex process of gastrulation culminates with blastopore closure, which occurs through the elongation of embryonic cells (blastoderm) from the animal to the vegetal pole until completely covering the yolk. Until now, there were no reports in O. mykiss that specifically quantified embryonic mortality during the gastrulation process; however, it has been found that this process lasts for various days due to the temperatures at which it occurs as well as to characteristics of the egg (abundant yolk). Nevertheless, this prolonged epiboly time overlaps with other processes, such as tail formation and the start of somitogenesis, suggesting that trout undergo a heterochronic process (Finch et al. 2009).
The expression levels of pou2 and zorba were significantly different between the high and low quality egg groups, with higher expression in the low quality eggs. A highly significant, negative correlation between transcript abundance and egg quality was previously reported in O. mykiss eggs for the maternal mRNA prohibitin 2 under conditions of natural ovulation (Bonnet et al. 2007b). This protein is a negative regulator of the cell cycle, impeding its progression and exerting antiproliferative functions (Manjeshwar et al. 2003). Considering these findings, the differential expression of prohibitin 2 in rainbow trout eggs of various qualities could act as an indicative molecular marker for the potential developmental success of eggs (Bonnet et al. 2007b). This negative correlation it has been described for others 5 transcripts during post - ovulatory ageing, where egg quality is diminished (Aegerter et al. 2004, 2005). In this way, another 4 transcripts increased their expression in eggs from females hormonally induced to ovulate and in response to photoperiod manipulation, artificial conditions that decrease eggs quality (Bonnet et al. 2007b). To evaluate if the negative correlation determined for pou2 and zorba occurred in other transcripts, maternal RNA expressions for PHB2 and IGF-1 were also assessed (Aegerter et al. 2005; Bonnet et al. 2007b). The expression levels for both transcripts did not evidence significant differences between high and low quality groups (Exp1 PHB2 p = 0.4233; Exp2 PHB2 p = 0.373; IGF-1 p = 0.4093). Furthermore, no statistically significant differences were found for correlations between expression levels and survival at blastopore closure (Exp1 PHB2 r = −0.39, p = 0.215; Exp2 PHB2 r = 0.262, p = 0.410; IGF-1 r = −0.303, p = 0.292). These results indicate that the determined correlations do not reflect overall egg responses or an effect influenced by the used housekeeping genes.
Both Pou2 and Zorba are maternally-derived mRNA fundamental for early embryogenesis in diverse organisms (Suzuki et al. 2009; Skjærven et al. 2011; Liu et al. 2015; O’Connell et al. 2014), but these have not previously been studied in rainbow trout during development. A recent study did evaluate the expression of pou2 in various O. mykiss tissues, with results showing significantly higher expression of transcripts in ovarian tissue as compared to other evaluated tissues. This suggests that this protein would participate in the growth of this tissue as well as of synthesized oocytes (Bellaiche et al. 2014). Similarly, D. rerio temporally and spatially expresses pou2 mRNA from the one-cell stage until the gastrulation phase uniquely in the epiblast, which suggests the participation of this protein in the epiboly event and posterior blastopore closure (Takeda et al. 1994; Reim and Brand 2006). Moreover, defects in the expression of this gene impact the formation of the dorsal-ventral axis and stop the epiboly process, thereby affect correct thinning and spreading of the blastoderm over the yolk (Reim and Brand 2006; Abrams and Mullins 2009). Additionally, induced overexpression of this mRNA results in delayed zebrafish development, principally due to incomplete gastrulation (Takeda et al. 1994).
Regarding Zorba, no previous studies have reported on its expression in O. mykiss eggs or tissues. However, studies on the function of this protein have been performed in D. rerio. In this model, Zorba is a CPEB protein specific to eggs, with expression restricted to the early blastula stages. The function of Zorba is to bind to other maternally-derived mRNAs and modulate their storage/repression or translation in a spatial-temporal manner through polyadenylation (Richter 2007; O’Connell et al. 2014). For example, Zorba is able to regulate the spatial-temporal translation of genes fundamental for zebrafish embryogenesis and the formation of the dorsal-ventral axis. This function suggests a critical role of Zorba in the embryonic development of different vertebrate species, including fish. Considering this and the present results, it is possible that increased Zorba levels in rainbow trout eggs would lead to a dysregulation in the translation/storage of some maternal transcripts, such as has been demonstrated for the maternal RNA ElrA (O’Connell et al. 2014). ElrA is able to translate other maternal mRNAs, such as hnRNPab, homolog to the Drosophila gene squid (Kelley 1993), which itself is fundamental for establishing dorsoventral patterning in flies (Norvell et al. 2005).
The increased expression of some transcripts under conditions that diminish egg quality suggest that poor egg quality is not a simple product of mRNA degradation, but rather arises from complex transcript regulations (Sullivan et al. 2015). Furthermore, the present results, together with prior knowledge in other species, suggest that not only is the presence of maternal pou2 and/or zorba mRNA necessary, but that expressions should also follow a defined spatial/temporal pattern. To date, such a pattern has not been described in O. mykiss. The current experimental approximations do not eliminate this possible expressional dynamic or that alterations between fertilization and blastopore closure could influence embryonic viability. Due to these points, further studies are required for clarification.
Finally, the present results regarding abundances of maternally-derived pou2 and zorba in O. mykiss eggs form a basis for complementary studies, in addition to suggesting that these maternal mRNA could serve as potential quality indicators. Indeed, these expressions could be used in combination with other molecular markers to develop a predictive tool for identifying eggs with the qualities needed for sustainable O. mykiss production.
References
Aas GH, Refstie T, Gjerde B (1991) Evaluation of milt quality of Atlantic Salmon. Aquaculture 95:125–132
Abrams EW, Mullins MC (2009) Early zebrafish development: It’s in the maternal genes. Curr Opin Genet Dev 19:396–403
Aegerter S, Jalabert B, Bobe J (2003) mRNA stockpile and egg quality in rainbow trout (Oncorhynchus mykiss). Fish Physiol Biochem 28:317–318
Aegerter S, Jalabert B, Bobe J (2004) Messenger RNA stockpile of cyclin B, insulin-like grow factor I, insulin-like grow factor II, insulin-like grow factor receptor Ib, and p53 in the rainbow trout oocyte in relation with development competence. Mol Reprod Dev 67:127–135
Aegerter S, Jalabert B, Bobe J (2005) Large scale real-time PCR analysis of mRNA abundance in rainbow trout eggs in relationship with egg quality and post-ovulatory ageing. Mol Reprod Dev 72:377–385
Andeol Y (1994) Early transcription in different animal species: implications for transition from maternal to zygotic control of development. Roux arch. Dev Biol 204:3–10
Babiak I, Glogowski J, Luczynski M, Goryczko K, Dobosz S, Kuzminski H (1998) The effect of individual male potency on fertilization ability of fresh and cryopreserved milt of rainbow trout, Oncorhynchus mykiss (Walbaum). Aquac Res 29:337–340
Bally-Cuif L, Schatz WJ, Ho RK (1998) Characterization of the zebra-fish Orb/CPEB-related RNA binding protein and localization of maternal components in the zebrafish oocyte. Mech Develop 77:31–47
Bellaiche J, Lareyre JJ, Cauty C, Yano A, Allemand I, LeGac F. (2014) Spermatogonial stem cell quest: nanos2, marker of a subpopulation of undifferentiated a spermatogonia in trout testis. Biol Reprod 90(4):79, 1–14
Bobe J (2015) Egg quality in fish: present and future challenges. Animals. Frontiers 5(1):66–72
Bobe J, Labbé C (2010) Egg and sperm quality in fish. Gen Comp Endocr 165:535–548
Bonnet E, Montfort J, Esquerre D, Hugot K, Fostier A, Bobe J (2007a) Effect of photoperiod manipulation on rainbow trout (Oncorhynchus mykiss) egg quality: a genomic study. Aquaculture 268:13–22
Bonnet E, Fostier A, Bobe J (2007b) Microarray-based analysis of fish egg quality after natural or controlled ovulation. BMC Genomics 8
Bromage NR, Randall JJ, Thrush M, Davies B, Springate J, Duston J, Barker G (1992) Broodstock management, fecundity, egg quality and the timing of egg production in the rainbow trout (Oncorhynchus mykiss). Aquaculture 100:141–166
Brooks S, Tyler CR, Sumpter JP (1997) Egg quality in fish: what makes a good egg? Rev Fish Biol Fisher 7:387–416
Christerson LB, McKearin DM (1994) Orb is required for anteroposterior and dorsoventral patterning during Drosophila oogenesis. Gen. Dev 8(5):614–628
Craik JCA, Harvey SM (1984) Egg quality in rainbow trout. The relation between egg viability, selected aspects of eggs composition, and time of stripping. Aquaculture 40:115–134
Dosch R, Wagner DS, Mintzer KA, Runke G, Wiemelt AP, Mullins MC (2004) Maternal control of vertebrate development before the midblastula transition: mutants from the zebrafish I. Dev Cell 6:771–780
Finch E, Cruz C, Sloman K, Kudoh T (2009) Heterochrony in the germ ring closure and tail bud formation in embryonic development of rainbow trout (Oncorhynchus mykiss). JExpZool (Mol.Dev. Ecol.) 314B:187–195
Gilbert, S. (2010) Developmental Biology. Sinauer Associates, Inc. Sunderland, MA
Gile SR, Ferguson MM (1995) Factors affecting male potency in pooled gamete crosses of rainbow trout, Oncorhynchus mykiss. Envir Biol Fish 42:267–275
Hake LE, Richter JD (1997) Translational regulation of maternal mRNA. Biochim Biophys Acta 1332:M31–M38
Hashimoto Y, Maegawa S, Nagai T, Yamaha E, Suzuki H, Yasuda K, Inoue K (2004) Localized maternal factors are requerid for zebrafish germ cell formation. Dev Biol 268:152–161
Kelley RL (1993) Initial organization of the Drosophila dorsoventral axis depends on an RNA-binding protein encoded by the squid gene. Gen Dev 6:948–960
Khan A, Nakamoto A, Okamoto S, Tai M, Nakayama Y, Kobayashi K, Kawamura A, Takeda H, Yamasu K (2012) Pou2, a class V POU-type transcription factor in zebrafish, regulates dorsoventral patterning and convergent extension movement at different blastula stages. Mech Develop 129(9–12):219–235. doi:10.1016/j.mod.2012.07.007
Kjorsvik E, Pitman K, Pavlov D (2004) From fertilization to the end of metamorphosis. Functional Development. In: Moksness, Kjorsvik, Olsen (eds) Culture of Cold-Water Marine Fish Blackwell Publishing Oxford pp 204–278
Kjørsvik E, Mangor-Jensen A, Holmefjord I (1990) Egg quality in fishes. Adv Mar Biol 26:71–113
Kunz, YW (2004) Developmental Biology of Teleost Fishes, Springer, Dublin
Lantz V, Ambrosio L, Schedl P (1992) The Drosophila orb gene is predicted to encode sex-specific germline RNA-binding proteins and has localized transcripts in ovaries and early embryos. Development 115(1):75–88
Liu R, Li M, Li Z, Hong N, Xu H, Hong Y (2015) Medaka Oct4 is essential for pluripotency in blastula formation and ES cell derivation. Stem Cell Rev and Rep 11:11–23
Manjeshwar S, Branam DE, Lerner MR, Brackett DJ, Jupe ER (2003) Tumor suppression by the prohibitin gene 3’untranslated region RNA in human breast cancer. Cancer Res 63(17):5251–5256
Marandel L, Labbe C, Bobe J, Jammes H, Lareyre JJ, Le Bail PY (2013) Do not put all teleosts in one net: focus on the sox2 and pou2 genes. Comp BiochemPhysiol B Biochem MolBiol 164(2):69–79
McKearin D, Christerson L (1994) Molecular genetics of the early stages of germ cell differentiation during Drosophila oogenesis. Ciba F Symp 182:210–219
Nagler JJ, Parsons JE, Cloud JG (2000) Single pair mating indicates maternal effects on embryo survival in rainbow trout, Oncorhynchus mykiss. Aquaculture 184:177–183
Norvell A, Debec A, Finch D, Gibson L, Thoma B (2005) Squid is required for efficient posterior localization of oskar mRNA during Drosophila oogenesis. Dev Genes Evol 215(7):340–349
O’Connell ML, Cavallo WC Jr, Firnberg M (2014) The expression of CPEB proteins is sequentially regulated during zebrafish oogenesis and embryogenesis. Mol Reprod Dev 81(4):376–387
Ramachandra RK, Salem M, Gahr S, Rexroad CE III, Yao J (2008) Cloning and characterization of microRNAs from rainbow trout (Oncorhynchus mykiss): their expression during early embryonic development. BMC Dev Biol 8
Reim G, Brand M (2006) Maternal control of vertebrate dorsoventral axis formation and epiboly by the POU domain Spg/Pou2/Oct4. Development 133:2757–2770
Richter JD (2007) CPEB: a life in translation. Trends Bio Chem Sci 32(6):279–285
Ridelman J, Hardy R, Brannon E (1984) The effect of short-term starvation on ovarian development and egg viability in rainbow trout. Aquaculture 37:133–140
Roberts W, White G (1992) Effects of angler wading on survival of trout eggs and pre-emergent fry. North Am J Fish Mana 12:450–459
Schier AF (2001) Axis formation and patterning in zebrafish. Curr Opin Genet Dev 11(4):393–404
Schier AF (2007) The maternal-zygotic transition: death and birth of RNAs. Science 316:–406
Shelton J L (1994) Trout production. Cooperative extension service, The University of Georgia College of Agricultural and Environmental Sciences. Aquaculture Technical Series: 1–15.
Skjærven KH, Olsvik PA, Finn RN, Holen E, Hamre K (2011) Ontogenetic expression of maternal and zygotic genes in Atlantic cod embryos under ambient and thermally stressed conditions. Comp Biochem. Phys A 159(2):196–205
Springate JRC, Bromage NR, Elliott JAK, Hudson DL (1984) The timing of ovulation and stripping and their effects on the rates of fertilization and survival to eyeing, hatch and swim-up in the rainbow trout (Salmo gairdneri) R. Aquaculture 43:313–322
Stebbins-Boaz B, Hake LE, Richter JD (1996) CPEB controls the cytoplasmic polyadenylation of cyclin, cdk2, and c-Mos mRNAs and is necessary for oocyte maturation in Xenopus. EMBO J 15:2582–2592
Steeger H, Freitag J, Michl S, Wiemer M, Paul R (2001) Effects of UV-B radiation on embryonic, larval and juvenile stages of north Seaplaice (Pleuronectes platessa) under simulated ozone-hole conditions. Helgoland Mar Res 55:56–66
Strahle U, Jesuthasan S (1993) Ultraviolet irradiation impairs epibolyin zebrafish embryos: evidence for a microtubule-dependent mechanismof epiboly. Development 119:909–919
Sullivan C, Chapman R, Reading B, Anderson P (2015) Transcriptomics of mRNA and egg quality in farmed fish: some recent developments and future directions. Gen Comp Endocr 221:23–30
Suzuki H, Tsukahara T, Inoue K (2009) Localization of c-mos mRNA around the animal pole in the zebrafish oocyte with Zor-1/Zorba. BioSci Trends 3(3):96–104
Takeda H, Matsuzaki T, Oki T, Miyagawa T, Amanuma H (1994) A novel POU domain gene, zebrafish pou2: expression and roles of two alternatively spliced twin products in early development. Genes Dev 8:45–59
Tata JR (1986) Coordinated asembly of the developing egg. Bio Essays 4:197–200
Wagner DS, Dosch R, Mintzer KA, Wiemelt AP, Mullins MC (2004) Maternal control of development at the midblastula transition and beyond: mutants from the zebrafish II. Dev Cell 6:781–790
Yang BY, Green M, Chen TT (1999) Early embryonic expression of the growth hormone family protein genes in the developing rainbow trout, Oncorhynchus mykiss. Mol Reprod Dev 53:127–134
Acknowledgments
This research was funded by the COPAS-Sur Austral Program (PFB-31/2007) awarded by the Universidad de Concepción, Chile; by Project 213.114.001-1AP awarded by the Universidad de Concepción, Chile, and by the projects INNOVA 12.178 (R. Aliaga), INNOVA Bio Bio 10-CHS2-690 F11 (D. Bravo), and INNOVA Bio Bio 102 (J.P. Alvarez). We also thank Ricardo Quiroz and Pablo Baldebenito from Salmones Pangue for access to the sampling facilities and aid in each of the experiments.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Cruzat, F., Bravo, D., Alvarez, J. et al. Abundance of the maternal mRNAs Pou2 and Zorba and their relation to events in the embryonic development of Oncorhynchus mykiss (Walbum). Environ Biol Fish 99, 845–855 (2016). https://doi.org/10.1007/s10641-016-0525-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10641-016-0525-6